Abstract
The Runx3 transcription factor is a key regulator of lineage-specific gene expression in several developmental pathways and could also be involved in autoimmunity. We report that, in dendritic cells (DC), Runx3 regulates TGFβ-mediated transcriptional attenuation of the chemokine receptor CCR7. When Runx3 is lost, i.e., in Runx3 knockout mice, expression of CCR7 is enhanced, resulting in increased migration of alveolar DC to the lung-draining lymph nodes. This increased DC migration and the consequent accumulation of activated DC in draining lymph nodes is associated with the development of asthma-like features, including increased serum IgE, hypersensitivity to inhaled bacterial lipopolysaccharide, and methacholine-induced airway hyperresponsiveness. The enhanced migration of DC in the knockout mice could be blocked in vivo by anti-CCR7 antibodies and by the drug Ciglitazone, known to inhibit CCR7 expression. The data indicate that Runx3 transcriptionally regulates CCR7 and that, when absent, the dysregulated expression of CCR7 in DC plays a role in the etiology of asthmatic conditions that recapitulate clinical symptoms of the human disease. Interestingly, human RUNX3 resides in a region of chromosome 1p36 that contains susceptibility genes for asthma and hypersensitivity against environmental antigens. Thus, mutations in RUNX3 may be associated with increased sensitivity to asthma development.
Keywords: transcription regulation, autoimmunity, Ciglitazone, human chromosome 1p36
One of the challenges encountered by the immune system is to distinguish between pathogenic and innocuous antigens and to respond accordingly by developing active immunity or tolerance, respectively (1). Allergic asthma is a multifactorial disease characterized by a T cell-mediated immune reaction and chronic airway inflammation in response to inhaled allergen (2). The disease is often accompanied by an increase in serum IgE and involves airway hyperresponsiveness (AHR) (3, 4). Recent studies point at a primary role of dendritic cells (DC) in regulating in vivo allergic responses (5, 6). The respiratory tract DC are responsible for orchestrating the immune reaction against inhaled antigens through initiation of a T helper 2 (Th2) response and by stimulating cytokine-producing T cells during the ongoing airway inflammation (7, 8). Additionally, DC play an important role in the maintenance of self-tolerance in the periphery (9–11). How do these two functions converge?
Lung alveolar DC, which constantly sample inhaled antigens, are normally maintained at an immature state by immunosuppressive cytokines, such as TGFβ, secreted by the surrounding cellular environment, including lung macrophages and epithelial cells (12–15). At this immature state, alveolar DC have a poor capacity to present antigens and/or to migrate to the draining lymph nodes (LN) and function in maintenance of peripheral tolerance (9, 11, 16). Detection of pathogen-associated molecular patterns (PAMPs) by the immature tissue-resident DC initiates their maturation and migration to the regional LN where they prime naive T cells. This maturation process involves up-regulation of specific chemokine receptors (5) that mediate DC migration to the T cell zones of the draining LN. This DC trafficking is directed by the secondary lymphoid tissue chemokine (SLC)/CCL21 through binding to its cognate receptor, the chemokine receptor 7 (CCR7) (5). SLC/CCL21 is constitutively produced in the high endothelium venules (HEVs) of LN and Peyer's patches (PP) and in the lymphatic endothelium of multiple tissues (17). Upon up-regulation of CCR7 expression, DC respond to these homing signals and migrate to draining LN (18–20). Because the migratory capacity of DC is important for their function in both immunity and tolerance, the regulation of CCR7 expression constitutes a crucial checkpoint (20), tightly regulated by the antiinflammatory cytokine TGFβ (21).
Runx3 belongs to the runt domain family of transcription factors, which are key regulators of lineage-specific gene expression, and when mutated are associated with human diseases (22). Interestingly, recent findings raised the possibility of RUNX involvement in autoimmunity (23). In the developing mouse embryo, Runx3 displays a distinct tissue-specific expression pattern. It is expressed in hematopoietic organs, developing bones, peripheral nervous system, and skin appendages (24). Studies in knockout (KO) mice have delineated several cell-autonomous functions of Runx3. In neurogenesis, Runx3 is required for the development and survival of dorsal root ganglia TrkC neurons (25, 26). In thymopoiesis, Runx3 is required for silencing of CD4 during T cell lineage decisions (27–29), and, in DC, Runx3 functions as a component of the TGFβ-signaling cascade (30). In the absence of Runx3, KO DC do not respond to TGFβ; their maturation is accelerated and accompanied by an increased efficacy to stimulate T cells (30).
Runx3 KO mice also develop lesions in the gastrointestinal tract (GIT). Li et al. (31) have reported that Runx3 KO newborn mice exhibit hyperplasia of gastric epithelium (31, 32). They have attributed this defect to the loss of Runx3 function in GIT epithelium and suggested that Runx3 is a novel tumor suppressor involved in stomach cancer (31). Brenner et al. (33), on the other hand, have reported that, at ≈4 weeks of age, Runx3 KO mice develop colitis and only at a much older age of 8 months also develop gastric mucosal hyperplasia (33). Because Runx3 could not be detected in GIT epithelium (24, 33, 34), but is readily detected in the resident leukocytic population, the current conclusion is that the GIT ailments of the KO mice are due to the loss of an intrinsic cell function of Runx3 in leukocytes (33).
