Abstract
Background
Primary cilia facilitate cellular signalling and play critical roles in development, homeostasis, and disease. Their assembly is under the control of Tau-Tubulin Kinase 2 (TTBK2), a key enzyme mutated in patients with spinocerebellar ataxia. Recent work has implicated TTBK2 in the regulation of cilia maintenance and function, but the underlying molecular mechanisms are not understood.
Methods
To dissect the role of TTBK2 during cilia growth and maintenance in human cells, we examined disease-related TTBK2 truncations. We used biochemical approaches, proteomics, genetic engineering, and advanced microscopy techniques to unveil molecular events triggered by TTBK2.
Results
We demonstrate that truncated TTBK2 protein moieties, unable to localize to the mother centriole, create unique semi-permissive conditions for cilia assembly, under which cilia begin to form but fail to elongate. Subsequently, we link the defects in cilia growth to aberrant turnover of a microtubule-depolymerizing kinesin KIF2A, which we find restrained by TTBK2 phosphorylation.
Conclusions
Together, our data imply that the regulation of KIF2A by TTBK2 represents an important mechanism governing cilia elongation and maintenance. Further, the requirement for concentrating TTBK2 activity to the mother centriole to initiate ciliogenesis can be under specific conditions bypassed, revealing TTBK2 recruitment-independent functions of its key partner, CEP164.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-025-02072-8.
Keywords: Cilia, Ciliogenesis, TTBK2, KIF2A, Basal body
Background
Primary cilia are evolutionarily conserved signalling organelles found on the surface of a broad spectrum of vertebrate cells [1, 2]. They play a major role in intercepting extracellular stimuli, which helps to govern complex processes in multicellular organisms, including humans [3, 4]. Even though these organelles have not attracted much attention in the past, their biomedical importance became apparent following the discovery of ciliopathies, an expanding group of human diseases caused by ciliary dysfunction [5, 6].
The primary cilium typically appears as a hair-like protrusion of the cell membrane. It consists of the mother centriole (MC)-derived basal body, the transition zone allowing sorting of ciliary components, and the microtubule-based axoneme enclosed within an ARL13B-rich ciliary membrane [7, 8]. The MC distal end is decorated by two sets of appendage proteins. The distal set of appendages is especially important for the initiation of cilia formation – it helps to dock the MC to preciliary vesicles and/or cell membrane and recruits pro-ciliogenesis factors to the base of the growing cilium [9, 10]. To initiate the outgrowth of ciliary axoneme, capping complexes consisting of CP110 and CEP97 proteins are removed from MC distal end [11]. Subsequently, the axoneme extends with the assistance of intraflagellar transport (IFT) machinery [12].
Perhaps the most prominent regulator of the cilium assembly pathway outlined above is Tau-Tubulin Kinase 2 (TTBK2) [13], a serine/threonine kinase from the CK1 superfamily [14, 15]. To initiate ciliogenesis, TTBK2 is recruited to MC distal appendages via interaction with CEP164 [16]. Thus, TTBK2 gets in a prime position to govern key events at the base of the future cilium by its kinase activity [13]. The processes regulated by TTBK2 include the recruitment of Golgi-derived preciliary vesicles [17] and IFT proteins [13], or the removal of CP110/CEP97 [13]. However, the full scope of TTBK2-regulated mechanisms and their relation to ciliogenesis remains to be discovered.
TTBK2 has been shown to phosphorylate a number of MC-related (e.g. CEP164, CEP83, CEP89 [17, 18]) or cell signalling-related proteins (e.g. Dishevelled, a component of the WNT signaling pathway [18, 19]), but the functional relevance of these interactions is often not clear. Furthermore, recent work highlights the possibility of TTBK2 maintaining the length and integrity of primary cilia [20, 21], but molecular mechanisms that would link TTBK2 to axoneme length control remain elusive. Finally, truncating mutations in TTBK2 have been shown to trigger the onset of spinocerebellar ataxia 11 (SCA11), a rare progressive neurodegenerative disease [21–23]. While the genetic link between TTBK2 and SCA11 has been well described, the molecular pathology of SCA11 is unclear.
In this study, we aimed to elucidate the role TTBK2 plays beyond initiating cilia formation in human cells. As targeting TTBK2 through conventional loss-of-function approaches completely blocks cilia formation [13, 18], we examined TTBK2 variants similar in length and sequence to SCA11 mutants [22]. We show that expressing truncated TTBK2 in hTERT RPE-1 TTBK2 KO cells creates unique semi-permissive conditions for ciliogenesis, bypassing TTBK2 recruitment to the MC. In turn, we utilize this model system to reveal a TTBK2-mediated regulatory mechanism involving kinesin KIF2A, a member of kinesin-13 family with microtubule-depolymerizing activity [24, 25]. KIF2A has been previously shown to aid the resorption of primary cilia following cell cycle re-entry [26], but its role during cilia assembly was not clear. Here, we demonstrate that the interplay between KIF2A and TTBK2 represents an important regulatory mechanism governing primary cilia formation in human cells.
Materials and methods
RPE-1 cell culture and stable line derivation
All hTERT RPE-1 cell lines were cultivated in DMEM/F12 cultivation media (Thermo Fisher Scientific, Cat.N.31331028), supplemented by 10% fetal bovine serum (Biosera), 1% penicillin/streptomycin and 1% L-glutamine. When perfoming 12/24-well format experiments, culture media was changed daily. To induce primary cilia formation, the cells were cultivated in serum-free complete media for the last 24 h of the experiment. Plasmid transfection (200ng plasmid DNA/well in a 24-well format) was carried out using the Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, Cat.N. L3000001, 0.2 µl P3000 / 100ng DNA, 0.3 µl Lipofectamine / 100ng DNA) and following the manufacturer’s manual. Paclitaxel treatment was carried out by adding paclitaxel (Merck, Cat.N. T7402, final concentration 5µM) to culture media 4 h prior to fixation.
To generate DOX-inducible transgenic cell lines, hTERT RPE-1 Flp-In T-Rex cells (a gift from Erich A. Nigg) or hTERT RPE-1 Flp-In T-REX TTBK2 KO cells [18] were seeded on 5 cm dishes, grown to 90% confluency, and co-transfected with pOG44 (5µg total DNA) and a donor vector (pgLap1/2, 500ng total DNA) containing the gene of interest (GOI) coupled to a G418-resistance gene. Transfectants with stably integrated GOI were then selected based on their resistance to G418 (0.5 mg/ml, 1–2 weeks, Merck, Cat.N. G8168). To induce the expression of the GOI the cells were treated with doxycycline (2 µg/ml, Merck, Cat.N. 3072) for the duration of the experiment. For a list of plasmids used in this work see Supplementary Table 1.
For lentiviral transduction of hTERT RPE-1 cells with TTBK2 constructs we used plasmids listed in Supplementary Tables 1 and followed a previously published protocol [27].
For KIF2A knockdown, the cells were seeded on glass coverslips in a 24-well format and cultivated in complete media. After 24 h, the cells were transfected with KIF2A siRNA (50nM final concentration, for siRNA details, see Supplementary Table 1) using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Cat.N.13778100). Culture media was changed on the next day, the cells were cultivated for 48 h in complete media, serum-starved for 24 h, fixed, and analyzed.
HEK293T cell culture and transfection
HEK293T cells were cultivated in DMEM cultivation media (Thermo Fisher Scientific, Cat.N.31966047), supplemented by 10% fetal bovine serum (Biosera) and 1% penicillin/streptomycin. When performing 12/24-well format experiments, culture media was changed daily. To induce primary cilia formation, the cells were cultivated in serum-free complete media for the last 24 h of the experiment.
Transfection of HEK293T cells was carried out using polyethyleneimine (PEI, 2 mg/ml stock solution) in the following way: PEI was incubated in serum-free DMEM media for 10 min, plasmids (Supplementary Table 1) were equilibrated in serum-free DMEM media and then mixed with PEI in a 3µl of PEI to 1µg of plasmid ratio. The resulting mixes of plasmid and PEI in DMEM were then added to cells and left in the culture overnight, the media was changed for a fresh complete media on the next day. 48 h after transfection, the cells were processed (fixed with MetOH or lysed) and analyzed.
Western blot
Cells were lysed in 1x Laemli lysis buffer (62.5 mM Tris-HCl pH 6.8, 2% 2-mercaptoethanol, 10% glycerol, 0.01% bromphenol blue, 2% sodium dodecyl sulfate (SDS). SDS-PAGE and membrane transfer were performed using instrumentation by BioRad (Mini-PROTEAN tetra vertical electrophoresis cell, Mini Trans-blot module). Cell lysates were loaded to a 5% stacking gel combined with an 8% running gel and ran at 150 V in a running buffer (0.1% SDS, 0.192 M glycine, 0.025 M Tris-base). The proteins were transferred to an Immobilon-PVDF membrane (Merck, Cat.N. IEVH00005) at 100 V for 75 min in a transfer buffer (20% methanol, 0.192 M glycine, 0.025 M Tris-base). The membranes were then blocked in 5% solution of skimmed milk in wash buffer (20 mM Tris-base, 0.1% Tween 20, 150 mM NaCl) and incubated with primary antibodies (see Supplementary Table 1) diluted in the same solution overnight at 4 °C. The next day the membranes were washed 3 times for 10 min in wash buffer, incubated with secondary antibodies (Supplementary Table 2) diluted in 5% milk/wash buffer for 2 h at room temperature, and then washed in wash buffer 3 times for 10 min. The membranes were then developed using ECL Prime (Merck, Cat.N. GERPN2236) and Chemidoc Imaging System (BioRad, Cat.N. 12003154). For an easier analysis of membranes, a labelled protein ladder (Thermo Fisher Scientific, Cat.N. 26625) was used in every SDS-PAGE performed.
Immunoprecipitation and MS/MS analysis
To isolate KIF2A for MS/MS analysis, HEK293T cells grown on 15 cm plates were transfected with 2 µg of Flag-KIF2A plasmid + 8 µg of the corresponding GFP-TTBK2 construct. 48 h post-transfection the cells were scraped into lysis buffer (0.5% Triton X-100, 0.5% NP40, 150mM NaCl, 20mM Tric-HCl pH 7.4) containing PhosSTOP (Roche, Cat.N. 4906837001, 1 tablet/10 ml buffer), Complete Mini Protease Inhibitor Cocktail (Roche, Cat.N. 11836153001, 1 tablet/10 ml buffer) and lysed for 15 min on ice. The cell lysates were centrifuged at 16,000x g for 10 min at 4 °C and the supernatants were then incubated overnight at 4 °C with anti-Flag M2 affinity gel beads (Merck, Cat.N. A2220). M2 beads were then pelleted and washed three times with lysis buffer containing protease inhibitors, resuspended in 1x Laemli lysis buffer, and boiled at 95 °C for 10 min. The resulting samples were separated in an 8% acrylamide gel and stained with Coomassie stain. Prominent bands of the correct size were cut out and subjected to protein extraction followed by MS/MS analysis [16]. The final plotted values represent mean relative phosphointensities from 3 independent experiments, only phosphosites detected with an absolute intensity above 106 units were taken into consideration.
