Abstract
Rapid evolution of RNA viruses with mRNA-sense genomes is a major concern to health and economic welfare because of the devastating diseases these viruses inflict on humans, animals, and plants. To test whether host genes can affect the evolution of RNA viruses, we used a Saccharomyces cerevisiae single-gene deletion library, which includes ≈80% of yeast genes, in RNA recombination studies based on a small viral replicon RNA derived from tomato bushy stunt virus. The genome-wide screen led to the identification of five host genes whose absence resulted in the rapid generation of new viral RNA recombinants. Thus, these genes normally suppress viral RNA recombination, but in their absence, hosts become viral recombination “hotbeds.” Four of the five suppressor genes are likely involved in RNA degradation, suggesting that RNA degradation could play a role in viral RNA recombination. In contrast, deletion of four other host genes inhibited virus recombination, indicating that these genes normally accelerate the RNA recombination process. A comparison of deletion strains with the lowest and the highest recombination rate revealed that host genes could affect recombinant accumulation by up to 80-fold. Overall, our results demonstrate that a set of host genes have a major effect on RNA virus recombination and evolution.
Keywords: host factors, plus-strand RNA virus, tombusvirus, yeast, evolution
Rapid evolution of RNA viruses with mRNA-sense genomes, which include severe acute respiratory syndrome coronavirus, hepatitis C virus, and West Nile virus, makes controlling RNA viruses a difficult task. The emergence of new pathogenic RNA viruses is frequently due to RNA recombination (1, 2), which can lead to dramatic changes in viral genomes by creating novel combinations of genes, motifs, or regulatory RNA sequences. Thus, RNA recombination can change the infectious properties of RNA viruses and render vaccines and other antiviral methods ineffective (2). RNA recombination likely contributed to outbreaks with denguevirus (3, 4), poliovirus (5), calicivirus (6), astrovirus (7), enterovirus (8, 9), influenzavirus (10), pestivirus (11, 12), and severe acute respiratory syndrome coronavirus, a newly emerged viral pathogen of humans (13–15). RNA recombination also is important in viral RNA repair, which likely increases the fitness of RNA viruses that lack proofreading polymerases (1, 16–18).
Current models of RNA recombination are based on a template-switching mechanism driven by the viral replicase (1, 16) or RNA breakage and ligation (19). The more common template-switching RNA recombination is thought to occur as an error during the replication process (1, 16). Because viral RNA replication depends not only on viral proteins but also on host factors (20), it is likely that host factors could affect the recombination process, too. However, despite the significance of RNA recombination in viral evolution, the possible roles of host genes in the viral RNA recombination process are currently unknown.
Tombusviruses, including tomato bushy stunt virus (TBSV) and cucumber necrosis virus, are nonsegmented, small model positive-strand RNA viruses (21). Because of their robust replication and ability to generate novel RNA recombinants in whole plants and single cells, tombusviruses are used extensively to dissect the roles of cis-acting RNA elements during virus infections (21). In vivo and in vitro replication/recombination studies with a small replicon RNA, termed defective interfering 72 (DI-72) RNA (21, 22), established a role for RNA sequences/structures and viral replicase proteins in RNA recombination. Coexpression of the replicon RNA with the two essential tombusviral replicase proteins (see Fig. 1A) resulted in robust DI RNA replication in Saccharomyces cerevisiae (23, 24), which is a model eukaryotic host. Yeast also supported viral RNA recombination, giving rise to recombinants similar to those in plants and plant protoplasts (23). Therefore, yeast could be a useful host to study viral RNA recombination and to identify host proteins involved in this process.
Fig. 1.
Absence of CTL1, MET22/HAL2, HUR1, XRN1, and UBP3 host genes leads to enhanced recombination of TBSV DI-72 RNA replicon in yeast. (A) Plasmid-based expression of p33 and p92 replicase proteins and DI-72 RNA replicon in yeast. (B) Total RNA extracts from the shown yeast strains (two independent samples are shown for each strain to illustrate the reproducibility of recombinant accumulation) were visualized with ethidium bromide or probed with a radiolabeled RNA that was complementary with either RIII (C) or RI (D) of DI-72. Arrow points at the replicon, whereas the previously uncharacterized recombinant RNAs (recRNA) are bracketed. Various recombinants in hur1Δ are depicted with arrowheads. Samples from hur1Δ yeast were overloaded (≈×5) to facilitate visualization of viral RNAs. Short, 5′ truncated viral RNAs are marked with asterisks.