Here, we report that, in addition to their lung inflammation (30), Runx3 KO mice spontaneously develop physiological conditions characteristic of asthma, including AHR, increased levels of serum IgE, and hypersensitivity to inhaled LPS. In the KO mice, the trafficking of DC from airways to the regional LN is accelerated due to dysregulated expression of the chemokine receptor CCR7. We show that Runx3 regulates the TGFβ-mediated transcription attenuation of CCR7, and, when it is lost, CCR7 expression on KO DC is enhanced. The enhanced CCR7 expression results in increased migration of alveolar DC and accumulation of mature activated DC in lung-draining LN. The accelerated DC migration could be blocked by anti-CCR7 antibodies and by the drug Ciglitazone, known to inhibit DC CCR7 expression (35). The data delineate Runx3 as a transcriptional regulator of CCR7 expression and indicate that dysregulation of CCR7 in DC could lead to development of asthmatic conditions recapitulating clinical symptoms of the human disease. Interestingly, human RUNX3 resides in a region of chromosome 1p36 (36) that contains susceptibility genes for asthma and hypersensitivity against environmental antigens (37, 38).
Materials and Methods
Mice and Treatments. Runx3-KO mice were generated as described (25) and bred on ICR and MF1 backgrounds. KO mice and WT littermates of both backgrounds were used for the experiments. Mice were maintained in individually ventilated cages in a specific pathogen-free (SPF) facility free of known viral and bacterial pathogens. For AHR measurements, Runx3 KO and WT littermates were placed in a whole-body plethysmograph (Buxco, Winchester, U.K.) after anesthesia with i.p. injection of 10 μl/g solution containing ketamine (10 mg/ml) and xylazine (2 mg/ml). Recording of baseline respiratory patterns was followed by measurements after 1 min inhalation of the muscarinic agonist methacholine (40 mg/ml) (Sigma), by using an ultrasonic nebulizer. For the analysis of serum and bronchoalveolar lavage (BAL) IgE, mice were bled from the retroorbital plexus and serum was prepared and kept at -70°C until further analyzed. BAL was prepared as described (30) and kept at -70°C. In vivo cell labeling with carboxyfluorescein diacetate-succinimidyl ester (CFSE, Molecular Probes) was carried out by intranasal instillation of 50 μl per mouse of 8 mM CFSE in RPMI medium 1640 to isoflurane-anesthetized mice. For analysis of BAL and LN, mice were killed by CO2 asphyxiation 16 h after CFSE labeling, tracheae were cannulated, and lungs washed by gentle infusion of 2–4 aliquots of 1 ml PBS. Subsequently, mediastinal and axillary LN were removed, incubated with 1 mg/ml collagenase type VIII (C-2139, Sigma) for 45 min at 37°C, minced, and pressed through an 80-μm nylon mesh to obtain a single cell suspension. The administration of reagents affecting DC migratory capacity was carried out by intranasal instillation of 50 μg of LPS (Sigma) per mouse 5 h after CFSE treatment, or anti-CCR7 antibodies (purified goat anti-mouse CIO131, Capralogics, Hardwick, MA) 8 μg per mouse, 1 h after CFSE treatment. For the Ciglitazone treatment, mice were subjected to inhalation of 50 μg per mouse of Ciglitazone (71730, Cayman Chemical, Ann Arbor, MI) for 20 min before CFSE treatment.
RT-PCR Analysis. RT-PCR products were generated by using the following primers: 5′-CATCAGCATTGACCGCTACGT-3′ and 5′-GGTACGGATGATAATGAGGTAGCA-3′ for murine CCR7; 5′-GTGTTCATCATTGGAGTGGTG-3′ and 5′-GGTTGAACAGGTAGATGCTGGTC-3′ for murine CCR1; 5′-GGGACATCATCAAACCAGACC-3′ and 5′-GCCAACCAAGCAGAAGACAGC-3′ for murine IL-12.
Culturing Bone Marrow DC. Bone marrow-derived DC (BMDC) were prepared as described (39). Briefly, mice were killed, and bone marrow was extracted from femurs and tibias by flushing the shaft with 5 ml of RPMI medium 1640. RBC were lyzed in 1.66% NH4Cl, and cells were seeded into non-tissue culture plates at a density of 1 × 106 cells per ml in medium (RPMI medium 1640/5% FCS/50 μM 2-mercaptoethanol/penicillin/streptomycin) containing 10 ng/ml murine recombinant granulocyte/macrophage colony-stimulating factor (GM-CSF, PeproTech, Rehovot, Israel). Medium was replenished every three days, and the loosely adherent DC were collected and used for further studies at designated time points. To induce DC maturation, day 7–12 cultures were treated with LPS (1 μg/ml) and analyzed 1 day later.
Flow Cytometry. Single cells were suspended in FACS buffer (PBS/1 mM EDTA/1% BSA/0.05% sodium azide). Immunostaining (1–2 × 106 cells) was performed in the presence of rat anti-mouse FcγRIII/II receptor (CD16/32; clone 2.4G2, American Type Culture Collection), by incubating the cells with monoclonal antibodies for 30 min on ice (100 μl per 1 × 106 cells). Flow cytometry was performed with a FACSCalibur (Becton Dickinson) and cellquest software (Becton Dickinson). Staining reagents included CD11c APC/PE and IA/IE PE. For analysis of IgE Receptors (FcεR), cells were treated by acid elution before specific staining. Specifically, cells were suspended in an acidic buffer (0.05 M sodium acetate/acetic acid, pH3.5/0.085 M NaCl/0.005 M KCl/1% FCS) and incubated at room temperature for 3 min. After neutralizing with PBS/BSA 1%, cells were washed twice with FACS buffer and subjected to further staining with IgE-FITC antibodies.
