The article by Löytynoja and Goldman (1) in this issue of PNAS describes a novel and useful method of handling gaps in progressive multiple sequence alignments. Gaps are the bits that get left behind when you try to align DNA or protein sequences and have to use padding or null characters to match homologous residues. These could get placed at sites where one sequence has apparently lost some residues (caused by a deletion), and you simply pad out the sequence with gap characters such as hyphens or blanks to make it match up with the sequences that have not lost anything. Similarly, if one or more sequences have some extra residues (caused by an insertion) then these will need to be matched by gap characters in the other sequences. It is the placement of these gaps that creates all of the problems when you try to automatically generate alignments. If insertions and deletions never happened, then sequences could easily be matched by sliding them past each other and taking the alignment that best matched the residues. When gaps are needed, things get complicated and much of the first 20 years of bioinformatics was devoted to how these should be placed and why (e.g., refs. 2-4).
When you have just two sequences, there are fast and relatively simple algorithms that can guarantee the best alignment between the sequences, given a scoring function that gives a score for each pair of aligned residues. The most familiar of these is the famous dynamic programming algorithm, first described for sequence alignment by Needleman and Wunsch (5). Gaps can be placed all over both sequences to get the best score so a “gap penalty” function is used to penalize for gaps of different sizes. These scores are used to give a balance between gaps and matches. In an ideal world, if you use appropriate values for the residue match scores such as from a blosum matrix (6) and a sensible form for the gap penalty function, then you might end up with an alignment where the gaps are placed at or near the actual sites of insertions or deletions and as many homologous residues as possible would be lined up. With just two sequences you have no way of knowing if any of the resulting gaps are caused by insertions or deletions or a combination. They simply correspond to places where the sequences are aligned better by using gaps to get the best overall score. Hence these gap positions are sometimes referred to as “indels” or simply as gaps. Until the early 1990s, these alignments were usually carried out by using dynamic programming and simple deterministic scoring schemes or an approximation to them. These days, you can also use probabilistic scoring schemes (7) and hidden Markov models to carry out alignments as done by Löytynoja and Goldman (1).
With multiple alignments, things are more complicated. Here, in principle,
One can also use probabilistic scoring schemes and hidden Markov models to carry out alignments.
you can sometimes tell whether a particular gap was caused by an insertion or a deletion and in which sequences. In practice, this is complicated to do routinely, and the programs that were most commonly used to make multiple alignments [e.g., clustalw (8) and t-coffee (9)] simply ignored this nicety. The direct application of dynamic programming to more than a handful of sequences is an extremely demanding task, and, more or less, all programs that are routinely used use heuristics. The most common heuristic is what Feng and Doolittle (10) referred to as “progressive alignment” but which has been described in different ways by a number of authors (e.g., refs. 11 and 12). This heuristic involves building the multiple alignment up gradually according to the branching order in an initial approximate tree. Progressive alignment is behind the widely used clustalw programs (8) and many of the most successful multiple alignment programs that have been developed over the past 5 years or so (9, 13-15).
When clustalw runs, the gaps that are produced do not necessarily contain any direct phylogenetic information. Insertions that occur in early subalignments get penalized again in later alignments because gaps have to be inserted in all sequences that get joined to the earlier alignment (illustrated in Fig. 1). clustalw attempts to compensate by using an elaborate scoring scheme to encourage gaps to end up on top of each other. Position-specific gap penalties are used to reduce the gap opening penalty at these positions so that new gaps prefer to end up over old gaps. This process results in alignments that are very “block-like” with sections of gap-free alignment separated by sections that are full of gaps. The results look good with protein alignments, and when it works well the blocks correspond to sections of core secondary structure, whereas the more gap-rich sections correspond to the less conserved loops that connect them. We know empirically that this strategy works well because we can take sets of structurally aligned proteins [e.g., homstrad (16)] and compare the performance of clustalw to the reference sets. Although clustalw is by no means the most accurate program in use, it performs well in a wide variety of situations. As an added bonus, the alignments can look aesthetically pleasing and simple.
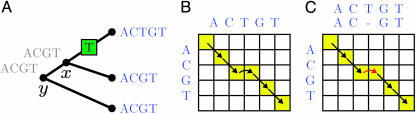
Fig. 1.
An example of an insertion in a sequence that is dealt with correctly by the new algorithm. (A) Progressive alignment is performed on the tree. Note the insertion of T. (B) The dynamic programming that occurs at node x is shown. (C) The dynamic programming that occurs at node y is shown. The red arrow indicates that this insertion has already been penalized (at node x) and is not penalized again.