In this paper, we have tested the effect of ≈80% of all yeast genes on TBSV recombination based on screening the entire yeast single-gene knockout (YKO) library for the occurrence of viral RNA recombinants. Using the TBSV-derived replicon RNA, we identified five YKO strains that supported unusually high levels of new recombinant RNAs. We also identified four yeast deletion strains that showed reduced viral recombinant accumulation. Therefore, a selected set of host genes could either suppress or accelerate viral RNA recombination, demonstrating that host genes play significant roles in virus recombination and evolution.
Materials and Methods
Yeast Strains and Expression Plasmids. S. cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the haploid-deletion series (BY4741 strain background) were from Open Biosystems (Huntsville, AL). The expression plasmids pGBK-His-33 (carrying cucumber necrosis virus p33 gene behind the ADH1 promoter), pGAD-His-92 (containing cucumber necrosis virus p92 gene behind the ADH1 promoter), and pYC/DI-72 (expressing TBSV DI-72 RNA under the control of the GAL1 promoter) were described in refs. 23 and 25. Each yeast strain was cotransformed with all three plasmids by using the lithium acetate/ssDNA/polyethylene glycol method (26), and transformants were selected by complementation of auxotrophic markers. Of 4,848 strains, we found that 71 were not transformable and 229 strains did not grow on galactose-containing medium. Therefore, a total of 4,548 strains were tested for RNA recombination below.
Yeast Cultivation. Each transformed yeast strain from the YKO library was cultured under two different conditions during the genome-wide screen for RNA recombinants. The first screen included yeast strains grown in 96-deep-well plates at 23°C in selective medium (SC-ULH-) (23) with 2% galactose until reaching cell density of 0.8–1.0 OD600. For the second screen, the yeast strains were grown in 96-deep-well plates at 23°C for 6 h in SC-ULH- medium containing 2% galactose, followed by 1:10 dilution with SC-ULH- medium containing 5% glucose. Then, the cells were grown for 24 h at 23°C, followed by additional dilution (1:10) and subsequent culturing until cell density reached 0.8–1.0 OD600. Yeast cells were harvested by centrifugation at 1,100 × g for 5 min.
High-Throughput RNA Analysis. We performed two separate genome-wide screens of the YKO library that included total of four to six independent samples per each strain. Total RNA isolation and Northern blot analysis were done as described in ref. 23, except by using a high-throughput approach. Briefly, yeast cells in 96-deep-well plates were resuspended in RNA extraction buffer (50 mM sodium acetate, pH 5.2/10 mM EDTA/1% SDS) and phenol, followed by incubation for 4 min at 65°C. After removal of phenol, the RNA was recovered by precipitation with ethanol. Agarose gel electrophoresis (1.5%) and Northern blotting were done as described in refs. 23 and 27. For negative-strand detection, total yeast RNA obtained from selected strains was separated in denaturing 5% polyacrylamide/8 M urea gels as described in ref. 23. The RNA was quantified by using a PhosphorImager (Molecular Dynamics) as described in ref. 28.
RT-PCR Analysis of the Junction Sites in the Recombinants. We have used both total yeast RNA extracts and gel-isolated recombinants for RT-PCRs to specifically amplify regions covering junction sites. First, the reverse-transcription reaction included primer 14 (GTAATACGACTCACTATAGGGTTCTCTGCTTTTACGAAG) for cDNA synthesis, followed by PCR with primers 168 (TCGTCTTATTGGACGAATTCCTGTTTACGAAAG) and 270 (TTGGAAATTCTCCTTCAGTCTGAGTTTGTGGA). The PCR products were cloned into pGEM-T Easy Vector (Promega) and sequenced by using M13 reverse primer (29).
5′ Rapid Amplification of Complementary Ends (RACE) and 3′RACE of Recombinants. The 5′ and 3′ sequences of recombinants were determined by using 5′RACE and 3′RACE, respectively, as described in ref. 29. To enrich for recombinants, RNA bands were gel-isolated as described in ref. 29. The resulting products were cloned and sequenced.
In Vitro Tombusvirus Replicase Assay. The in vitro replicase assay was performed with the copurified (endogenous) RNA as described in ref. 23.