DC Migration Assays. BMDC were grown in the presence or absence of TGFβ (10 ng/ml). On day 6, cells were treated overnight with LPS (1 μg/ml) to induce maturation. BMDC (5 × 105) were placed in a Transwell migration chamber (Costar 3421, Corning) and allowed to migrate through a polycarbonate mesh (pore size 5 μm) at 37°C. SLC (100 ng/ml) (457-6C, R & D Systems) or control buffer was placed in the lower chamber to induce CCR7-dependent chemotaxis. After 3 h, cells that migrated to the lower chamber were counted, and SLC-dependent migration was calculated as the ratio of the number of cells migrating with/without SLC. For the dermal DC migration assay, mice were killed, and dermal sheaths were prepared by splitting the ears. The sheaths were incubated dermal side down at 37°C, in medium (RPMI medium 1640/10% FCS/20 μM 2-mercaptoethanol/penicillin/streptomycin), with or without TGFβ (10 ng/ml). Twenty-four hours later, SLC (100 ng/ml) or buffer was added. After an additional 48 h, migrated cells were collected and stained, and the number of CD11c+MHCIIbright DC were determined by FACS analysis.
Results
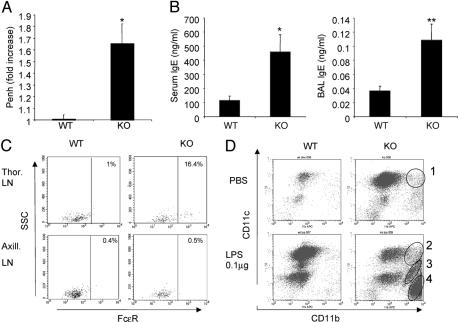
Runx3 KO Mice Display Major Hallmarks of Human Asthma. In an earlier study, we reported that Runx3 KO mice develop eosinophilic lung inflammation associated with airway remodeling and mucus hypersecretion (30). However, the question of whether the spontaneous development of these features in the KO mice was associated with other symptoms characteristic of human asthma remained open. AHR is a cardinal pathophysiological feature of human asthma, clinically defined by an increase in airway sensitivity to inhaled histamine or methacholine (40). In mouse models of asthma, AHR could be assessed by using the noninvasive enhanced pause (Penh) method (41). Examination of 4- to 9-week-old Runx3 KO mice and WT littermates revealed significantly increased methacholine-induced AHR in the KO mice (Fig. 1A). Of note, increasing the dosage of methacholine from 40 mg/ml to the reported 200 mg/ml (42) resulted in suffocation of the KO mice, whereas the WT mice remained unaffected. Interestingly, however, this AHR was transient and no longer observed in 5- to 6-month-old KO mice.
Fig. 1.
Human asthma hallmarks in Runx3 KO mice. (A) Increase of AHR in the KO mice. AHR was determined in 4- to 9-week-old KO mice and WT littermates as described in Materials and Methods. Results are presented as fold increase in enhanced pause (Penh) [a parameter related to pulmonary resistance (41)] relative to baseline value. Data represent the mean ± SEM of 15 mice per group and disclose a significant difference between WT and KO mice (Student's two-tailed t test; *, P = 0.0005). (B) Increase in serum and BAL IgE in the KO mice. Total IgE in serum and BAL of 4- to 6-week-old KO mice (n = 7) and WT littermates (n = 6) was quantitated by ELISA (Mouse IgE ELISA Quantization Kit, E90-115, Bethyl Laboratories, Montgomery, TX) according to the manufacturer's recommended conditions. Values in ng/ml disclose significant differences between WT and KO mice (Student's two-tailed t test; *, P = 0.026; **, P = 0.014). (C) Increased expression of FcεR receptors on DC from KO thoracic LN. Single-cell suspension was obtained from LN of WT and KO mice (n = 3), subjected to acid elution of occupied FcεR receptor, incubated with IgE-FITC conjugate and with anti CD11c, and subjected to FACS analysis. CD11c+ cells were gated, and FcεR level was determined (D) Increased responsiveness of KO mice to inhaled LPS. Runx3 KO mice and WT littermates (n = 4) were anesthetized and subjected to intranasal inhalation of 25 μl of PBS containing 0.1 μg of LPS or 25 μl of PBS alone. Sixteen hours later mice were killed and BAL was obtained and analyzed by FACS and, after cytospin, by Giemsa staining. Circled are specific populations: CD11c+ DC (circles 1 and 2), neutrophils (circle 3), and eosinophils (circle 4), as was also confirmed by Giemsa staining (data not shown).
Human asthma is characterized by a pronounced elevation in serum IgE (43, 44). In the KO mice, concentrations of IgE in both BAL and blood serum were 3- and 4-fold higher, respectively, than in WT littermate mice (Fig. 1B). High surface expression of the high-affinity receptor for IgE, FcεR, on LN DC is another important feature of human asthma (45, 46). FcεR expression on DC increases the efficiency of allergen presentation up to 1,000-fold through an antigen-focusing mechanism that enhances MHC presentation (45). We used FACS analysis to assess surface expression of FcεR on DC derived from the lung-draining thoracic LN. Significantly, as much as 16% of the DC in the thoracic LN of the KO mice expressed this allergy-mediating receptor, compared with a mere 1% in WT littermates (Fig. 1C). This latter low abundance of FcεR-expressing WT thoracic LN DC was similar to that in non-lung-draining LN (i.e., axillary LN) of either WT or Runx3 KO mice (Fig. 1C). These data are also consistent with findings that, in humans, increased serum IgE was associated with elevated FcεR expression on LN DC (46).
The severity of asthma in humans is influenced by exposure to LPS (47). We next assessed whether the KO mice are more susceptible than WT mice to minute amounts of inhaled LPS. Even without LPS (i.e., inhalation of PBS), a significant increase in the proportion of the highly allergenic (48) CD11c+/CD11b+ alveolar DC was observed in KO mice compared with WT littermates (Fig. 1D). Inhalation of LPS (0.1 μg) led to a further accumulation of the CD11c+/CD11b+ DC subset and to a pronounced recruitment of CD11c-/CD11b+ eosinophils and neutrophils to the lung of KO mice (Fig. 1D). This influx of eosinophils and neutrophils to the lungs of the KO mice indicates acute inflammation and underscores the oversensitivity of these mice to exogenous inducers of lung inflammation.