Software and data analysis
All statistical analyses were performed in GraphPad Prism software (version 8.0.1), standard deviations (SDs) are shown in all graphs, unless otherwise stated in the figure legend. The ACDC [28] Matlab script (version 0.9) or the CiliaQ [29] Fiji plugin (version 0.1.4) were used for measuring cilia number and length. 3D reconstructions of expansion microscopy images were generated using Imaris software (version 9.8.2). Alphafold structure predictions were generated using the Alphafold multimer model (Alphafold version 2.3.1, DB preset: full DBS, model preset: multimer, 5 predictions per seed with model relaxation), protein structures were visualized using ChimeraX (version 1.8).
Immunofluorescence microscopy
hTERT RPE-1 cells were grown on glass coverslips in a 24-well format, washed with PBS, and fixed using ice-cold methanol at -20 °C for 20 min. The coverslips were then briefly washed 2 times with PBS, incubated with primary antibodies overnight at 4 °C, washed 3 × 10 min with PBS before being incubated with secondary antibodies for 2 h in a dark chamber at room temperature, washed 3 × 10 min with PBS again and mounted using glycergel (Dako, Cat.N.C0563) or the ProLong Glass Antifade Mountant (ThermoFisher, Cat.N. P36980). The imaging was performed with the use of ZEISS microscopes, either with AxImager A2 (Plan-Apochromat 100x/1.40 Oil DIC, Hamatsu camera) or LSM-800 (Plan-Apochromat 63x/1.40 Oil DIC M27, Hamatsu camera). Raw images (.czi files) were acquired as z-stacks, processed with the maximum intensity projection feature in Fiji, saved as 16-bit.tif files, and analysed further (e.g. signal measurements). For a list of antibodies used during imaging see Supplementary Table 2.
Image analysis and signal intensity measurements were done in Fiji (version 2.0). Centriolar signal intensity of the protein of interest (POI) and the corresponding centriolar marker (CAP350, CETN1, gTUB) was measured by drawing an ellipsoidal region of interest (ROI) around the centrioles and measuring mean signal intensities inside the ROI (= centriolar signal), then slightly moving the ROI next to the centrioles and measuring mean signal intensities again (= background signal). The final plotted values are equal to:
 |
Expansion microscopy
Following cultivation on glass slides in a 24-well format, the cells were briefly washed with PBS and fixed with fixation buffer (PBS, 4% parafolmaldehyde, 4% acrylamide) for 48 hours at room temperature, then briefly washed 2 times with PBS. For each sample, a droplet of polymerizing acrylamide gel (PBS, 19% sodium acrylate, 10% acrylamide, 0.1% N, N’-Methylenebisacrylamide, 0.5% ammonium persulfate, 0.5% temed) was prepared, the coverslips were quickly put on top of the gel droplet, with the cells facing the droplet. The samples were then incubated at 4 °C for 10 min, then at 37 °C for 30 min. The resulting coverslips covered in polymerized gel were transferred to a denaturation buffer (50mM Tris-base, 200mM NaCl, 200mM SDS) and the glass coverslips were gently removed from the gels using flat forceps. The gels were then incubated in denaturation buffer at 95 °C for 2 h and let to expand for 1 h in ddH2O at room temperature. The gels were then cut into smaller pieces and incubated overnight at room temperature in primary antibodies (Supplementary Table 2) diluted 1:50 in blocking buffer (PBS, 2% BSA, 0.02% sodium azide). The next day the samples were washed 2 × 30 min with ddH2O and then incubated at room temperature overnight in secondary antibodies (Supplementary Table 2) diluted 1:500 in blocking buffer. The following day the samples were washed 2 × 30 min with ddH2O, placed in a glass-bottom microscopy dish (Ibidi), and imaged using the LSM-880 Airy2 microscope (alpha Plan-Apochromat 100x/1.46 Oil DIC M27 Elyra, AiryScan detector) by ZEISS. Raw images (.czi files) were processed with the AiryScan and Orthogonal Projection (maximum intensity) post-processing features in ZenBlack.
Centriole/axoneme lengths (3C) were measured by first reconstructing the AcTUB signal in 3D using the Surfaces function in Imaris, then using the Elipsoid Axis Length function in Imaris to calculate the length of 3D-reconstructed centrioles/axonemes.
Transmission electron microscopy
hTERT RPE-1 cells were grown on 5 cm dishes for 48 h, then fixed with 3% glutaraldehyde solution (G5882-100mL, Sigma Aldrich) in 0.1 M cacodylate buffer (C0250-100G, Sigma Aldrich) at 4 °C. After rinsing in 0.1 M cacodylate buffer, the samples were postfixed by 1% OsO4 (05500-1G Sigma Aldrich), dehydrated using ascending ethanol grade (50, 70, 96, and 100% 71250 − 11002 Penta), embedded in LR White resin (AGR1281 Agar Scientific), polymerized 3 days at 65 °C, and processed by the standard protocol for electron microscopy. Ultrathin sections were imaged using transmission electron microscope (Morgagni 268D, ThermoFisher Scientific, Netherlands).
CEP164 phosphoantibody generation
Antibody against pS201 in CEP164 was custom-derived by Moravian Biotechnology Ltd (https://www.moravian-biotech.com). In brief, rabbits were immunized using a 13mer peptide TKGLLGpSIYEDKT during 3 rounds of immunization, the IgG was then purified using a gel matrix.
Live-cell imaging
Reporter hTERT-RPE-1 Flp-In T-Rex TTBK2 KO cell lines with DOX-inducible expression of Flag-TTBK2 constructs and constitutive mNeonGreen-ARL13B reporter expression were prepared as described before [27]. For the time-lapse live imaging experiment cells were seeded in DMEM/F12 medium, 10%FBS, 1%L-glutamine, and Penicilin/streptomycin, supplemented with 1 µg/mL DOX on a 10-well glass-bottom CELLVIEW CELL CULTURE SLIDE, PS, 75/25 MM (Greiner Bio-One) at a high density (∼30.000 cells per well). 72 h after seeding the medium was replaced with FluoroBrite DMEM with 1%L-glutamine and Penicilin/Streptomycin supplemented with 1 µg/mL DOX to start the starvation of cells and induce cilia growth. Slides with cells were equilibrated in the microscope environmental chamber for at least 30 min before imaging started. We used Elyra7 inverted microscope equipped with super-resolution structured illumination microscopy (SIM) module with Plan-Apochromat 40x/1.4 Oil DIC M27. Z-stack images were taken every 15 min, the resulting multi-scene.czi file was processed in Zen Black Software by the SIM2 Method. The processed z-stacks were projected in one layer by Maximal Orthogonal Projection in Zen Blue Software and individual scenes were saved as.tif files using Bio-Formats Importer plugin in Fiji. Cilia length was measured with the Segmented line tool in Fiji.
FRAP
hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-tagged TTBK2 constructs were seeded in µSlide 8 Well High (Ibidi) chambers. The next day the cells were transfected with GFP-KIF2Awt, cultivated for another 24 h, and then subjected to FRAP analysis using the LSM-880 microscope system by Zeiss – centriolar GFP signal was bleached with a strong laser pulse (100% laser intensity, 500ms) and the region of interest was imaged over the following 60s before measuring GFP signal intensity using the ZenBlack software. Thalf was calculated from individual FRAP recovery curves (single-fit algorithm) using the EasyFRAP web tool [30].
RT-qPCR
hTERT RPE-1 cell lines were seeded in a 12-well format, cultivated for 48 h (with 2 µg/ml DOX where indicated), washed with PBS and immediately stored at -80 °C overnight. The samples were then processed with the RNAeasy kit (QIAgen, Cat.N. 74104) in compliance with the manufacturer’s protocol to isolate mRNA. cDNA synthesis was performed using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Cat.N. 04379012001). To compare gene expression (cDNA level), real-time PCR was performed with the use of primers listed in Supplementary Tables 1 and LightCycler SYBR Green I Master (Roche, Cat.N. 04887352001). Relative gene expression was then calculated using the delta-delta Ct method [31].
Site-directed mutagenesis
Site-directed mutagenesis of TTBK2 and KIF2A constructs was performed using the Agilent QUIKChange II XL kit (Agilent Technologies, cat.n. 200522) according to manufacturer’s instructions, except 25ng (instead of 10ng) of parental template/plasmid was used in each PCR reaction. PCR reaction products were used to transform XL10 Gold chemo-competent bacteria (Agilent Technologies, cat.n. 200315) and successful mutagenesis of target sequences was confirmed by DNA sequencing.
GFP-KIF2A lysates preparation
HEK293T cells were seeded on 15 cm dishes, transfected with 15ug of plasmid DNA / dish, 30 h post transfection they were scraped down into PBS and cenrifuged at 200g / 5 min. Cell pellets were resuspended in 0.5 pellet volumes of lysis buffer (BRB20 (20mM PIPES pH 6.9, 1mM EGTA,1mM MgCl2) supplemented with 1x phosphatase inhibitors (4906845001, Sigma Aldrich), 1x protease inhibitors (04693159001, Sigma Aldrich) and 0.05% Triton X-100 (# X100, Sigma). The mixture was sonicated on ice with three short pulses using the MS1 sonotrode (Hielscher Ultrasonics), setting “cycle” 1, “amplitude” 100% (30 kHz). The solution was then transferred to 270µl Beckman ultracentrifuge tubes and ultra-centrifuged in the Beckman 42.2 Ti rotor at 35000 x g, 4 °C for 30 min in the Beckman Coulter Optima XPN-90 ultracentrifuge. The supernatant was used directly for experiments or snap frozen in liquid nitrogen and stored at -80 °C.
Microtubule Assembly
Tubulin, Biotin-labelled tubulin as well as HiLyte647-labelled tubulin were purchased from Cytoskeleton Inc. (T240, T333P and TL670M, respectively). Biotinylated Hilyte647-labelled microtubules were polymerized from 4 mg/ml tubulin in the BRB80 (80mM PIPES, 1mM EGTA, 1mM MgCl2, pH 6.9) supplemented with 1 mM MgCl2 and 1 mM GTP (Jena Bioscience, Jena, Germany) for 30 min at 37 ºC. The polymerized microtubules were diluted in BRB80T (BRB80 with 10µM taxol) and centrifuged for 30 min at 21380g and room temperature in a Hettich® Universal 320R centrifuge, rotor 1420-A. After centrifugation, the pellet was resuspended and kept in BRB80T at room temperature.