Results
Systematic Analysis of Yeast Single-Gene Deletion Strains for Enhanced Level of Viral RNA Recombination. To facilitate identification of host genes involved in RNA virus evolution/recombination, we took advantage of the advanced genomics tools available for yeast and the ability of yeast to support TBSV recombination (23). The recombination assay was based on a replication-competent TBSV DI-72 RNA replicon, which, when coexpressed with the two essential tombusviral replicase proteins (p33 and p92; Fig. 1A), undergoes robust replication and also generates a small amount of RNA recombinants (23). The 621-nt DI-72 RNA replicon contains four noncontiguous segments (Fig. 1A), including the cis-acting replication elements, derived from the full-length genomic RNA (21). It is important to note that in this assay, replication/evolution of DI-72 RNA and the de novo generated recombinant RNAs take place in the absence of artificial selection markers in all viral RNAs.
To systematically test the effect of each host gene on viral RNA recombination, we have developed a high-throughput method based on the available YKO library. Briefly, the collection of 4,848 YKO yeast strains representing ≈80% of yeast genes (those that are nonessential for yeast growth) was cotransformed with three plasmids expressing DI-72 RNA replicon in addition to p33 and p92 replicase proteins (Fig. 1A). We successfully transformed 4,548 strains that grew on galactose-containing medium (see Materials and Methods) and cultured them in 96-deep-well plates, followed by total RNA extraction and agarose gel electrophoresis. Under these conditions, the replication-competent DI-72 RNA is easily detectable in yeast cells, and its amount is similar to the yeast ribosomal RNAs (Fig. 1B). In contrast, recombinant RNAs carrying rearranged RNA sequences accumulate inefficiently in the parental yeast strain as demonstrated by Northern blotting (≈1–2% of the level of the replicating DI-72 RNA) (Fig. 1C). Thus, under the above conditions, the screening approach is expected to favor the identification of those YKO strains that support increased levels of viral RNA recombinants, compared with the parental strain.
Identification of Five Host Genes Whose Absence Leads to Increased Frequency of Viral RNA Recombination. Using the above high-throughput genome-wide screen, we identified a total of five YKO strains that generated 10- to 50-fold higher levels of recombinant viral RNAs than did the parental yeast strain (Fig. 1 B and C). Four of these deletion strains, ctl1Δ, met22/hal1Δ, xrn1Δ, and ubp3Δ, accumulated one major type of recombinant RNA at levels comparable with that of the wild-type viral replicon (Fig. 1 B and C), whereas hur1Δ generated four recombinant RNAs, which were ≈10- to 50-fold more abundant than the WT replicon (Fig. 1 B and C). Northern blot analysis with a probe specific for an internal RIII sequence in the DI-72 RNA replicon demonstrated the viral origin of these previously uncharacterized recombinant RNAs (Fig. 1C). In contrast, a probe specific for the 5′ RI sequence detected only the WT DI-72 replicon in total RNA samples from all five strains but not the recombinant-like RNAs (Fig. 1D), suggesting that RNA recombination might have led to dramatic rearrangement of the viral RNA. These recombinant viral RNAs accumulated in the presence of the WT DI-72 replicon, suggesting that they were generated efficiently and/or competed efficiently with the WT DI-72 RNA replicon.
To determine the sequence of the previously uncharacterized recombinant-like RNAs, we gel-isolated them, followed by RT-PCR, 3′RACE, 5′RACE, cloning, and sequencing (27, 29). We found that the most common recombinant RNAs obtained from xrn1Δ, ctl1Δ, met22Δ, and ubp3Δ strains were similar, partially dimeric RNAs (Fig. 2A and data not shown). They contained various duplicated 3′ sequences (part of RII and complete RIII and RIV) and had deletions of 5′ DI-72 RNA sequences (i.e., RI and part of RII) (Fig. 2A). Most recombinants differed slightly in their junction sequences, a feature shared with TBSV recombinants arising in planta (5). Recombinants in hur1Δ contained two to five incomplete copies of DI-72 RNA sequences with highly variable 5′ truncations (Fig. 8, which is published as supporting information on the PNAS web site). The origin of the 1–13 extra nucleotides at the 5′ end or at the junctions is currently unknown. Extra nucleotides also are frequently detected at the junctions in tombusvirus recombinants in plant protoplasts (22, 27) and in vitro with purified tombusvirus replicase (29), supporting the model that the tombusviral replicase adds extra sequences to the ends of viral RNAs (16).
Fig. 2.
Schematic presentation of the DI-72 replicon with four regions (RI–RIV) and the recombinants with duplicated 3′ sequences (3′ part of RII, RIII, and RIV) and 5′ deletions (RI and 5′ part of RII). The actual sequences of the recombinants (shown for xrn1Δ) at the 5′ ends (Left) and at the junctions are shown. Δ indicates the number of deleted nucleotides, whereas virus-templated and nonviral sequences are shown in uppercase and lowercase letters, respectively. The 3′ end in RIV (both at the internal and 3′ terminal locations) contained the authentic sequence.