The results of methacholine-induced AHR, the increased serum and BAL IgE, the elevated DC surface expression of FcεR, and the hyperresponsiveness to inhaled LPS demonstrate that the pathophysiological features found in Runx3 KO mice recapitulate the clinical symptoms characterizing human asthma.
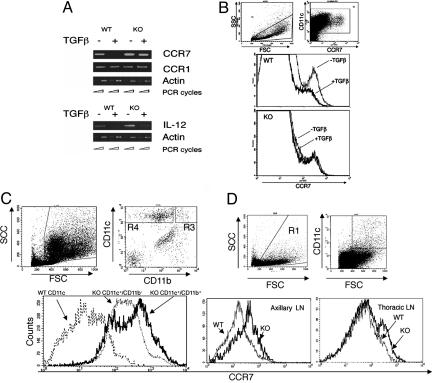
Runx3 Is Required for TGFβ-Mediated Attenuation of CCR7 Expression. Transcription and surface expression of CCR7 in DC are upregulated by maturation inducers (49) and down-regulated by TGFβ (21). Because our previous findings indicate that Runx3 mediates TGFβ signaling in DC, we addressed whether Runx3 is also involved in TGFβ-directed inhibition of CCR7 expression. A marked decrease in CCR7 transcription and surface expression occurred when WT BMDC were treated with TGFβ during maturation (Fig. 2 A and B). However, no such inhibition of CCR7 expression was noted in KO BMDC (Fig. 2 A and B). Of note, expression of another chemokine receptor, CCR1, known to be refractory to TGFβ inhibition (21), was not reduced in either KO or WT BMDC, whereas expression of IL-12, known to respond to TGFβ (50), was reduced in both KO and WT BMDC (Fig. 2A). The data indicate that Runx3 is specifically involved in TGFβ-dependent transcriptional attenuation of CCR7 expression in BMDC.
Fig. 2.
Dysregulated expression of CCR7 in Runx3 KO DC. (A) Impaired TGFβ-dependent inhibition of CCR7 transcription in KO BMDC. WT and Runx3 KO BMDC were grown in the presence or absence of TGFβ (10 ng/ml). At day 6, cells were induced to undergo maturation by LPS, and, at day 7, RNA was prepared and analyzed by RT-PCR. (B) Impaired TGFβ-dependent inhibition of surface expression of CCR7 in KO BMDC. WT and Runx3 KO BMDC were grown and treated as in A. At day 7, cells were analyzed by FACS by using anti-CCR7 antibodies (goat anti-mouse CI0131, Capralogics). FSChighCD11c+ DC were gated, and their CCR7 expression was determined. Reduction in the level of surface CCR7 and in the number of cells expressing it was noted only in WT and not in Runx3 KO BMDC. (C and D) Increased CCR7 expression on alveolar and LN DC of Runx3 KO mice. BAL and peripheral LN cells of KO and WT mice (n = 3) were obtained and analyzed. (C) Alveolar DC (FSChigh/CD11chigh) were gated and analyzed for CCR7 expression. Of note, expression of CCR7 on the DC subpopulation CD11c+/CD11b+ present only in KO lungs is shown along with that of KO CD11c+/CD11b- DC. (D) FSChigh/CD11chigh DC of axillary and thoracic LN were gated and analyzed for CCR7 expression.
Elevated CCR7 Expression on Alveolar and LN DC of Runx3 KO Mice. The fact that, in the absence of Runx3, transcriptional regulation of CCR7 expression in BMDC is impaired led us to examine the in vivo levels of CCR7 in WT and KO DC. Lung DC isolated from BAL of Runx3 KO mice express significantly higher levels of CCR7 than the corresponding DC of WT littermates (Fig. 2C). Of note, the highest level of CCR7 was recorded on the allergenic CD11c+/CD11b+ DC subset, which are not present in WT lung but are abundant in BAL of Runx3 KO mice (30). These results indicate that alveolar DC of the KO mice, including the CD11c+/CD11b+ allergenic subset, overexpress CCR7 and should thus possess an increased propensity to migrate. Significantly, increased surface expression of CCR7 was also noted on DC derived from either thoracic or axillary LN of KO mice, compared with WT littermates, (Fig. 2D).
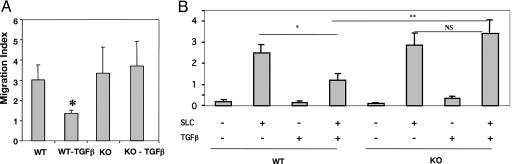
Runx3 KO DC Display Increased Migration Ability. Because CCR7 and its ligands (ELC/CCL19 and SLC/CCL21) are important components of the DC migration and navigation system (51), we asked whether increased CCR7 due to the loss of Runx3 indeed affects DC migratory properties. WT and KO BMDC treated or untreated with TGFβ were induced to undergo maturation by LPS and then subjected to an SLC/CCL21-dependent transmigration assay. TGFβ treatment significantly reduced the migration rate of WT BMDC (Fig. 3A), consistent with the inhibition of CCR7 expression by this cytokine (Fig. 2A). No such inhibition of SLC/CCL21-mediated chemotaxis was noted in TGFβ-treated KO BMDC (Fig. 3A).
Fig. 3.