TIRF Microscopy
Total internal reflection fluorescent (TIRF) microscopy experiments were performed on Zeiss Elyra PS.1 microscope using 100x/1.46 oil immersion objective and EM CCD Andor PALM camera. Fluorescence-labelled microtubules and KIF2A proteins were visualized using 647 nm and 488 nm lasers, respectively. The microscope was controlled by ZEN software (black edition). All experiments were performed at room temperature. For the TIRF experiments, the chambers were prepared by attaching two cleaned and silanized (0.05% dichlorodimethylsilane - DDS, Sigma Aldrich, 440272) glass coverslips (22 × 22 mm2 and 18 × 18 mm2; Corning, Inc.) with melted thin strips of parafilm. Chambers were incubated with 20 µg/mL anti-biotin antibody (Sigma Aldrich, B3640) in PBS for 5 min followed by incubation with 1% Pluronic (F127, Sigma Aldrich, P2443) for at least 30 min. The chambers were then washed with BRB80T, 5uL of in vitro prepared microtubules were added to the chamber and allowed to adhere to the antibodies for 30 s. Unbound microtubules were washed away with BRB80T and chambers were pre-incubated with TIRF assay buffer (BRB20 supplemented with 1 mM EGTA, 2mM MgCl2, 75mM KCl, 10mM dithiothreitol, 0.02 mg/ml casein, 10µM taxol, 1mM Mg-ATP, 20mM D-glucose, 1% Tween, 0.22 mg/ml glucose oxidase and 20 µg/ml catalase) before the experiments. All experiments were performed in TIRF assay buffer (AB). All experiments were quantified by pooling data from three different days.
KIF2A lysate imaging
Chambers were prepared as described above. For intensity and depolymerisation rate analysis, KIF2A lysates were diluted in AB buffer as follows: the WT lysate was diluted 1000x and the other lysates were all diluted to match the intensity of WT GFP signal in the epifluorescence (indicating similar concentration of GFP). For experiments shown in supplementary data, 10x higher concentrations were used to confirm the observed effects. Diluted lysates were added to surface-immobilized microtubules and imaged for 3 min with 5 s intervals. Microscopy data were analyzed using ImageJ 2.3.0/1.53q (FIJI). KIF2A density on the microtubules was measured by drawing a line through the entire microtubule and measuring the Mean Grey Value. For background subtraction, the line was then moved to an area close to the microtubule where no microtubule is present, and the Mean Grey Value was measured again and subtracted from the Mean Grey Value on the microtubule. To measure the depolymerisation rate, the length of the microtubule was measured at the beginning and then at the end of the video. The depolymerisation rate was calculated and normalized to wild-type KIF2A.
Results
Truncated TTBK2 proteins display reduced biochemical activity (Fig. 1)
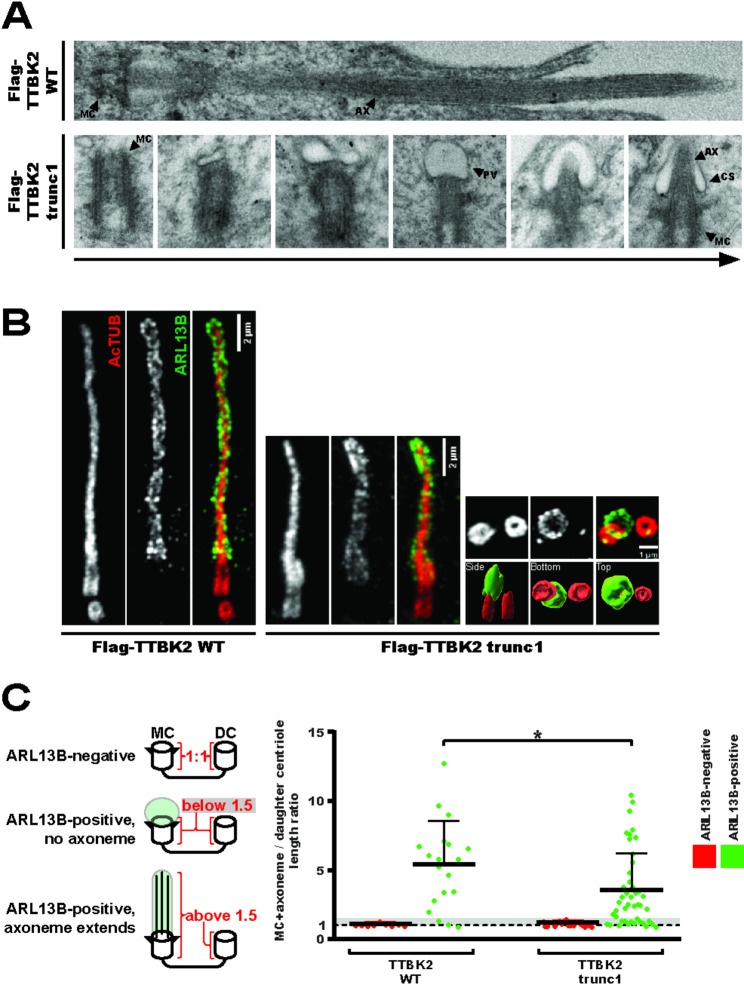
Fig. 1.
Truncated TTBK2 proteins display reduced biochemical activity (1A) study outline; (1B) TTBK2 constructs used in this study; (1C-D) Western blot analysis of lysates from HEK293T cells transfected with Myc-CEP164NT and Flag-TTBK2 constructs; (1E-F) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were fixed and stained for pCEP164 (scale bars: 0.5μm); (1F) The intensity of pCEP164 centriolar signal was quantified from the images (4 independent experiments, n≥79 cells per condition, normalized to gTUB, one-way ANOVA, ****P<0.0001); (1G-I) Western blot analysis of lysates from HEK293T cells transfected with DVL3 and Flag-TTBK2 constructs
As outlined above, a complete lack of TTBK2 activity blocks the cilium assembly cascade at the very beginning. This poses a considerable challenge when studying molecular events that only occur later, after cilium assembly has been triggered by TTBK2 (1A). To tackle this issue, we aimed to establish a system with reduced TTBK2 activity, rather than completely blocking this key kinase. To this end, we focused on TTBK2 truncating mutations which lead to SCA11 pathology in human [22], while the corresponding truncated protein moieties showed significantly reduced or no activity in mice [13, 21]. We prepared two truncated TTBK2 constructs (1B, see also Supplementary Table 1): TTBK2trunc1 (1-450 AA protein truncated shortly after the kinase domain) and TTBK2trunc2 (adenosine insertion at nucleotide 1329 shifts the reading frame in the last 6 AAs and creates a premature STOP codon [22]) and examined their biochemical activity. CEP164, a bona fide substrate of TTBK2, undergoes a mobility shift in response to TTBK2-induced phosphorylation [16, 18]. Indeed, we observed that Flag-tagged wild-type TTBK2 (Flag-TTBK2wt) induced a mobility shift of Myc-tagged N-terminal part of CEP164 (1-467 AA, CEP164NT) when co-expressed in HEK293T cells. In contrast, the expression of Flag-TTBK2trunc1 (1C) or Flag-TTBK2trunc2 (1D) showed no effect on Myc-CEP164NT mobility, indicating that phosphorylation of Myc-CEP164NT was hampered upon TTBK2 truncation. To corroborate our findings, we generated an antibody against the TKGLLGpSIYEDKT peptide of CEP164 corresponding to the phosphorylated S201 residue targeted by TTBK2 [18] (we termed the antibody „pCEP164“, see S1A-B for antibody validation). Having confirmed the specificity of the pCEP164 antibody to CEP164 protein, we in turn examined pCEP164 levels at MCs in hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing individual Flag-tagged variants of TTBK2 (see S1C for cell lines validation). Using this system, we found pCEP164 MC signal markedly increased in cells expressing Flag-TTBK2wt, compared to Flag-tagged kinase-dead TTBK2 (Flag-TTBK2kd), Flag-TTBK2trunc1, or Flag-TTBK2trunc2 (1E-F), in agreement with our WB data. We note that while the results clearly show the induction of the pCEP164 antibody epitope by TTBK2wt, the ability of this antibody to recognize specifically the phosphorylated S201 needs to be confirmed in the future.
Next, we examined the phosphorylation status of Dishevelled-3 (DVL3), another TTBK2 substrate [18]. As expected, DVL3 was phosphorylated and up-shifted when co-expressed with Flag-TTBK2wt, but not Flag-TTBK2kd. However, we observed a mobility shift of DVL3 when co-expressed with Flag-TTBK2trunc1 (1G) or GFP-TTBK2trunc2 (1H). We titrated TTBK2 plasmids and confirmed that both TTBK2wt and TTBK2trunc1 also induced DVL3 mobility shift when expressed at comparable levels (1I). This intriguing result suggested that truncated TTBK2 proteins indeed possessed a residual biochemical activity towards a subset of TTBK2 substrates rather than being completely inactive.
Truncated TTBK2 triggers cilia assembly (Fig. 2)
Fig. 2.
Truncated TTBK2 triggers cilia assembly (2A-C) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were fixed and stained for ARL13B and the indicated antibodies (scale bars: 2μm); (2B) shows percentage of ciliated cells in the corresponding cell line (n=4 independent experiments, one-way ANOVA, ***P<0.001); (2C) shows cilia length quantification (4 independent experiments, n≥153 cilia per condition, one-way ANOVA, ***P<0.001); (2D-E) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2trunc1 were transfected with siRNA targetting CEP164 (2E) shows cilia number upon siCEP164 treatment (n=3 independent experiments, unpaired T-test, ****P<0.0001)
If the diminished activity of TTBK2trunc1 (we elected it over TTBK2trunc2 due to slightly more pronounced defects in phosphorylating CEP164) was to be considered to model the role of TTBK2 in later stages of ciliogenesis/cilia maintenance, it had to support at least some degree of cilia formation. To test this, we again turned to hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing TTBK2 constructs. First, we found that TTBK2 KO RPE-1 cells were devoid of ARL13B-positive primary cilia altogether and expressing Flag-TTBK2wt rescued the frequency of ciliated cells to ~ 34% (2A-B), as expected. Remarkably, expressing Flag-TTBK2trunc1 rescued cilia formation in ~ 11% of cells, in contrast to Flag-TTBK2kd which completely failed to rescue (2A-B). At the same time, we noticed a marked reduction of cilia length in cells expressing Flag-TTBK2trunc1 compared to Flag-TTBK2wt, hinting at a possible defect in cilia assembly and/or maintenance (2C). The C-terminal part of wild-type TTBK2 interacts with CEP164, which targets TTBK2 to the MC [16]. As expected, Flag-TTBK2trunc1, which lacks the CEP164-binding region (1B), failed to be recruited to the MC and instead localized dispersely throughout the cytoplasm (2A, S2A). Interestingly, re-targeting TTBK2trunc1 to the MC by fusing it to the C-terminal part of CEP164 (468–1460 AA, we termed the construct „CEP164 chimera”) rescued both cilia number and cilia length to the level of TTBK2wt (2A-C), suggesting that the observed defects in cilia assembly were caused by TTBK2trunc1 mislocalization. Noteworthy, we confirmed that different cilia length in cells expressing Flag-TTBK2wt vs Flag-TTBK2trunc1 stemmed from qualitative differences between these constructs, rather than different expression levels (S2B-F).