To gain insights into the dynamics of recombinant formation, we performed time-course experiments by analyzing total RNA samples at given time points after induction of RNA transcription from the GAL1 promoter in hur1Δ and xrn1Δ strains. We found that the recombinants emerged as early as 2 h after induction (Fig. 3A) in the absence of artificial selection to facilitate their appearance, suggesting that their formation is an efficient process. The amount of recombinants increased over time because of either new recombination events and/or replication of the recombinant RNAs (Fig. 3A). Moreover, we found that the recombinant RNAs replicated and evolved further in yeast cells over 10 serial dilutions in suppressive media (Fig. 3B).
Fig. 3.
Rapid formation of replication-competent viral RNA recombinants. (A) Time-course experiment with hur1Δ and xrn1Δ coexpressing p33 and p92 reveals rapid generation of recombinants [probed with RIII(-)] after induction of DI-72 RNA transcription from plasmid pYC/DI-72. (B) Recombinant RNAs are still present after 10 serial dilutions in glucose-containing medium, which suppresses transcription of DI-72 RNA from the GAL1 promoter. (C) The new viral recombinants are replication-competent. The in vitro replicase assay is based on the tombusvirus replicase/viral RNA complex present in the isolated membrane-enriched fraction of yeast (Left). The presence of minus-stranded RNA replication intermediates for the recombinant RNAs was detected in total RNA extracts by using a minus strand-specific probe (Right).
To demonstrate that the recombinant DI RNAs are replication-competent, we isolated membrane fractions containing tombusvirus replicase/viral RNA complexes from xrn1Δ and hur1Δ cells. These replicase preparations were used for in vitro replicase assays in the presence of added ribonucleotides, including 32P-labeled UTP. These experiments led to in vitro labeling of the recombinantsized RNAs in the replicase assay, suggesting that the recombinant RNAs were part of the replicase complexes (Fig. 3C). Their replication competence also was confirmed by detection of minus-stranded replication intermediates for the recombinant RNAs (Fig. 3C). Taking all these results together, we conclude that the recombinant RNAs, similar to the WT DI-72 RNA replicon, are replication-competent and are maintained on suppressive media for an extended period in yeast.
To test whether the deletion of the host gene altered recombination frequency vs. recombinant selection, we analyzed the stability of four cloned recombinants and the WT DI-72 replicon RNA in the parental and xrn1Δ strain. Fig. 4 demonstrates that the stability of the recombinants and the WT replicon was comparable in the parental strain, whereas the recombinants and the WT replicon RNA showed 2- to 3-fold increased stability in the xrn1Δ strain (Fig. 4). This increased RNA stability suggests that viral RNA degradation is hindered in the xrn1Δ strain. Importantly, however, all of the recombinants and the WT replicon RNA showed similar level of increase in stability in the xrn1Δ strain, suggesting that these RNAs have comparable stability. Overall, selective RNA degradation of the WT replicon vs. recombinants cannot explain the increased accumulation of recombinants over the WT replicon in xrn1Δ strain.
Fig. 4.
Deletion of Xrn1p increases the stability of recombinant RNAs and DI-72 replicon RNA. Four representative recombinant RNAs containing partially duplicated sequences (first four bars on the left) and the DI-72 replicon RNA (the dark gray bar on the right) were separately expressed in the parental (A) and xrn1Δ (B) strains from GAL1 promoter. After repression of transcription with glucose (time points of 0, 2, 4, and 6 h), the residual viral RNAs were measured by Northern blotting and quantified by using a PhosphorImager. The data are shown in % (the amount of viral RNA at 0 time point is 100%) derived from four independent experiments.