SLC-directed chemotaxis of Runx3 KO DC is not attenuated by TGFβ. (A) BMDC from WT and Runx3 KO littermates were cultured as above with or without TGFβ and, at day 6, were treated overnight with LPS. BMDC (5 × 105) were placed in a Transwell migration chamber, and SLC-dependent chemotaxis was measured as described in Materials and Methods. The mean ± SEM of three separate experiments is presented. Migration inhibition of WT DC by TGFβ was significant (*) by using the paired Student t test (P = 0.05). (B) Dermal sheaths of WT and KO mice (n = 3) were prepared, and SLC-mediated chemotactic migration was assessed. Inhibition of WT DC migration by TGFβ (*) was significant (P = 0.029) as were the responses of WT and KO DC to SLC and TGFβ (**)(P = 0.019).
DC migration was also assessed by using another in vitro assay. SLC is produced at two points along DC migration route from skin to regional LN, the lymphatic endothelium and the LN T cell zone (17). We thus examined the migratory capability of dermal DC, comparing WT with KO mice. Dermal sheaths were prepared and incubated, dermal side down, with or without TGFβ and SLC. In both WT and Runx3 KO sheaths, a pronounced SLC-dependent migration of CD11c+/MHCIIhigh DC was recorded (Fig. 3B). However, whereas the migration of WT dermal DC was inhibited by TGFβ, migration of the KO DC was not. The results of BMDC and dermal DC migration assays indicate that loss of Runx3 not only impairs TGFβ-mediated transcription regulation of CCR7, but also leads to an impairment of TGFβ-dependent migration inhibition of DC toward the CCR7 ligand SLC.
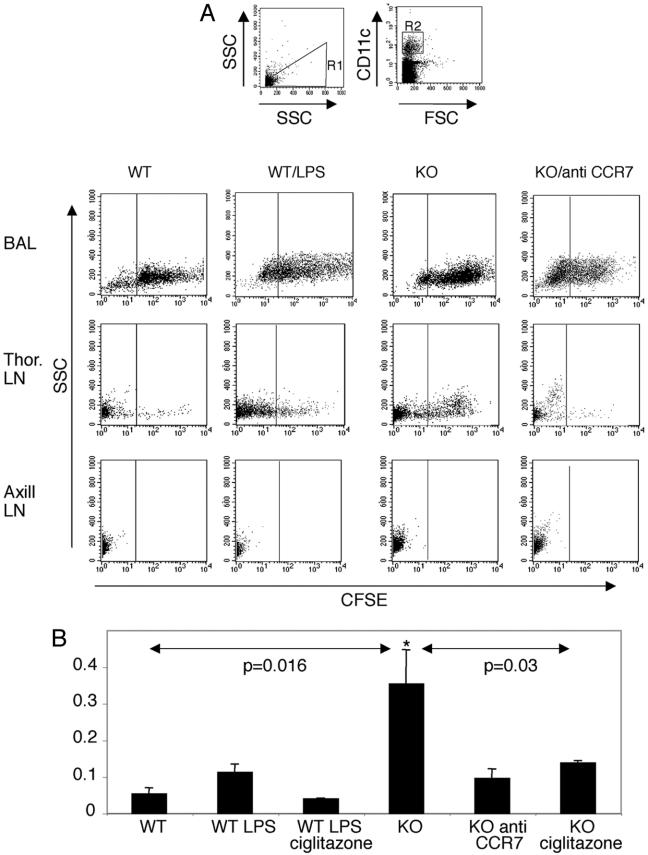
Enhanced CCR7-Dependent Migration of Respiratory DC to Draining LN in Runx3 KO Mice. Migration of respiratory DC to draining LN is a key step in the initiation of immune response in the lung (52). The results of the transmigration in vitro assays led us to examine the in vivo trafficking of respiratory DC in KO and WT mice. In vivo migration was assessed by monitoring the accumulation of CFSE-labeled respiratory DC in the regional LN. In WT mice, respiratory CFSE+CD11c+ DC accumulate primarily in the draining thoracic LN, amounting to 1–3% of the total LN DC (Fig. 4). These data correspond with previously reported findings (52) and are highly specific because no CFSE+ respiratory DC were detected in the nondraining axillary LN, used as a reference (Fig. 4A). In comparison with WT, significantly more respiratory CFSE+ DC were found in thoracic LN of KO mice (Fig. 4), which correlates well with the higher migratory capacity of KO DC in vitro and the elevated CCR7 on KO DC. Treatment of WT mice by intranasal instillation of LPS caused an increase in migration of respiratory DC, although to a much lower extent compared with the untreated KO mice (Fig. 4B). These results indicate that the increased expression of CCR7 on Runx3 KO DC results in increased CCR7-dependent migration of respiratory DC to the draining LN. This conclusion is supported by the demonstration that blocking CCR7 by receptor-specific antibodies markedly reduced the migration of respiratory DC in the KO mice (Fig. 4B).
Fig. 4.
Elevated CCR7 mediated in vivo trafficking of alveolar DC to the draining LN in the KO mice. (A and B) Runx3 KO (n = 7) and WT (n = 5) mice were treated by intranasal administration of CFSE to label in vivo the respiratory DC. When indicated, WT mice were treated with LPS (n = 4), and Runx3 KO mice were treated with anti-CCR7 antibody (n = 4) or with buffer only (n = 4). KO mice (n = 4) and LPS-treated WT mice (n = 3) were also treated by inhalation of Ciglitazone. Eighteen hours later, mice were killed, and single-cell suspensions of BAL, thoracic LN, and axillary LN were prepared and analyzed by FACS. (A) FSChigh/CD11c+ DC were gated (R1 and R2). Shown is representative side scatter (SSC) versus CFSE staining of DC populations in BAL and LN after the various treatments. (B) Migration index of alveolar DC to thoracic LN represents the ratio between the percentage of CFSE+ cells within the CD11c+ population in the thoracic LN and the respective value in BAL cells. Results are presented as mean ± SEM. Analysis of variance showed that the migration index of untreated KO DC was significantly higher than that of WT (*, P = 0.016). Notably, the anti-CCR7-treated KO DC migration index was similar to basal migration of WT, and Ciglitazone significantly (*, P = 0.03) reduced the migration of KO DC.