Further, the partial rescue of cilia formation by Flag-TTBK2trunc1 (2B) suggested that TTBK2 enrichment at the MC was not strictly necessary for triggering cilia assembly. To test this, we examined hTERT RPE-1 CEP164 KO cells (a kind gift from Ciaran Morrison), which displayed prominent ciliogenesis defects, yet occasionally formed primary cilia [32]. First, we confirmed that CEP164 and TTBK2 signals were absent from the MCs of CEP164 KO cells (S2G-J), in line with the reported role of CEP164 in TTBK2 recruitment [16]. Remarkably, the rare primary cilia we found forming in the absence of CEP164 lacked the CEP164-recruited TTBK2 pools at their MCs (S2K). This data supports the notion that TTBK2 might trigger cilia formation even when localizing outside the MC, albeit with very low efficiency.
Considering the established role of CEP164 in ciliogenesis and TTBK2 recruitment, it was interesting to see that Flag-TTBK2trunc1 could trigger cilia assembly despite lacking the CEP164-interacting motif, prompting us to ask whether CEP164 was required for cilia formation in these cells. We found that depleting MC-associated CEP164 through siRNA (see S2L-M for siRNA validation) disrupted cilia assembly in Flag-TTBK2trunc1 cells (2D-E), suggesting that CEP164 mediated additional processes besides recruiting TTBK2 to the MC.
TTBK2trunc1 cilia are short, but display normal architecture (Fig. 3)
Fig. 3.
TTBK2trunc1cilia are short, but display normal architecture (3A) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were analyzed using TEM, upper panel shows a mature cilium in Flag-TTBK2wt, bottom panels show consecutive stages of early ciliogenesis in Flag-TTBK2trunc1 (MC=mother centriole, PV=preciliary vesicle, CS=ciliary sheath, AX=axoneme); (3B) expansion microscopy reveals ARL13B-positive ciliary membrane (green chanel) and AcTUB-positive ciliary microtubules (red chanel), 6 small panels on the right show an ARL13B-positive vesicle docked to the MC with the bottom panels showing 3D-reconstruction of the same image viewed from different perspectives (3C) MC+axoneme and DC lengths were measured using the AcTUB signal from expansion microscopy images, the ratio of each MC+axoneme to DC was quantified (4 independent experiments, n=19 for TTBK2wt, n=45 for TTBK2trunc1, unpaired T-test, *P<0.05), grey area in the graph indicates values between 1 and 1.5
Having established that truncated TTBK2 could not rescue ciliogenesis as effectively as wild-type TTBK2 (2B-C), we wanted to distinguish early and late defects in cilia formation. Cilia assembly in RPE-1 cells starts with the formation of a preciliary vesicle at the distal end of the MC, which later elongates into a ciliary sheath once ciliary microtubules extend towards it [33]. Using transmission electron microscopy (TEM), we detected various, consecutive stages of preciliary vesicle formation in Flag-TTBK2trunc1 cells, including cells with an elongated ciliary sheath (3A). In contrast to Flag-TTBK2wt, we struggled to detect fully assembled cilia in Flag-TTBK2trunc1, in agreement with their lower occurrence in our IF microscopy experiment (2B).
To corroborate our findings, we took advantage of expansion microscopy, which offered vastly improved cilia detection over TEM. Primary cilia formed in the Flag-TTBK2wt condition showed a typical arrangement of acetylated tubulin (AcTUB)-positive axonemal microtubules, enclosed within a ciliary membrane labelled by ARL13B (3B, left panel). Many cilia found in the Flag-TTBK2trunc1 condition were similar to that but with a shorter axoneme (3B, middle panel). Further, in Flag-TTBK2trunc1 we often noticed structures consisting of ARL13B-positive vesicles docked to the MC but with no apparent sign of axonemal microtubules extending (3B, right panel).
To quantify this phenotype, we measured the length of AcTUB signal of both centrioles in a pair. Using these measurements, we determined the ratio of combined MC + axoneme length relative to the length of the daughter centriole (3C, see left panel). ARL13B was used simply to determine whether a given cell initiated cilium assembly. For centriole pairs with no ARL13B signal the MC/DC length ratio was around 1 (3C, red datapoints), indicating that no ciliary axoneme was extending. In ARL13B-positive centriole pairs (3C, green datapoints), the MC/DC length ratio shifted towards higher values, as the corresponding mother centrioles templated cilia. However, the average MC/DC length ratio was smaller in Flag-TTBK2trunc1 compared to Flag-TTBK2wt (3C), in agreement with the observed cilia length defect (2C). What is more, ARL13B-positive Flag-TTBK2trunc1 centriole pairs frequently displayed a length ratio between 1 and 1.5 (3C, grey area in the graph), indicating that their axonemal microtubules did not extend (14/45 cells in Flag-TTBK2trunc1 compared to 3/19 cells in Flag-TTBK2wt).
In sum, our data show that cilia forming in Flag-TTBK2trunc1 cells displayed no obvious ultrastructural defects, but were simply shorter than their Flag-TTBK2wt counterparts.
Early ciliogenesis progression in TTBK2trunc1 (Fig. 4)
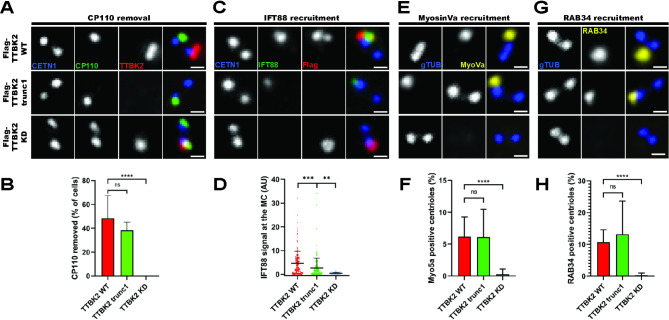
Fig. 4.
Early ciliogenesis progression in TTBK2trunc1(4A, 4C, 4E, 4G) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were stained with the indicated antibodies (scale bars: 0.5μm) (4B) shows percentage of cells with CP110 removed from the MC (n=3 independent experiments, one-way ANOVA, ****P<0.0001); (4D) shows IFT88 signal intensity at the MC (normalized to CETN1, 6 independent experiments, n≥59 for each condition, one-way ANOVA, ***P<0.001, **P<0.01); (4F) shows percentage of cells with MyosinVa signal present at the MC (n=4 independent experiments, one-way ANOVA, ****P<0.0001); and (4H) shows percentage of cells with RAB34 signal present at the MC (n=4 independent experiments, one-way ANOVA, ****P<0.0001)
To examine how efficiently the truncated TTBK2 triggered individual early steps of ciliogenesis, we first stained for CP110, a protein removed from the MC in response to TTBK2 activity [13]. We found both Flag-TTBK2wt and Flag-TTBK2trunc1, but not Flag-TTBK2kd, to efficiently induce the MC removal of CP110 in TTBK2 KO background (4A-B).
Next, we examined the MC recruitment of IFT88, a protein involved in shuttling of molecules inside cilia [12]. We found that while Flag-TTBK2kd failed to promote IFT88 recruitment to the MC, Flag-TTBK2wt and to a lesser extent also Flag-TTBK2trunc1 showed a rescue effect on IFT88 MC levels (4C-D).
Finally, we tested if MyosinVa and RAB34, two markers of early membrane structures [34–36], localized to the MC in TTBK2trunc1. Flag-TTBK2wt and Flag-TTBK2trunc1 showed comparable frequencies of MyosinVa- and RAB34-positive MCs, whereas Flag-TTBK2kd did not rescue the MC recruitment of these two markers (4E-H).
To capitalize on the ability of TTBK2trunc1 to trigger cilia formation in a CEP164-dependent manner (2D), but independently of CEP164-mediated TTBK2 recruitment to the MC (2A), we examined the effects of CEP164 depletion on the above-tested markers. Intriguingly, our data revealed a clear dependency of all markers on CEP164 presence (S3A-D). To our knowledge, our model system is the first to decouple the role of CEP164 in recruiting TTBK2 from its other roles during cilia assembly, which warrants further investigation.
To conclude, our results showed that truncated TTBK2 could indeed reasonably well trigger canonical early steps of the cilium assembly cascade.
Short cilia in TTBK2trunc1 are linked to KIF2A accumulation (Fig. 5)
Fig. 5.