Systematic Analysis of Yeast Single-Gene Deletion Strains for Decreased Level of Viral RNA Recombination. Because the above genome-wide screen was suitable only for testing for increased levels of TBSV recombination, we modified the screening approach to allow the identification of YKO strains supporting reduced levels of virus recombinants, compared with the parental strain. To this end, we induced DI-72 RNA transcription in all 4,548 YKO strains (also coexpressing p33/p92) for 6 h, followed by growing them in glucose-containing medium before total RNA extraction and analysis by Northern blotting. Under these conditions, detectable amounts of recombinant RNAs accumulated in the parental strain (10–18% of the standard WT DI-72 RNA, Fig. 5A). These recombinant RNAs included complete dimers (two copies of full-length DI-72 replicons joined head-to-tail) and incomplete dimeric DI-RNAs (5′ truncated monomers joined head-to-tail, see Fig. 5A). Northern blot analysis of total RNA extracts from all transformants (two to four samples per strain) revealed that the ratio of recombinant RNA vs. WT DI-72 RNA was 3- to 5-fold lower in only four YKO strains (Fig. 5A). Note that we did not measure the absolute amounts of RNA recombinants but instead estimated the ratio of recombinants vs. nonrecombinant DI-72 RNA. The reasoning is that DI-72 RNA accumulation level could be different in various YKO strains (30), which could affect the amount of RNA substrates available for recombination.
Fig. 5.
Absence of PEP7, IPK1, CHO2, and DCI1 host genes leads to low frequency of recombination of TBSV DI-72 RNA replicon in yeast coexpressing p33 and p92 replicase proteins. (A) The relative amounts of recombinants T, M, and B (compared with the DI-72 RNA replicon, which is set as 100%) are shown. The sequences of the dimeric recombinant RNAs are shown schematically at the bottom. Deletion within RII usually included from 65 to 170 5′ nucleotides. The junction sites are circled. (B) A similar recombination experiment was performed with DI-AU-FP replicon RNA. The sequences of DI-AU-FP and the recombinant RNAs are shown schematically on the Top and Bottom, respectively. See further details in the Fig. 1 legend.
Characterization of Four Host Genes Whose Absence Leads to Decreased Frequency of Viral RNA Recombination. To further test the recombination deficiency of the above-identified four YKO strains, we examined whether they could support RNA recombination with a modified viral replicon, DI-AU-FP. This replicon contains a 186-nt-long heterologous sequence including a 46-nt-long AU-rich stretch (Fig. 5B). Previous work in plant protoplasts demonstrated that DI-AU-FP induced recombination with high efficiency in the presence of the WT helper virus (27). Northern blot analysis of total RNA obtained from the four YKO strains coexpressing DI-AU-FP and p33/p92 proteins revealed that RNA recombinants accumulated poorly (3- to 5-fold decrease) in these strains, compared with the parental strain (Fig. 5B). The sequences of the generated recombinants isolated from the parental and the selected YKO strains were comparable (shown schematically in Fig. 5B), indicating that the mechanism of their generation was likely similar. Overall, we conclude that the four identified YKO strains supported recombination with reduced frequency and/or accumulation rate of recombinants was lower in these strains than in the parental yeast.
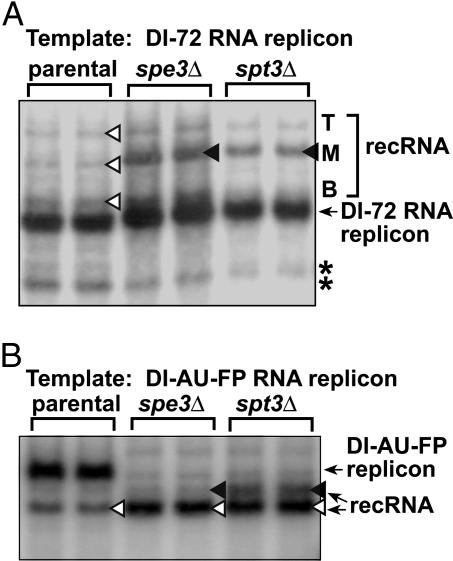
It is worth noting that the above genome-wide screen also identified five additional YKO strains that supported a 2-fold reduced level of recombinant accumulation in comparison with the parental strain (see “weak accelerators” in Table 1). In addition, we found that spe3Δ and spt3Δ generated a different recombinant profile from the parental strain (Fig. 6A). The notable difference was the accumulation of a single dominant recombinant RNA in these strains (see M in Fig. 6A). To confirm that the lack of SPE3 and SPT3 genes indeed affected RNA recombination, we analyzed recombinant formation during DI-AU-FP RNA replication. This experiment demonstrated that (i) spe3Δ and spt3Δ showed 2- to 4-fold increased levels of recombinants, and (ii) the profiles of recombinants generated were somewhat different from the profile observed with the parental strain (Fig. 6B).