Hammad et al. (35) have reported that activation of the peroxisome proliferator-activated receptor γ (PPARγ) by selective PPARγ agonists such as Rosiglitazone and Ciglitazone decreased CCR7 expression on DC and inhibited their migratory properties. Consequently, this treatment resulted in reduced DC-induced eosinophilic lung inflammation (35). We addressed whether PPARγ activation would inhibit the migration of respiratory KO DC, as did the anti-CCR7 antibodies. When mice inhaled Ciglitazone, before instillation of CFSE, a pronounced inhibition of KO DC migration to draining LN was observed (Fig. 4B). Of note, Ciglitazone was almost as effective as the anti-CCR7 antibodies in inhibiting DC migration to LN, and also caused migration inhibition of WT DC in LPS-treated mice (Fig. 4B). Taken together, the results of elevated CCR7, RNA, and protein, in KO DC and the in vitro and in vivo DC migration studies, demonstrate that Runx3 functions as a negative regulator of CCR7 transcription in response to TGFβ signaling and thereby regulates the trafficking of alveolar DC to the draining LN. The data may also suggest that Ciglitazone could be useful in treatment of DC-induced immunity to self-antigens.
Discussion
Respiratory tract DC are an important population of regulatory cells that orchestrate immunity against inhaled antigens (5, 7). These cells are normally maintained at their immature state through interaction with antiinflammatory cytokines, such as TGFβ, secreted by the surrounding cellular environment (5). At this immature state, respiratory DC have a low propensity to migrate to the draining LN and a poor capacity to present antigens to T cells (1, 9, 11, 16, 52). We previously reported that DC in Runx3 KO mice do not respond to TGFβ-mediated maturation inhibition and that the KO mice spontaneously develop lung inflammation associated with mucus hypersecretion and airway remodeling (30). The data presented here show that these histopathological features have pathophysiological consequences and that the KO mice exhibit major hallmarks of human asthma (53). These features include high sensitivity to methacholine-induced AHR (40, 42), elevation of BAL and serum IgE, increased FcεR1 expression on DC (5, 46, 54) and hypersensitivity to inhaled LPS (47). The data indicate that the pathophysiological features found in Runx3 KO mice recapitulate clinical symptoms of human asthma.
The process of DC maturation involves the up-regulation of CCR7, whose expression enables the activated DC to leave sites of inflammation and migrate, by means of afferent lymphatics, to draining LN (5, 19, 20). Accordingly, up-regulation of CCR7 is required for inducing allergic lung inflammation (55). TGFβ attenuates the expression of CCR7 by down-regulation of its transcription (21). We show here that Runx3 mediates this TGFβ-directed transcriptional attenuation of CCR7. Accordingly, when Runx3 is lost, alveolar DC in the KO mice express higher levels of CCR7 compared with WT mice. This occurrence caused marked increase in the migratory capacity of KO DC in response to the CCR7 ligand SLC and led to enhanced migration of airway DC that have not been exposed to pathogen-associated molecular pattern “danger signals” to the draining LN. In the LN, these promiscuously mature KO DC could present innocuous antigens to naive T cells and elicit clonal proliferation against otherwise “not dangerous” or self-antigens. Further propagation of this induced immune response leads to the spontaneous development of asthmatic conditions in the KO mice.
These data are particularly relevant to previous findings demonstrating the important role of immature DC in peripheral tolerance (9, 11, 16). Under steady-state conditions, immature tolerance-inducing DC do not elicit an inflammatory immune response, but are nevertheless able to phagocytose cells dying by apoptosis, migrate to regional LN, and stimulate T cell proliferation (9). Importantly, the steady-state migration of these immature tolerance-inducing DC depends on the expression of CCR7, indicating a role for CCR7 expression in maintenance of peripheral tolerance (51). In Runx3 KO mice, the dysregulated CCR7 expression, which leads to enhanced migration and unscheduled maturation of what should otherwise be immature tolerance-inducing DC, could impair peripheral tolerance and elicit immunity to self-antigens (11). Interestingly, an autoimmune disease-associated regulatory SNP affecting RUNX-binding sites that are known to be recognized by all three RUNX proteins was found in several autoimmune diseases (23). Thus, the potential involvement of RUNX3 deficiency in autoimmunity, through affecting the balance of steady-state trafficking of tolerogenic and mature activated DC, is an intriguing possibility (56).
Acknowledgments
We thank Judith Chermesh and Rafi Saka for help in animal husbandry; Dorit Nathan and Tamara Berkuzki for technical assistance; Dr. Alon Harmelin and Dr. Ori Brenner for helpful discussions; Drs. Ditsa Levanon, Joseph Lotem, and Steffen Jung for insightful comments; and Dr. Israel Pecht (The Weizmann Institute of Science) for the generous gift of IgE-FITC conjugate. This work was supported by grants from the Commission of the European Union, the Israel Science Foundation, the Minerva Foundation Germany, the Philip Morris External Research Program, and the Weizmann Institute Shapell Family Biomedical Research Foundation.
Abbreviations: AHR, airway hyper-responsiveness; DC, dendritic cell; CCR7, chemokine receptor 7; LN, lymph node; BAL, bronchoalveolar lavage; KO, knockout; CFSE, carboxyfluorescein diacetate-succinimidyl ester; BMDC, bone marrow-derived DC; SLC, secondary lymphoid-tissue chemokine.