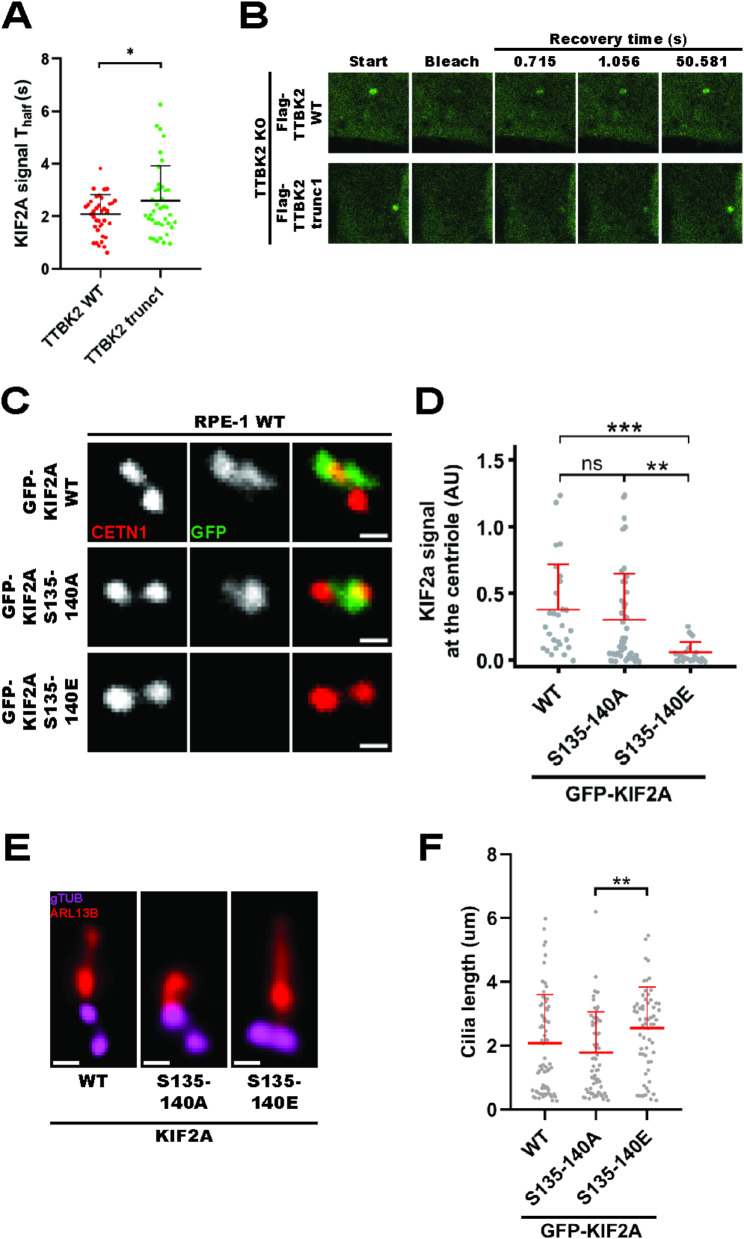
Short cilia in TTBK2trunc1are linked to KIF2A accumulation (5A-C) hTERTRPE-1 TTBK2 KO cells expressing mNeonGreen-ARL13B and Flag-TTBK2 constructs were subjected to live-cell imaging (images show the NeonGreen signal, scale bars: 5μm); (5B) shows the mean length of cilia measured over the course of 5 hours (+/- standard error of mean, 4 independent experiments, n≥90 cilia per condition); (5C) Flag-TTBK2trunc1 cells exhibited frequent cilia breakage events; (5D-E) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were fixed and stained for KIF2A and the indicated antibodies (scale bars: 0.5μm); (5E) shows KIF2A signal intensity at the MC (4 independent experiments, n≥54 cells per condition, normalized to CETN1, one-way ANOVA, ****P<0.0001). (5F-G) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were transfected with mock or KIF2A siRNA (scale bars: 2μm); (5G) shows cilia length quantification(4 independent experiments, n≥189 cilia per condition, one-way ANOVA, **P<0.01, ***P<0.001)
Given that Flag-TTBK2trunc1 cells could initiate ciliogenesis (Figs. 3 and 4), we speculated that the observed defects in cilia formation (2A-C) might arise later, during axoneme extension. To test our hypothesis, we examined cilia growth dynamics using live-cell imaging of cells expressing mNeonGreen-ARL13B. Of note, the expression of exogenous ARL13B led to an overall increase in cilia length (if compared to experiments without exogenous ARL13B), in agreement with previous observations [27, 37]. We found that RPE-1 cells expressing Flag-TTBK2wt formed steadily growing cilia during the 5 h period of the experiment (5A-B). In contrast, cilia in Flag-TTBK2trunc1-expressing cells elongated very slowly, sometimes even shortened towards the end of the experiment. Moreover, we noted an increase in cilia breakage events (5C) in the Flag-TTBK2trunc1 condition (0.355 breaks per cilium) compared to Flag-TTBK2wt (0.222 breaks per cilium). Taken together with the rather modest defect in IFT88 MC levels in TTBK2trunc1-expressing cells (4C-D), we hypothesized that TTBK2 might regulate axoneme length by an additional, unknown mechanism and sought to identify it.
We focused on KIF2A, a microtubule(MT)-depolymerizing kinesin implicated in cilia resorption upon cell cycle re-entry [26], which interacts with TTBK2 [24]. We found that KIF2A MC levels were elevated in RPE-1 cells expressing Flag-TTBK2trunc1 compared to Flag-TTBK2wt (5D-E), confirming and extending previous observations from mutant mouse embryonic fibroblasts [13]. In addition, we found a modest negative correlation between KIF2A basal body presence and cilia length (S4A). Given that KIF2A acts as a negative regulator of cilia length [26], we next tested the possible causality between KIF2A MC accumulation and axoneme extension defects in Flag-TTBK2trunc1 cells by depleting KIF2A through siRNA (see S4B-C for depletion efficiency validation). Remarkably, even though KIF2A depletion did not significantly change the percentage of ciliated cells (S4D), it fully rescued the cilia length defect in Flag-TTBK2trunc1-expressing cells (5F-G). Importantly, KIF2A depletion showed no additive effect on cilia length in Flag-TTBK2wt, suggesting that KIF2A-mediated inhibition of axoneme growth was specific to Flag-TTBK2trunc1 cells.
KIF2A overactivation phenocopies TTBK2 truncation (Fig. 6)
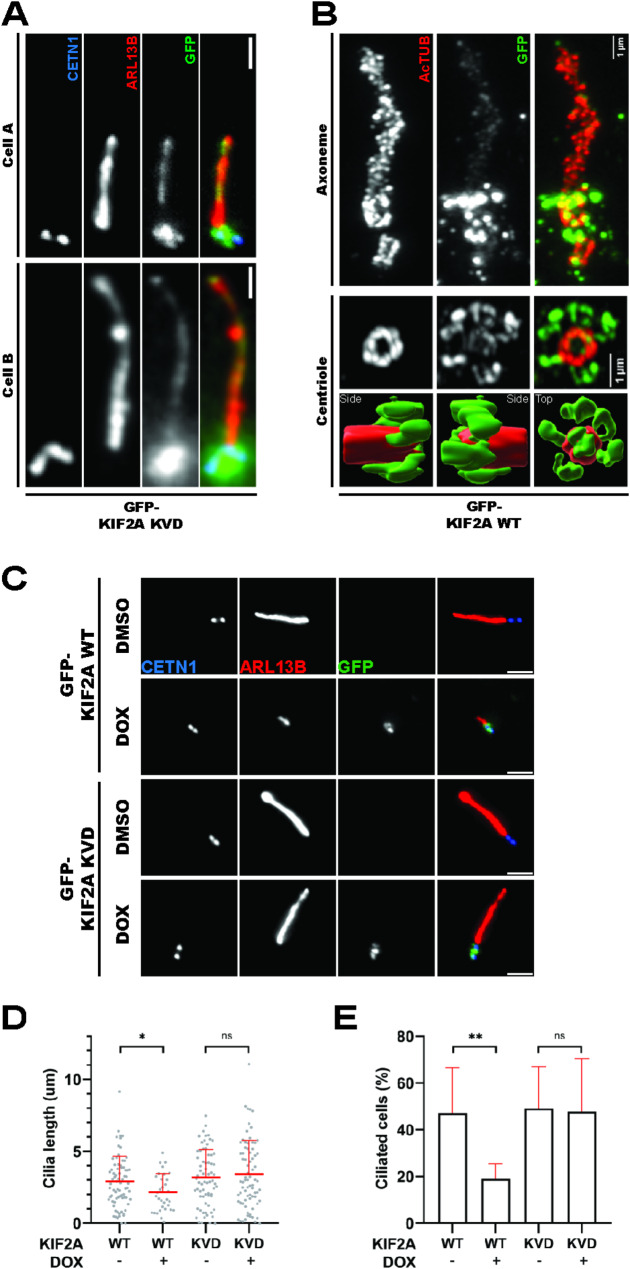
Fig. 6.
KIF2A overactivation phenocopies TTBK2 truncation (6A) hTERT RPE-1 cells expressing GFP-KIF2AKVD were fixed and stained for ARL13B and CETN1 (scale bars: 1μm), the images show two representative cells with GFP-KIF2AKVD signal decorating the cilium (6B) hTERT RPE-1 cells expressing GFP-KIF2Awt were fixed, subjected to expansion microscopy, and stained for AcTUB and GFP, GFP-KIF2Awt (green channel) localized to the basal body and also showed a faint axonemal signal, bottom panels show Imaris 3D reconstructions of a centriole viewed from different perspectives (6C-E) hTERT RPE-1 cells expressing GFP-KIF2A constructs were treated with DMSO or DOX, fixed, and stained for ARL13B and the indicated antibodies (scale bars: 2μm); (6D) shows cilia length quantification (2 independent experiments, n≥32 cilia per condition, unpaired T-test, *P<0.05); (6E) shows percentage of ciliated cells (n=2 independent experiments, unpaired T-test, **P<0.01)
Having established the functional link between truncated TTBK2 and KIF2A, we examined in detail the effects of KIF2A on cilia length. First, we generated hTERT RPE-1 cell lines DOX-inducibly expressing GFP-KIF2Awt or GFP-KIF2AKVD, a mutant defective in MT depolymerization [26, 38]. We found both constructs to preferentially localize to centrioles and/or their proximity (6A-B). Intriguingly, we could detect GFP-KIF2AKVD decorating the ciliary axoneme, suggesting that KIF2A could localize directly inside cilia (6A). In addition, we resolved the signal of GFP-KIF2Awt using expansion microscopy and found it to form ring-like structures near the MC distal end, closely resembling the localization pattern of distal or subdistal appendage proteins (6B). Furthermore, our expansion microscopy protocol revealed a faint signal of GFP-KIF2Awt decorating the ciliary axoneme. We also detected endogenous KIF2A localizing to centrioles and their proximity and its centriolar localization was abolished by paclitaxel treatment (5µM, 4 h), suggesting a dependency on intact microtubules (S5A-B).
Upon investigating KIF2A localization pattern, we examined its effects on primary cilia formation. Following DOX-induced expression of GFP-KIF2Awt, but not GFP-KIF2AKVD, we could readily observe a reduction of cilia length (6C-D), in line with previous work [26]. In addition, we found that GFP-KIF2Awt, but not GFP-KIF2AKVD, reduced the percentage of ciliated cells (6E).
To conclude, our data established that the activity of KIF2A in primary cilia interferes with cilia assembly and phenocopies the defects seen in Flag-TTBK2trunc1 cells.