Table 1. Names and functions of the identified host genes.
| Gene | Molecular function/biological process |
|---|---|
| Supressors | |
| CTL1 | Polynucleotide 5′-phosphatase |
| MET22/HAL2 | 3′(2′),5′-Bisphosphate nucleotidase |
| HUR1 | Unknown |
| XRN1 | 5′-3′ exoribonuclease |
| UBP3 | Ubiquitin-specific protease |
| Accelerators | |
| PEP7/VPS19 | Unknown/Golgi to vacuole transport |
| IPK1 | Inositol/phosphatidylinositol kinase |
| CHO2/PEM1 | Phosphatidylethanolamine N-methyltransferase |
| DCI1 | Dodecenoyl-CoA Δ-isomerase |
| Weak accelerators* | |
| VPS43/VAM7 | Golgi to vacuole transport |
| PTH1/VAM3 | Golgi to vacuole transport |
| VPS29 | endosome to Golgi transport |
| VPS35 | endosome to Golgi transport |
| NGG1 | transcription cofactor |
| Modifiers | |
| SPE3 | Spermidine synthase |
| SPT3 | Transcription cofactor |
Deletion of these genes decreased viral recombinant accumulation by ≈2-fold
Fig. 6.
Absence of SPE3 and SPT3 genes results in altered recombination profile with DI-72 RNA (A) and DI-AU-FP (B) replicon. See further details in the Fig. 1 legend.
Major Contribution of Host Genes to Viral RNA Recombination. To demonstrate the full extent of contribution by host genes to viral RNA recombination, we compared recombinant accumulation in the xrn1Δ and pep7Δ strains carrying DI-AU-FP and p33/p92 expression plasmids. These experiments revealed that the xrn1Δ strain accumulated viral RNA recombinants up to an 80-fold higher level than did the pep7Δ strain (Fig. 7). Because these yeast strains differed only in the two deleted genes, the above experiment demonstrated that host genes could play major roles in viral RNA recombination.
Fig. 7.
Comparison of recombination activity of DI-AU-FP replicon in xrn1Δ, pep7Δ, and parental yeast strains. See further details in the Fig. 1 legend.
Discussion
Viruses are known to evolve rapidly in selected hosts, yet the roles of host genes in RNA virus recombination/evolution are currently unknown. This work, based on a high-throughput genetic screen in yeast, a model host, has led to the identification of 11 host genes that significantly affected tombusvirus recombination. We found that single deletion of the identified genes had three types of effects on tombusvirus recombination: (i) five genes increased, whereas (ii) four genes decreased recombinant accumulation, and (iii) two genes changed the profile of recombinants. An additional five genes had a lesser effect (≈2-fold) on RNA recombination.
Suppressors of RNA Virus Recombination. The observation that the accumulation of viral RNA recombinants increased 10- to 50-fold in the absence of five host genes (Table 1) suggests that these genes, when present, can suppress RNA virus recombination. Interestingly, three of the identified genes, namely XRN1, CTL1, and MET22/HAL2, are involved in RNA metabolism/degradation. It is plausible that these genes could affect viral recombination by influencing the 5′–3′ RNA degradation pathway (31). The proposed connection between RNA degradation and viral RNA recombination is supported by the following findings: (i) the recombinants had deletions within their 5′ sequences (Fig. 2); (ii) 5′ truncated viral RNAs accumulated in these yeast strains (Fig. 1 B–C); and (iii) identification of Xrn1p, which is the key enzyme in the 5′–3′ RNA degradation pathway (31, 32), as one of the viral recombination affecting proteins; and (iv) the increased stability of both recombinant and DI-72 RNA replicon in the xrn1Δ strain (Fig. 4). Moreover, three of the five identified host genes are predicted and/or known to affect the activity of Xrn1p. For example, Met22p/Hal2p has been shown to affect the activity of Xrn1p through regulating the level of pAp, an inhibitor of Xrn1p (33). Also, Ctl1p is known to modify the 5′ end of the RNA by removing a phosphate group that could potentially facilitate Xrn1p-driven 5′–3′ RNA degradation (34). In addition, Ubp3p has been shown to increase stability of Xrn1p in cells (35). Finally, the 5′–3′ exoribonuclease activity of Xrn1p could be inhibited in the absence of one of these genes. In contrast, the role/function of Hur1p is currently unknown. The profile of recombinants generated in hur1Δ, however, is different (Fig. 8) from the profile of the recombinants identified in the other four YKO strains, indicating that hur1Δ might use a different mechanism during viral RNA recombination. Overall, this genome-wide screen indicates a close connection between viral RNA recombination and RNA metabolism/RNA degradation. Interestingly, a 5′–3′ exoribonuclease, similar to Xrn1p, is present in Arabidopsis (36), and the Hal2p homolog has been cloned from rice (37), suggesting that similar genes are functional in plants, too.