However, there may be a price for this prettiness and detachment from phylogenetic reality. clustalw (and other programs) may be guilty of “over-alignment” (17), that is where sequences that should not go together are forced into neat-looking blocks. These over-aligned regions may be neat looking but misleading. With DNA and RNA, the situation is possibly more serious. In general, the developers of alignment programs have always found DNA to be a bit of a nuisance compared with proteins. Amino acids come in nice groups of related amino acids with lots of interesting properties. With nucleotide sequences, you have just four, equally uninteresting, residues. Further, depending on what the DNA or RNA codes for or does, gaps might occur in a haphazard manner. It will depend on the situation.
Löytynoja and Goldman's new algorithm attempts to keep track of each gap that is introduced in a multiple alignment.
Löytynoja and Goldman (1) give an example in their article of some genomic sequences that are clearly mis-aligned by clustalw but correctly aligned when gaps are treated properly. Their new algorithm attempts to properly keep track of each gap that is introduced in a multiple alignment and especially whether it appears to have come from an insertion or a deletion. In contrast to normal progressive alignment algorithms, insertions are only penalized once (see Fig. 1). This process makes progressive alignment try to correctly reflect what actually happened the sequences. It should help to give alignments that have gaps placed correctly with regard to where insertions and deletions have actually occurred rather than some aesthetic notion. The downside is that alignments may start getting ugly, compared with how we have learned to appreciate neatly colored multiple alignments when they are reproduced on journal pages.
As more and more whole genomes get sequenced, there is an increasing need to align more and more nucleotide sequences of different kinds. These include sequences that code for functional RNAs and noncoding sequences that may contain regulatory motifs. With protein alignments, sets of test cases such as those from homstrad or balibase (18) have helped enormously in the development of new algorithms and in providing sanity checks against outlandish claims by algorithm developers. With DNA or RNA, such test cases are much more difficult to make, and there are many different types of situations that would need to be covered. Structural or functional RNAs may be straightforward enough that test cases can be made (19), but noncoding genomic DNA could be much more difficult.
There is an understandable tendency for users of multiple alignment software to want their residues neatly aligned in blocks and columns. This is fine when such blocks are biologically accurate as will happen in parts of protein alignments. In cases where insertions or deletions have happened in a less organized manner, as will happen in many noncoding DNA sequences and in less organized parts of protein sequences, such block-like alignments may be biologically meaningless. Perhaps we need to reeducate our eyes to see beauty in what actually happened rather than what looks nice on paper.
See companion article on page 10557.
References
- 1.Löytynoja, A. & Goldman, N. (2005) Proc. Natl. Acad. Sci. USA 102, 10557-10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sankoff, D. & Kruskal, J. (1983) Time Warps, String Edits, and Macromolecules: The Theory and Practice of Sequence Comparison (Addison-Wesley, Reading, MA).
- 3.Smith, T. F. & Waterman, M. S. (1981) J. Mol. Biol. 147, 195-197. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F. (1989) J. Theor. Biol. 138, 297-309. [DOI] [PubMed] [Google Scholar]
- 5.Needleman, S. B. & Wunsch, C. D. (1970) J. Mol. Biol. 48, 443-453. [DOI] [PubMed] [Google Scholar]
- 6.Henikoff, S. & Henikoff, J. G. (1994) J. Mol. Biol. 243, 574-578. [DOI] [PubMed] [Google Scholar]
- 7.Krogh, A., Brown, M., Mian, I. S., Sjolander, K. & Haussler, D. (1994) J. Mol. Biol. 235, 1501-1531. [DOI] [PubMed] [Google Scholar]
- 8.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205-217. [DOI] [PubMed] [Google Scholar]
- 10.Feng, D. F. & Doolittle, R. F. (1987) J. Mol. Evol. 25, 351-360. [DOI] [PubMed] [Google Scholar]
- 11.Taylor, W. R. (1988) J. Mol. Evol. 28, 161-169. [DOI] [PubMed] [Google Scholar]
- 12.Hogeweg, P. & Hesper, B. (1984) J. Mol. Evol. 20, 175-186. [DOI] [PubMed] [Google Scholar]
- 13.Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) Nucleic Acids Res. 30, 3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar, R. C. (2004) Nucleic Acids Res. 32, 1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do, C. B., Mahabhashyam, M. S., Brudno, M. & Batzoglou, S. (2005) Genome Res. 15, 330-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuguchi, K., Deane, C. M., Blundell, T. L. & Overington, J. P. (1998) Protein Sci. 7, 2469-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cline, M., Hughey, R. & Karplus, K. (2002) Bioinformatics 18, 306-314. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. D., Plewniak, F. & Poch, O. (1999) Nucleic Acids Res. 15, 87-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner, P. P., Wilm, A. & Washietl, S. (2005) Nucleic Acids Res. 33, 2433-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]