TTBK2-induced phosphorylations inhibit KIF2A MT binding and depolymerization in vitro (Fig. 7)
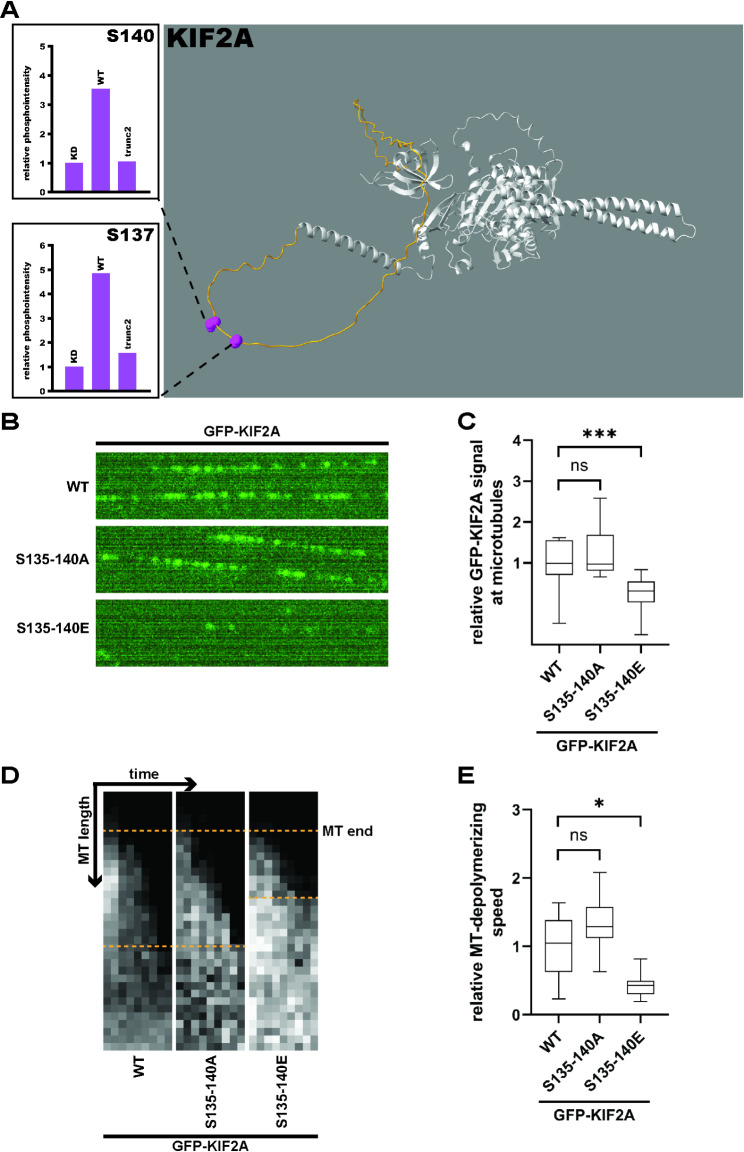
Fig. 7.
TTBK2-induced phosphorylations inhibit KIF2A MT binding and depolymerization in vitro (7A) HEK293T cells were transfected with Flag-KIF2A and GFP-TTBK2 constructs, lysed, incubated with anti-Flag affinity beads, and the isolates were subjected to MS/MS analysis. Phosphorylation intensities between TTBK2 WT, KD, and trunc2 were quantified as means of 3 independent experiments and normalized to TTBK2 KD (see S6A for complete phosphomap), TTBK2wt induced phosphorylation of KIF2A serines S137 and S140; (7B-E) lysates of HEK293T cells transfected with GFP-KIF2A constructs were incubated with in vitro stabilized microtubules; (7B) shows a representative picture of MT-bound GFP-KIF2A molecules; (7C) shows KIF2A signal intensity at MTs (n=3 independent experiments, normalised to GFP-KIF2Awt, Kruskal-Wallis test, ***P<0.001); (7D) representative kymograms showing MT shortening by different KIF2A constructs, gray signal shows biotinylated tubulin; (7E) shows the rate of MT depolymerization by KIF2A as a measure of MT shortening over time (n=3 independent experiments, normalised to GFP-KIF2Awt, Welch ANOVA test, *P<0.05)
To reveal how TTBK2 regulates KIF2A, we first examined KIF2A phosphorylation induced by TTBK2. Following co-expression of Flag-KIF2Awt with GFP-tagged TTBK2 variants in TTBK2 KO HEK293T, we immunopurified Flag-KIF2Awt, subjected the isolates to MS/MS analysis and identified several TTBK2-induced S/T phospho-sites (S6A). We further focused on S137 and S140 (7A) as phospho-sites that were specifically induced by GFP-TTBK2wt, but not by GFP-TTBK2trunc2 (we selected it over TTBK2trunc1 in this experiment due to its slightly higher activity towards CEP164) or GFP-TTBK2kd. Together with the closely neighbouring S135 which was identified, but not functionally characterized, in a study by Watanabe et al. [24], these 3 serine residues are located in an unstructured loop near the N-terminus of KIF2A, according to predicted KIF2A structure found in the AlphaFold DB [39] (7A, marked in yellow).
To test if phosphorylation of the S135-140 cluster regulated KIF2A function, we mutated the entire cluster to alanine (A) or glutamate (E) to block or mimic phosphorylation, respectively. Then, we incubated lysates from HEK293T cells transfected with GFP-tagged KIF2A phosphomutants with in vitro reconstituted and stabilized MTs and used total internal reflection fluorescence (TIRF) microscopy to examine KIF2A-MT interaction dynamics (7B). We found that mimicking phosphorylation of the S135-140 cluster (in GFP-KIF2AS135–140E) reduced the amount of KIF2A bound to MTs (7C, S6B).
Given that, we used a similar TIRF assay to examine whether S135-140 phosphorylation regulated MT depolymerization (7D). Intriguingly, mimicking phosphorylation of the S135-140 cluster inhibited MT depolymerization by GFP-KIF2AS135–140E (7E, S6C). Taken together, our data established that TTBK2-mediated phosphorylation of S135-140 regulates the ability of KIF2A to bind and depolymerize MTs in vitro.
In addition to the S135-140 cluster, we tested if phosphorylation of KIF2A residues S586/S604 (identified by MS/MS in S6A) regulated KIF2A function. Surprisingly, we found that both mimicking and blocking S586/S604 phosphorylation (in GFP-KIF2AS586/604E and GFP-KIF2AS586/604A respectively) enhanced KIF2A MT affinity (S6D) and depolymerization (S6E). Our modeling using Alphafold [40] showed that both mutants display stabilized alpha-helical structures in regions involved in KIF2A dimerization [38] (S6F). We note that while our in silico modelling results might explain the phenotype of S586/S604 mutants, we could not conclusively address the possible role of TTBK2-mediated phosphorylation of S586 and S604. Further work will be required to determine if these C-terminal phosphorylations truly regulate KIF2A under physiological conditions.
TTBK2 regulates KIF2A in cells to support cilia formation (Fig. 8)
Fig. 8.
TTBK2 regulates KIF2A in cells to support cilia formation (8A-B) hTERT RPE-1 TTBK2 KO cells DOX-inducibly expressing Flag-TTBK2 constructs were transfected with GFP-KIF2Awt and subjected to FRAP analysis; (8A) shows individual Thalf values calculated using the EasyFRAP web algorithm (3 independent experiments, n≥38 cells per condition, unpaired T-test, *P<0.05); (8C-D) hTERT RPE-1 cells were transfected with GFP-KIF2A constructs, fixed, and stained for CETN1 (scale bars: 0.5μm); (8D) shows GFP-KIF2A signal intensity at the mother centriole (3 independent experiments, n≥25 cellsper condition, normalized to CETN1, one-way ANOVA, **P<0.01, ***P<0.001);(8E-F) hTERT RPE-1 cells DOX-inducibly expressing GFP-KIF2A constructs were fixed and stained with the indicated antibodies; (8F) shows cilia length quantification (3 independent experiments, n≥61 cilia per condition, one-way ANOVA, **P<0.01)
Next, we examined the consequences of TTBK2-mediated KIF2A phosphorylation for its localization and effects on cilia. Since we observed changes in KIF2A MC levels between our RPE-1 cell lines (5D-E), but no difference in total levels of KIF2A (S4A), we reasoned that TTBK2 might specifically regulate KIF2A recruitment to MC rather than whole-cell KIF2A protein levels. To probe for changes in MC-associated KIF2A, we carried out a fluoresence recovery after photobleaching (FRAP) analysis of GFP-KIF2Awt MC pools in Flag-TTBK2wt and Flag-TTBK2trunc1 RPE-1 cells. We observed a small, but consistent reduction in recovery halftime (Thalf) of GFP-KIF2Awt MC signal in Flag-TTBK2wt compared to Flag-TTBK2trunc1 cells (8A-B), suggesting a faster KIF2A MC turnover in Flag-TTBK2wt.
Next, to examine if KIF2A MC recruitment was regulated by phosphorylation of the S135-140 cluster, we expressed GFP-KIF2Awt, GFP-KIF2AS135–140E or GFP-KIF2AS135–140A in RPE-1 cells. Remarkably, we found that mimicking S135-140 phosphorylation strongly diminished KIF2A MC localization in GFP-KIF2AS135–140E, whereas GFP-KIF2Awt and GFP-KIF2AS135–140A localized normally (8C-D). This result suggested that TTBK2 phosphorylation of S135-140 likely facilitates ciliogenesis by preventing the accumulation of KIF2A at MCs. However, our results also hinted at possible limitations of our experimental setup, as we were not able to resolve a difference in MC levels between GFP-KIF2Awt and GFP-KIF2AS135–140A (see also Discussion).
Finally, to test if phosphorylation of KIF2A at S135-140 regulated cilia formation, we compared ciliogenesis in hTERT RPE-1 cells DOX-inducibly expressing GFP-tagged KIF2A phosphomutants. We observed that cells overexpressing GFP-KIF2AS135–140E formed cilia that were longer compared to cells expressing GFP-KIF2AS135–140A (8E-F), indicating that phosphorylation of KIF2A at S135-140 supported axoneme elongation.
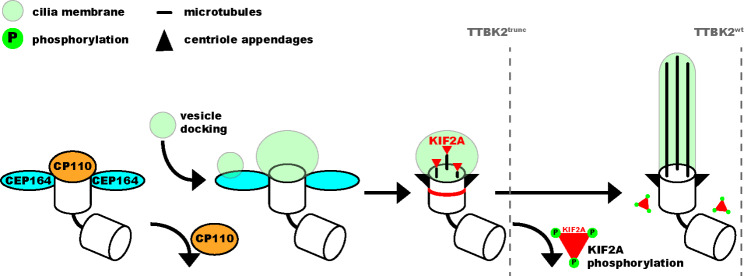
Taken together, our results suggest that TTBK2 phosphorylation destabilizes KIF2A basal body pools and prevents its accumulation at the base of the cilium, which supports efficient axoneme growth. In contrast, truncated TTBK2 inefficiently phosphorylates KIF2A, which seems to alter its turnover, leading to increased KIF2A levels at the ciliary base, and in turn to reduced ciliary length (Fig. 9).
Fig. 9.