References
- 1.Banchereau, J. & Steinman, R. M. (1998) Nature 392, 245-252. [DOI] [PubMed] [Google Scholar]
- 2.Umetsu, D. T., McIntire, J. J., Akbari, O., Macaubas, C. & DeKruyff, R. H. (2002) Nat. Immunol. 3, 715-720. [DOI] [PubMed] [Google Scholar]
- 3.Wills-Karp, M. (1999) Annu. Rev. Immunol. 17, 255-281. [DOI] [PubMed] [Google Scholar]
- 4.Wills-Karp, M. (2000) Immunopharmacology 48, 263-268. [DOI] [PubMed] [Google Scholar]
- 5.Upham, J. W. & Stumbles, P. A. (2003) Pharmacol. Ther. 100, 75-87. [DOI] [PubMed] [Google Scholar]
- 6.van Rijt, L. S., Jung, S., Kleinjan, A., Vos, N., Willart, M., Duez, C., Hoogsteden, H. C. & Lambrecht, B. N. (2005) J. Exp. Med. 201, 981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambrecht, B. N. & Hammad, H. (2003) Nat. Rev. Immunol. 3, 994-1003. [DOI] [PubMed] [Google Scholar]
- 8.Herrick, C. A. & Bottomly, K. (2003) Nat. Rev. Immunol. 3, 405-412. [DOI] [PubMed] [Google Scholar]
- 9.Steinman, R. M., Hawiger, D. & Nussenzweig, M. C. (2003) Annu. Rev. Immunol. 21, 685-711. [DOI] [PubMed] [Google Scholar]
- 10.Lutz, M. B. & Schuler, G. (2002) Trends Immunol. 23, 445-449. [DOI] [PubMed] [Google Scholar]
- 11.Banchereau, J., Pascual, V. & Palucka, A. K. (2004) Immunity 20, 539-550. [DOI] [PubMed] [Google Scholar]
- 12.Nathan, C. (2002) Nature 420, 846-852. [DOI] [PubMed] [Google Scholar]
- 13.Soltys, J., Bonfield, T., Chmiel, J. & Berger, M. (2002) J. Immunol. 168, 1903-1910. [DOI] [PubMed] [Google Scholar]
- 14.Takeuchi, M., Alard, P. & Streilein, J. W. (1998) J. Immunol. 160, 1589-1597. [PubMed] [Google Scholar]
- 15.Chelen, C. J., Fang, Y., Freeman, G. J., Secrist, H., Marshall, J. D., Hwang, P. T., Frankel, L. R., DeKruyff, R. H. & Umetsu, D. T. (1995) J. Clin. Invest. 95, 1415-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinman, R. M., Hawiger, D., Liu, K., Bonifaz, L., Bonnyay, D., Mahnke, K., Iyoda, T., Ravetch, J., Dhodapkar, M., Inaba, K. & Nussenzweig, M. (2003) Ann. N.Y. Acad. Sci. 987, 15-25. [DOI] [PubMed] [Google Scholar]
- 17.Gunn, M. D., Kyuwa, S., Tam, C., Kakiuchi, T., Matsuzawa, A., Williams, L. T. & Nakano, H. (1999) J. Exp. Med. 189, 451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baggiolini, M. (1998) Nature 392, 565-568. [DOI] [PubMed] [Google Scholar]
- 19.Hirao, M., Onai, N., Hiroishi, K., Watkins, S. C., Matsushima, K., Robbins, P. D., Lotze, M. T. & Tahara, H. (2000) Cancer Res. 60, 2209-2217. [PubMed] [Google Scholar]
- 20.Forster, R., Schubel, A., Breitfeld, D., Kremmer, E., Renner-Muller, I., Wolf, E. & Lipp, M. (1999) Cell 99, 23-33. [DOI] [PubMed] [Google Scholar]
- 21.Ogata, M., Zhang, Y., Wang, Y., Itakura, M., Zhang, Y. Y., Harada, A., Hashimoto, S. & Matsushima, K. (1999) Blood 93, 3225-3232. [PubMed] [Google Scholar]
- 22.Levanon, D. & Groner, Y. (2004) Oncogene 23, 4211-4219. [DOI] [PubMed] [Google Scholar]
- 23.Alarcon-Riquelme, M. E. (2004) Arthritis Res. Ther. 6, 169-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levanon, D., Brenner, O., Negreanu, V., Bettoun, D., Woolf, E., Eilam, R., Lotem, J., Gat, U., Otto, F., Speck, N. & Groner, Y. (2001) Mech. Dev. 109, 413-417. [DOI] [PubMed] [Google Scholar]
- 25.Levanon, D., Bettoun, D., Harris-Cerruti, C., Woolf, E., Negreanu, V., Eilam, R., Bernstein, Y., Goldenberg, D., Xiao, C., Fliegauf, M., et al. (2002) EMBO J. 21, 3454-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue, K., Ozaki, S., Shiga, T., Ito, K., Masuda, T., Okado, N., Iseda, T., Kawaguchi, S., Ogawa, M., Bae, S. C., et al. (2002) Nat. Neurosci. 5, 946-954. [DOI] [PubMed] [Google Scholar]
- 27.Ehlers, M., Laule-Kilian, K., Petter, M., Aldrian, C. J., Grueter, B., Wurch, A., Yoshida, N., Watanabe, T., Satake, M. & Steimle, V. (2003) J. Immunol. 171, 3594-3604. [DOI] [PubMed] [Google Scholar]
- 28.Taniuchi, I., Osato, M., Egawa, T., Sunshine, M. J., Bae, S. C., Komori, T., Ito, Y. & Littman, D. R. (2002) Cell 111, 621-633. [DOI] [PubMed] [Google Scholar]