Proposed Mechanism of Suppression of Viral RNA Recombination. Based on the identified host genes and the profile of generated viral RNA recombinants, we propose that four of five host genes, including XRN1, CTL1, MET22/HAL2, and UBP3, could suppress viral recombination through affecting the Xrn1p-dependent rapid and complete degradation of viral RNA. However, in the absence of Xrn1p, or because of the inhibition of Xrn1p activity in the absence of Ctl1p, Met22p, or Ubp3p, degradation of the viral RNA gets slower (Fig. 4). The resulting incompletely degraded viral RNA could then participate in RNA recombination efficiently, facilitating the accumulation of partly dimeric recombinant RNAs. The generated recombinants are also more stable in xrn1Δ strain, further facilitating the accumulation of recombinants. Moreover, abundance of 5′ truncated RNA species in these strains supports the model that these RNAs are intermediates (substrates) in the RNA recombination process. In addition, efficient recombination is likely due to “exposure” of the highly recombinogenic RII sequences (27) at the ends of the viral RNAs after their partial degradation. In contrast, the parental yeast cells could efficiently and completely degrade viral RNAs, thus reducing the chance for partly degraded RNAs to participate in RNA recombination.
Protein Accelerators of Viral RNA Recombination. The other set of host genes identified during this genome-wide screen includes four genes, PEP7, IPK1, CHO2, and DCI1, whose deletion resulted in reduced level of viral RNA recombination (Fig. 5). The viral replicon RNA, either DI-72 or DI-AU-FP, replicates efficiently in these strains, whereas the dimeric recombinant RNAs accumulate 3- to 5-fold less than in the parental strain. Therefore, these genes might directly influence the frequency of recombination. Based on the known functions of these genes (Table 1), we suggest that (i) intracellular transport of viral and/or host proteins (or possibly protein–viral RNA complexes) to the site of recombination (see genes PEP7 and DCI1), and/or (ii) the lipid content/structure of the membranous compartment, which contains the virus replicase, could be altered in the absence of these genes (IPK1, CHO2, and DCI1), resulting in reduced RNA recombination efficiency.
Although the current work has not addressed the mechanism of RNA recombination in the selected strains, a comparison of sequences at the recombination junctions suggests that the recombinants represent similarity-nonessential (nonhomologous) recombinants (16). The observed recombinants are likely generated through a viral replicase-driven template-switching mechanism, which has been shown for tombusviruses before (22, 29). Also, data presented in the online material (see Supporting Text, which is published as supporting information on the PNAS web site) exclude that DNA recombination or RNA recombination during RNA polymerase II-driven RNA transcription is the mechanism of viral RNA recombination (Fig. 9, which is published as supporting information on the PNAS web site).
General Conclusions. This genome-wide screen of yeast for host genes affecting viral RNA recombination demonstrates that a selected set of host genes can accelerate or suppress viral RNA recombination. We found that the majority of yeast single-deletion strains showed a low level of virus recombination, whereas five strains with particular genetic backgrounds were “hotbeds” for recombination, accelerating virus evolution. This finding implies that mutation(s) in host genes involved in suppression of virus recombination create “favorable” genetic backgrounds for virus RNA recombination, suggesting that such an individual(s) might contribute to RNA recombination and virus evolution more significantly than other individuals of the same species with less favorable genetic backgrounds. Our discovery promises to have a major influence on future thinking about the contribution of particular host genes and individual organisms to virus recombination and evolution.
Supplementary Material
Acknowledgments
We thank Drs. Tom Pirone, Judit Pogany, Saulius Serva, and John Shaw for valuable comments. This work was supported by National Institutes of Health–National Institute of Allergy and Infectious Diseases Grant R03 AI061437-01A1 and partly by the Kentucky Tobacco Research and Development Center and the University of Kentucky.
Abbreviations: TBSV, tomato bushy stunt virus; YKO, yeast knockout; DI, defective interfering; RACE, rapid amplification of complementary ends.