TTBK2 supports axoneme elongation by phosphorylation-mediated inhibition of KIF2A In our model, TTBK2 kinase activity is needed to prime the basal body for cilia outgrowth by removing CP110 and mediating the docking of ciliary vesicles to the mother centriole. Both wild-type and truncated TTBK2 are sufficient to support these initial processes. TTBK2 activity is then required to inhibit KIF2A and diminish its basal body pool, enabling full elongation of the ciliary axoneme in wild-type TTBK2. In contrast, truncated TTBK2 fails to inhibit KIF2A, leading to KIF2A accumulation at the basal body and shorter cilia
Discussion
We and others have demonstrated that the C-term region of TTBK2 is essential for its interaction with CEP164 [16, 41, 42]. Still, we were initially surprised to see the difference in TTBK2trunc1/2-induced phosphorylation between CEP164 and DVL3. We speculate that such selectivity in targeting individual TTBK2 substrates may be explained by hampered interaction capabilities of truncated TTBK2. Interestingly, our rescue experiment using the TTBK2-CEP164 chimera demonstrates that the diminished ciliogenesis-promoting activity of TTBK2trunc1 resides in its spatial properties (misslocalization), rather than a defective kinase activity per se. Importantly, the expression of TTBK2trunc1 in hTERT RPE-1 TTBK2 KO allowed for semi-permissive conditions for cilia assembly. Intriguingly, we found that initial events of primary cilia formation (CP110 removal, IFT recruitment, formation of a membrane vesicle), were rather moderately affected. In contrast, TTBK2trunc1 was not able to effectively mediate KIF2A turnover and support axoneme elongation. Based on these observations, we hypothesize that individual steps of ciliogenesis likely differ in their requirements for TTBK2 activity.
Earlier work established that the MC-associated pool of KIF2A is phosphorylated by mitotic kinase PLK1 to trigger primary cilia resorption [26]. Our data demonstrate that in addition to its MC-associated pool, KIF2A can also be detected along axonemal microtubules inside the cilium. This observation raises several intriguing possibilities. First, our expansion microscopy data suggest that the MC-associated pool of KIF2A is well within the reach of TTBK2 kinase activity at the MC [43, 44], given the flexibility of TTBK2’s long, highly unstructured C-terminal part [42]. Thus, KIF2A seems to be in a suitable position for direct regulation by TTBK2 phosphorylation, analogous to their interactions at microtubule plus ends [24]. Further, the presence of KIF2A inside the cilium helps to explain how KIF2A microtubule-depolymerizing activity mediates the resorption of primary cilia observed earlier [26]. However, we cannot exclude a contribution of additional mechanisms, e.g. KIF2A action towards microtubules anchored to the subdistal appendages of the MC [45, 46]. We also noted that the axoneme-associated pool of KIF2A was somewhat easier to detect in the case of KIF2AKVD, perhaps due to altered dynamics of tubulin-KIF2AKVD interaction [47, 48]. Furthermore, our data suggest that altered KIF2A turnover contributes to primary cilia defects related to the activities of truncated TTBK2 moieties. We showed that KIF2A accumulated at the MC of TTBK2 KO hTERT RPE-1 cells expressing Flag-TTBK2trunc1, and that primary cilia length can be rescued by KIF2A depletion. Noteworthy, a similar accumulation of KIF2A has also been observed in Ttbk2 hypomorphic mutant mice [21]. Nonetheless, differential phosphorylation of additional substrates by TTBK2wt vs. truncated TTBK2 may also play a role here – this notion is further supported by our observation that KIF2A depletion fully restored primary cilia length, but only showed a partial rescue on the number of cilia in TTBK2 KO hTERT RPE-1 Flag-TTBK2trunc1 cells (5G, S4B).
Our data demonstrate that TTBK2-induced phosphorylation of the S135-140 cluster in KIF2A represents a key regulatory mechanism, which inhibits KIF2A binding to microtubules and reduces KIF2A MC pools. Interestingly, the interaction between microtubules, which are negatively charged, and the positively charged neck region in KIF2C (MCAK), another member of the kinesin-13 family, has been shown to aid KIF2C microtubule binding [49]. It is therefore tempting to speculate that phosphorylation of the S135-140 cluster in KIF2A creates a local negative charge that inhibits KIF2A binding to microtubules by increasing the initial energy barrier between free-in-solution and MT-bound KIF2A states. Lastly, our in vitro and cell-based assays could not resolve a difference between KIF2Awt and KIF2AS135–140A, likely due to altered substrate-to-kinase stochiometry caused by KIF2A overexpression used in those experiments.
Our earlier work demonstrated that a CEP164 mutant defective in binding TTBK2 fails to rescue ciliogenesis following CEP164 depletion [16]. Indeed, in several systems TTBK2 is essential for the onset of ciliogenesis, with no alternative pathway available to compensate for TTBK2 MC recruitment defect [21, 23]. However, in light of our current data we speculate that concentrating TTBK2 activity at the MC is not strictly necessary for the phosphorylation of at least some of its basal body substrates, provided the levels and activity of the kinase outside of the MC are sufficiently high (e.g. in TTBK2trunc1-expressing TTBK2 KO hTERT RPE-1 cells). Under such conditions, there seems to be a way to bypass the requirement for MC-concentrated TTBK2 activity when triggering ciliogenesis. This hypothesis is further supported by our observations in CEP164 KO cells, which can form cilia (though rarely) even in the absence of detectable TTBK2 MC pools. The obvious question is if such regulation has any physiological relevance or merely represents a peculiarity of one particular cell culture system. We think the former is indeed a plausible scenario. In fact, our recent work has demonstrated that TTBK1, a kinase highly similar to TTBK2 but lacking the CEP164-binding motif, does not localize to the MC. Remarkably, we found TTBK1 expression upregulated during human pluripotent stem cell neuronal differentiation, in turn allowing it to exert a rescue effect on cilia formation in differentiated, but not undifferentiated TTBK2 KO cells [50], which is in line with the activities of truncated TTBK2 reported here.
Further, previous work by Schmidt et al. [51] demonstrated that CEP164 depletion severely disrupted docking of vesicles to the MC. However, it remained to be discovered if vesicles failed to dock due to TTBK2 misslocalization or because CEP164 played a more direct role during the docking process. Our data support the latter possibility, as cells expressing TTBK2trunc1 showed no defects in preciliary vesicle formation (3A-C). Moreover, depleting CEP164 from TTBK2trunc1 cells inhibited MC recruitment of membrane markers MyosinVa and RAB34 (S3C-D), and of IFT88 (S3B), favoring the notion that CEP164 might act as a common docking platform at the MC, independently of its TTBK2-recruitment function.
The last (but not least) observation we find pertinent to discuss relates to the TTBK2-mediated removal of CP110. As TTBK2trunc1 does not interact with CEP164, in principle it has no means of “distinguishing” between MC- and DC-associated CP110. However, while elevated levels of TTBK2trunc1 may compensate for its absence from the MC, our data clearly show that TTBK2trunc1 can specifically remove the MC-pool of CP110, but not the DC-pool of CP110. This decoupling of TTBK2 activity from the recruiting role of CEP164 challenges some of the recently proposed models of TTBK2-induced removal of CP110 [52] and suggests the existence of an additional mechanism ensuring MC-specific CP110 loss. We speculate that distal appendages may be plausible candidates for that function. In fact, depleting CEP164 from TTBK2trunc1 cells inhibited CP110 removal from the MC (S3A). Intriguingly, docking of vesicles to the distal appendages has been proposed to act upstream of CP110 removal [10, 17, 53, 54]. However, other studies reporting defective vesicle docking have found no concomitant defect in the removal of CP110 [34, 36]. Thus, the cooperation of distal appendages and TTBK2 to regulate primary cilia assembly warrants further investigation.
Conclusions
TTBK2 has emerged as a crucial regulator of primary cilia formation, but the full scope of its activities and the underlying mechanisms are only starting to become apparent. Herein, we present evidence that (i) TTBK2 supports axoneme growth by restraining KIF2A levels at the mother centriole – to the best our knowledge, our work provides the first functional validation of a TTBK2-mediated phosphorylation of a ciliary kinesin in the context of primary cilia assembly; and (ii) TTBK2 enrichment at the mother centriole, important to trigger ciliogenesis, might be under specific conditions bypassed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Erich Nigg, Peter Jackson, Randall Moon, Ciaran Morrison, Vítězslav Bryja, and Stephan Angers for sharing reagents with us. We are also grateful to Naděžda Vaškovicová for assistance with TEM. We acknowledge the core facility CELLIM supported by the Czech-BioImaging large RI project (LM2023050 funded by MEYS CR) for their support with obtaining scientific data presented in this paper. Furthermore, CEITEC Proteomics Core Facility is gratefully acknowledged for obtaining the scientific data presented in this paper. CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2023042 and European Regional Development Fund-Project „UP CIISB“(No. CZ.02.1.01/0.0/0.0/18_046/0015974), is acknowledged for the financial support of the measurements at the CEITEC Proteomics Core Facility. We also acknowledge the Centre of Imaging Methods core facility, Faculty of Science, Charles University, supported by the Czech-BioImaging through MEYS CR (LM2015062 and CZ.02.1.01/0.0/0.0/16_013/0001775). Lastly, computational resources were provided by the e-INFRA CZ project (ID:90254), supported by the Ministry of Education, Youth and Sports of the Czech Republic.
Author contributions
D.V. - performed and analyzed most of the experiments and assembled the figures, O.B.- contributed to stable cell lines generation and performed initial experiments, E.L.- performed in vitro TIRF experiments, T.R.- carried out experiments with pCEP164 antibody, L.B.- performed time lapse experiment with ARL13B reporter cell line in Fig. 5, A.L. - contributed to analysis of KIF2A mutants in Fig. 8, T.D.- contributed to WB analysis in Fig. 1, Z.L. - supervised in vitro TIRF experiments, L.C.- conceptualized and supervised the study and together with D.V. wrote the paper. All authors approved the final version of the manuscript.
Funding
This work was supported by grant 22–13277S from Czech Science Foundation to LC, the institutional support of the Czech Academy of Sciences (RVO: 86652036), and project LX22NPO5107 (MEYS): Financed by European Union – Next Generation EU. DV was supported by grant MUNI/C/0026/2020 from Masaryk University. TD was supported by grant MUNI/A/1738/2024 from Masaryk University. TR was supported by grant MUNI/R/1148/2021 from Masaryk University.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
N/A.
Consent for publication
N/A.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/29/2025
The original online version of this article was revised: Author would like to request that correct the supplementary files presented at the journal webpage.