- 29.Woolf, E., Xiao, C., Fainaru, O., Lotem, J., Rosen, D., Negreanu, V., Bernstein, Y., Goldenberg, D., Brenner, O., Berke, G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 7731-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fainaru, O., Woolf, E., Lotem, J., Yarmus, M., Brenner, O., Goldenberg, D., Negreanu, V., Bernstein, Y., Levanon, D., Jung, S. & Groner, Y. (2004) EMBO J. 23, 969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Q. L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X. Z., Lee, K. Y., Nomura, S., Lee, C. W., Han, S. B., et al. (2002) Cell 109, 113-124. [DOI] [PubMed] [Google Scholar]
- 32.Ito, Y. & Miyazono, K. (2003) Curr. Opin. Genet. Dev. 13, 43-47. [DOI] [PubMed] [Google Scholar]
- 33.Brenner, O., Levanon, D., Negreanu, V., Golubkov, O., Fainaru, O., Woolf, E. & Groner, Y. (2004) Proc. Natl. Acad. Sci. USA 101, 16016-16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levanon, D., Brenner, O., Otto, F. & Groner, Y. (2003) EMBO Rep. 4, 560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammad, H., de Heer, H. J., Soullie, T., Angeli, V., Trottein, F., Hoogsteden, H. C. & Lambrecht, B. N. (2004) Am. J. Pathol. 164, 263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levanon, D., Negreanu, V., Bernstein, Y., Bar-Am, I., Avivi, L. & Groner, Y. (1994) Genomics 23, 425-432. [DOI] [PubMed] [Google Scholar]
- 37.Haagerup, A., Bjerke, T., Schiotz, P. O., Binderup, H. G., Dahl, R. & Kruse, T. A. (2002) Allergy 57, 680-686. [DOI] [PubMed] [Google Scholar]
- 38.Yokouchi, Y., Shibasaki, M., Noguchi, E., Nakayama, J., Ohtsuki, T., Kamioka, M., Yamakawa-Kobayashi, K., Ito, S., Takeda, K., Ichikawa, K., et al. (2002) Genes Immun. 3, 9-13. [DOI] [PubMed] [Google Scholar]
- 39.Lutz, M. B., Suri, R. M., Niimi, M., Ogilvie, A. L., Kukutsch, N. A., Rossner, S., Schuler, G. & Austyn, J. M. (2000) Eur. J. Immunol. 30, 1813-1822. [DOI] [PubMed] [Google Scholar]
- 40.Koh, Y. Y., Park, Y. & Kim, C. K. (2002) Allergy 57, 1165-1170. [DOI] [PubMed] [Google Scholar]
- 41.Drazen, J. M., Finn, P. W. & De Sanctis, G. T. (1999) Annu. Rev. Physiol. 61, 593-625. [DOI] [PubMed] [Google Scholar]
- 42.Finotto, S., Neurath, M. F., Glickman, J. N., Qin, S., Lehr, H. A., Green, F. H., Ackerman, K., Haley, K., Galle, P. R., Szabo, S. J., et al. (2002) Science 295, 336-338. [DOI] [PubMed] [Google Scholar]
- 43.Wills-Karp, M. & Ewart, S. L. (2004) Nat. Rev. Genet. 5, 376-387. [DOI] [PubMed] [Google Scholar]
- 44.Cookson, W. (2004) Nat. Rev. Immunol. 4, 978-988. [DOI] [PubMed] [Google Scholar]
- 45.Maurer, D., Fiebiger, E., Reininger, B., Ebner, C., Petzelbauer, P., Shi, G. P., Chapman, H. A. & Stingl, G. (1998) J. Immunol. 161, 2731-2739. [PubMed] [Google Scholar]
- 46.Foster, B., Metcalfe, D. D. & Prussin, C. (2003) J. Allergy Clin. Immunol. 112, 1132-1138. [DOI] [PubMed] [Google Scholar]
- 47.Liu, A. H. (2004) Paediatr. Respir. Rev. 5, Suppl. A, S65-S71. [DOI] [PubMed] [Google Scholar]
- 48.Julia, V., Hessel, E. M., Malherbe, L., Glaichenhaus, N., O'Garra, A. & Coffman, R. L. (2002) Immunity 16, 271-283. [DOI] [PubMed] [Google Scholar]
- 49.Yanagihara, S., Komura, E., Nagafune, J., Watarai, H. & Yamaguchi, Y. (1998) J. Immunol. 161, 3096-3102. [PubMed] [Google Scholar]
- 50.Du, C. & Sriram, S. (1998) J. Leukocyte Biol. 64, 92-97. [DOI] [PubMed] [Google Scholar]
- 51.Ohl, L., Mohaupt, M., Czeloth, N., Hintzen, G., Kiafard, Z., Zwirner, J., Blankenstein, T., Henning, G. & Forster, R. (2004) Immunity 21, 279-288. [DOI] [PubMed] [Google Scholar]
- 52.Legge, K. L. & Braciale, T. J. (2003) Immunity 18, 265-277. [DOI] [PubMed] [Google Scholar]
- 53.Cotran, R., Kumar, V. & Collins, T. (1999) Robbins Pathologic Basis of Disease (Saunders, Philadelphia).
- 54.Cookson, W. & Moffatt, M. (2004) N. Engl. J. Med. 351, 1794-1796. [DOI] [PubMed] [Google Scholar]
- 55.Hammad, H., Lambrecht, B. N., Pochard, P., Gosset, P., Marquillies, P., Tonnel, A. B. & Pestel, J. (2002) J. Immunol. 169, 1524-1534. [DOI] [PubMed] [Google Scholar]
- 56.Alarcon-Riquelme, M. E. (2003) Nat. Genet. 35, 299-300. [DOI] [PubMed] [Google Scholar]