References
- 1.Lai, M. M. (1992) Microbiol. Rev. 56, 61-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worobey, M. & Holmes, E. C. (1999) J. Gen. Virol. 80, 2535-2543. [DOI] [PubMed] [Google Scholar]
- 3.Worobey, M., Rambaut, A. & Holmes, E. C. (1999) Proc. Natl. Acad. Sci. USA 96, 7352-7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes, E. C., Worobey, M. & Rambaut, A. (1999) Mol. Biol. Evol. 16, 405-409. [DOI] [PubMed] [Google Scholar]
- 5.Marturano, J. & Fiore, L. (2002) J. Clin. Microbiol. 40, 316-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang, X., Espul, C., Zhong, W. M., Cuello, H. & Matson, D. O. (1999) Arch. Virol. 144, 2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walter, J. E., Briggs, J., Guerrero, M. L., Matson, D. O., Pickering, L. K., Ruiz-Palacios, G., Berke, T. & Mitchell, D. K. (2001) Arch. Virol. 146, 2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukashev, A. N., Lashkevich, V. A., Koroleva, G. A., Ilonen, J. & Hinkkanen, A. E. (2004) J. Gen. Virol. 85, 463-470. [DOI] [PubMed] [Google Scholar]
- 9.Oprisan, G., Combiescu, M., Guillot, S., Caro, V., Combiescu, A., Delpeyroux, F. & Crainic, R. (2002) J. Gen. Virol. 83, 2193-2200. [DOI] [PubMed] [Google Scholar]
- 10.Khatchikian, D., Orlich, M. & Rott, R. (1989) Nature 340, 156-157. [DOI] [PubMed] [Google Scholar]
- 11.Becher, P., Orlich, M. & Thiel, H. J. (2001) J. Virol. 75, 6256-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fricke, J., Gunn, M. & Meyers, G. (2001) Virology 291, 77-90. [DOI] [PubMed] [Google Scholar]
- 13.Rest, J. S. & Mindell, D. P. (2003) Infect. Genet. Evol. 3, 219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stavrinides, J. & Guttman, D. S. (2004) J. Virol. 78, 76-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch, X. (2004) Lancet Infect. Dis. 4, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy, P. D. & Simon, A. E. (1997) Virology 235, 1-9. [DOI] [PubMed] [Google Scholar]
- 17.Allison, R., Thompson, C. & Ahlquist, P. (1990) Proc. Natl. Acad. Sci. USA 87, 1820-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan, H. & Simon, A. E. (2000) Proc. Natl. Acad. Sci. USA 97, 12451-12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chetverin, A. B., Chetverina, H. V., Demidenko, A. A. & Ugarov, V. I. (1997) Cell 88, 503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahlquist, P., Noueiry, A. O., Lee, W. M., Kushner, D. B. & Dye, B. T. (2003) J. Virol. 77, 8181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White, K. A. & Nagy, P. D. (2004) Prog. Nucleic Acid Res. Mol. Biol. 78, 187-226. [DOI] [PubMed] [Google Scholar]
- 22.White, K. A. & Morris, T. J. (1994) J. Virol. 68, 14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panavas, T. & Nagy, P. D. (2003) Virology 314, 315-325. [DOI] [PubMed] [Google Scholar]
- 24.Pantaleo, V., Rubino, L. & Russo, M. (2003) J. Virol. 77, 2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panaviene, Z., Panavas, T., Serva, S. & Nagy, P. D. (2004) J. Virol. 78, 8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gietz, R. D. & Woods, R. A. (2002) Methods Enzymol. 350, 87-96. [DOI] [PubMed] [Google Scholar]
- 27.Shapka, N. & Nagy, P. D. (2004) J. Virol. 78, 2288-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panavas, T., Pogany, J. & Nagy, P. D. (2002) Virology 296, 275-287. [DOI] [PubMed] [Google Scholar]
- 29.Cheng, C. P. & Nagy, P. D. (2003) J. Virol. 77, 12033-12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panavas, T., Serviene, E., Brasher, J. & Nagy, P. D. (2005) Proc. Natl. Acad. Sci. USA 102, 7326-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker, R. & Song, H. (2004) Nat. Struct. Mol. Biol. 11, 121-127. [DOI] [PubMed] [Google Scholar]
- 32.Sheth, U. & Parker, R. (2003) Science 300, 805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dichtl, B., Stevens, A. & Tollervey, D. (1997) EMBO J. 16, 7184-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez, C. R., Takagi, T., Cho, E. J. & Buratowski, S. (1999) Nucleic Acids Res. 27, 2181-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brew, C. T. & Huffaker, T. C. (2002) Genetics 162, 1079-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kastenmayer, J. P. & Green, P. J. (2000) Proc. Natl. Acad. Sci. USA 97, 13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng, Z. & Verma, D. P. (1995) J. Biol. Chem. 270, 29105-29110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.