References
- 1.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. 10.1146/annurev.physiol.69.040705.141236 [DOI] [PubMed] [Google Scholar]
- 2.Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol. 2019;15(4):199–219. 10.1038/s41581-019-0116-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426(6962):83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- 4.Basu B, Brueckner M. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr Top Dev Biol. 2008;85:151–174. 10.1016/S0070-2153(08)00806-5 [DOI] [PubMed] [Google Scholar]
- 5.Braun DA, Hildebrandt F. Ciliopathies. Cold Spring Harb Perspect Biol. 2017;9(3):a028191. Published 2017 Mar 1. 10.1101/cshperspect.a028191 [DOI] [PMC free article] [PubMed]
- 6.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18(9):533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed]
- 7.Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci. 2010;123(Pt 4):511–518. 10.1242/jcs.061093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12(5):767–778. 10.1016/j.devcel.2007.03.004 [DOI] [PubMed]
- 9.Tanos BE, Yang HJ, Soni R, et al. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27(2):163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanie T, Liu B, Love JF, Fisher SD, Gustavsson AK, Jackson PK. A hierarchical pathway for assembly of the distal appendages that organize primary cilia. Elife. Published online January 30, 2025. 10.7554/eLife.85999 [DOI] [PMC free article] [PubMed]
- 11.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130(4):678–690. 10.1016/j.cell.2007.06.027 [DOI] [PubMed] [Google Scholar]
- 12.Lechtreck KF, Van De Weghe JC, Harris JA, Liu P. Protein transport in growing and steady-state cilia. Traffic. 2017;18(5):277–286. 10.1111/tra.12474 [DOI] [PMC free article] [PubMed]
- 13.Goetz SC, Liem KF Jr, Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151(4):847–858. 10.1016/j.cell.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi M, Tomizawa K, Sato K, Ohtake A, Omori A. A novel tau-tubulin kinase from bovine brain. FEBS Lett. 1995;372(1):59–64. 10.1016/0014-5793(95)00955-9 [DOI] [PubMed] [Google Scholar]
- 15.Qi H, Yao C, Cai W, Girton J, Johansen KM, Johansen J. Asator, a tau-tubulin kinase homolog in Drosophila localizes to the mitotic spindle. Dev Dyn. 2009;238(12):3248–3256. 10.1002/dvdy.22150 [DOI] [PubMed]
- 16.Čajánek L, Nigg EA. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci U S A. 2014;111(28):E2841–E2850. 10.1073/pnas.1401777111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo CH, Lin IH, Yang TT, et al. Phosphorylation of CEP83 by TTBK2 is necessary for cilia initiation. J Cell Biol. 2019;218(10):3489–3505. 10.1083/jcb.201811142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernatik O, Pejskova P, Vyslouzil D, Hanakova K, Zdrahal Z, Cajanek L. Phosphorylation of multiple proteins involved in ciliogenesis by Tau Tubulin kinase 2. Mol Biol Cell. 2020;31(10):1032–1046. 10.1091/mbc.E19-06-0334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanáková K, Bernatík O, Kravec M, et al. Comparative phosphorylation map of Dishevelled 3 links phospho-signatures to biological outputs. Cell Commun Signal. 2019;17(1):170. Published 2019 Dec 23. 10.1186/s12964-019-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen A, Goetz SC. TTBK2 controls cilium stability by regulating distinct modules of centrosomal proteins. Mol Biol Cell. 2023;34(1):ar8. doi:10.1091/mbc.E22-08-0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowie E, Norris R, Anderson KV, Goetz SC. Spinocerebellar ataxia type 11-associated alleles of Ttbk2 dominantly interfere with ciliogenesis and cilium stability. PLoS Genet. 2018;14(12):e1007844. Published 2018 Dec 10. 10.1371/journal.pgen.1007844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houlden H, Johnson J, Gardner-Thorpe C, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11 [published correction appears in Nat Genet. 2008 Feb;40(2):255. Giunti, Paolo [corrected to Giunti, Paola]]. Nat Genet. 2007;39(12):1434–1436. 10.1038/ng.2007.43 [DOI] [PubMed] [Google Scholar]
- 23.Bouskila M, Esoof N, Gay L, et al. TTBK2 kinase substrate specificity and the impact of spinocerebellar-ataxia-causing mutations on expression, activity, localization and development. Biochem J. 2011;437(1):157–167. 10.1042/BJ20110276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe T, Kakeno M, Matsui T, et al. TTBK2 with EB1/3 regulates microtubule dynamics in migrating cells through KIF2A phosphorylation. J Cell Biol. 2015;210(5):737–751. 10.1083/jcb.201412075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96(1):69–78. 10.1016/s0092-8674(00)80960-5 [DOI] [PubMed]
- 26.Miyamoto T, Hosoba K, Ochiai H, et al. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Rep. 2015;10(5):664–673. 10.1016/j.celrep.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binó L, Mikulenková E, Štepánek L, et al. A protocol for generation and live-cell imaging analysis of primary cilia reporter cell lines. STAR Protoc. 2022;3(1):101199. Published 2022 Mar 2. 10.1016/j.xpro.2022.101199 [DOI] [PMC free article] [PubMed]
- 28.Lauring MC, Zhu T, Luo W, Wu W, Yu F, Toomre D. New software for automated cilia detection in cells (ACDC). Cilia. 2019;8:1. Published 2019 Aug 1. 10.1186/s13630-019-0061-z [DOI] [PMC free article] [PubMed]
- 29.Hansen JN, Rassmann S, Stüven B, Jurisch-Yaksi N, Wachten D. CiliaQ: a simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur Phys J E Soft Matter. 2021;44(2):18. Published 2021 Mar 8. 10.1140/epje/s10189-021-00031-y [DOI] [PMC free article] [PubMed]
- 30.Koulouras G, Panagopoulos A, Rapsomaniki MA, Giakoumakis NN, Taraviras S, Lygerou Z. EasyFRAP-web: a web-based tool for the analysis of fluorescence recovery after photobleaching data. Nucleic Acids Res. 2018;46(W1):W467–W472. 10.1093/nar/gky508 [DOI] [PMC free article] [PubMed]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed]
- 32.Daly OM, Gaboriau D, Karakaya K, et al. CEP164-null cells generated by genome editing show a ciliation defect with intact DNA repair capacity. J Cell Sci. 2016;129(9):1769–1774. 10.1242/jcs.186221 [DOI] [PubMed]
- 33.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15(2):363–377. 10.1083/jcb.15.2.363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu CT, Chen HY, Tang TK. Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat Cell Biol. 2018;20(2):175–185. 10.1038/s41556-017-0018-7 [DOI] [PubMed] [Google Scholar]
- 35.Stuck MW, Chong WM, Liao JC, Pazour GJ. Rab34 is necessary for early stages of intracellular ciliogenesis. Curr Biol. 2021;31(13):2887–2894.e4. 10.1016/j.cub.2021.04.018 [DOI] [PMC free article] [PubMed]
- 36.Ganga AK, Kennedy MC, Oguchi ME, et al. Rab34 GTPase mediates ciliary membrane formation in the intracellular ciliogenesis pathway. Curr Biol. 2021;31(13):2895–2905.e7. 10.1016/j.cub.2021.04.075 [DOI] [PMC free article] [PubMed]
- 37.Larkins CE, Aviles GD, East MP, Kahn RA, Caspary T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. Mol Biol Cell. 2011;22(23):4694–4703. 10.1091/mbc.E10-12-0994 [DOI] [PMC free article] [PubMed]
- 38.Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. 10.1146/annurev-cellbio-101512-122345 [DOI] [PubMed] [Google Scholar]
- 39.Varadi M, Anyango S, Deshpande M, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50(D1):D439–D444. 10.1093/nar/gkab1061 [DOI] [PMC free article] [PubMed]
- 40.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed]
- 41.Oda T, Chiba S, Nagai T, Mizuno K. Binding to Cep164, but not EB1, is essential for centriolar localization of TTBK2 and its function in ciliogenesis. Genes Cells. 2014;19(12):927–940. 10.1111/gtc.12191 [DOI] [PubMed]
- 42.Rosa E Silva I, Binó L, Johnson CM, et al. Molecular mechanisms underlying the role of the centriolar CEP164-TTBK2 complex in ciliopathies. Structure. 2022;30(1):114–128.e9. 10.1016/j.str.2021.08.007 [DOI] [PMC free article] [PubMed]
- 43.Yang TT, Chong WM, Wang WJ, et al. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat Commun. 2018;9(1):2023. Published 2018 May 22. 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed]
- 44.Bowler M, Kong D, Sun S, et al. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat Commun. 2019;10(1):993. Published 2019 Mar 1. 10.1038/s41467-018-08216-4 [DOI] [PMC free article] [PubMed]
- 45.Hall NA, Hehnly H. A centriole's subdistal appendages: contributions to cell division, ciliogenesis and differentiation. Open Biol. 2021;11(2):200399. 10.1098/rsob.200399 [DOI] [PMC free article] [PubMed]
- 46.Mazo G, Soplop N, Wang WJ, Uryu K, Tsou MF. Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev Cell. 2016;39(4):424–437. 10.1016/j.devcel.2016.10.006 [DOI] [PMC free article] [PubMed]
- 47.Trofimova D, Paydar M, Zara A, Talje L, Kwok BH, Allingham JS. Ternary complex of Kif2A-bound tandem tubulin heterodimers represents a kinesin-13-mediated microtubule depolymerization reaction intermediate. Nat Commun. 2018;9(1):2628. Published 2018 Jul 6. 10.1038/s41467-018-05025-7 [DOI] [PMC free article] [PubMed]
- 48.Wang W, Shen T, Guerois R, et al. New Insights into the Coupling between Microtubule Depolymerization and ATP Hydrolysis by Kinesin-13 Protein Kif2C. J Biol Chem. 2015;290(30):18721–18731. 10.1074/jbc.M115.646919 [DOI] [PMC free article] [PubMed]
- 49.Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17(1):77–82. 10.1038/nsmb.1728 [DOI] [PMC free article] [PubMed]
- 50.Binó L, Čajánek L. Tau tubulin kinase 1 and 2 regulate ciliogenesis and human pluripotent stem cells-derived neural rosettes. Sci Rep. 2023;13(1):12884. Published 2023 Aug 9. 10.1038/s41598-023-39887-9 [DOI] [PMC free article] [PubMed]
- 51.Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol. 2012;199(7):1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang N, Zhang D, Li F, et al. M-Phase Phosphoprotein 9 regulates ciliogenesis by modulating CP110-CEP97 complex localization at the mother centriole. Nat Commun. 2018;9(1):4511. Published 2018 Oct 30. 10.1038/s41467-018-06990-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Q, Insinna C, Ott C, et al. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat Cell Biol. 2015;17(4):531. 10.1038/ncb3155 [DOI] [PubMed] [Google Scholar]
- 54.Walia V, Cuenca A, Vetter M, et al. Akt Regulates a Rab11-Effector Switch Required for Ciliogenesis. Dev Cell. 2019;50(2):229–246.e7. 10.1016/j.devcel.2019.05.022 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.