Abstract
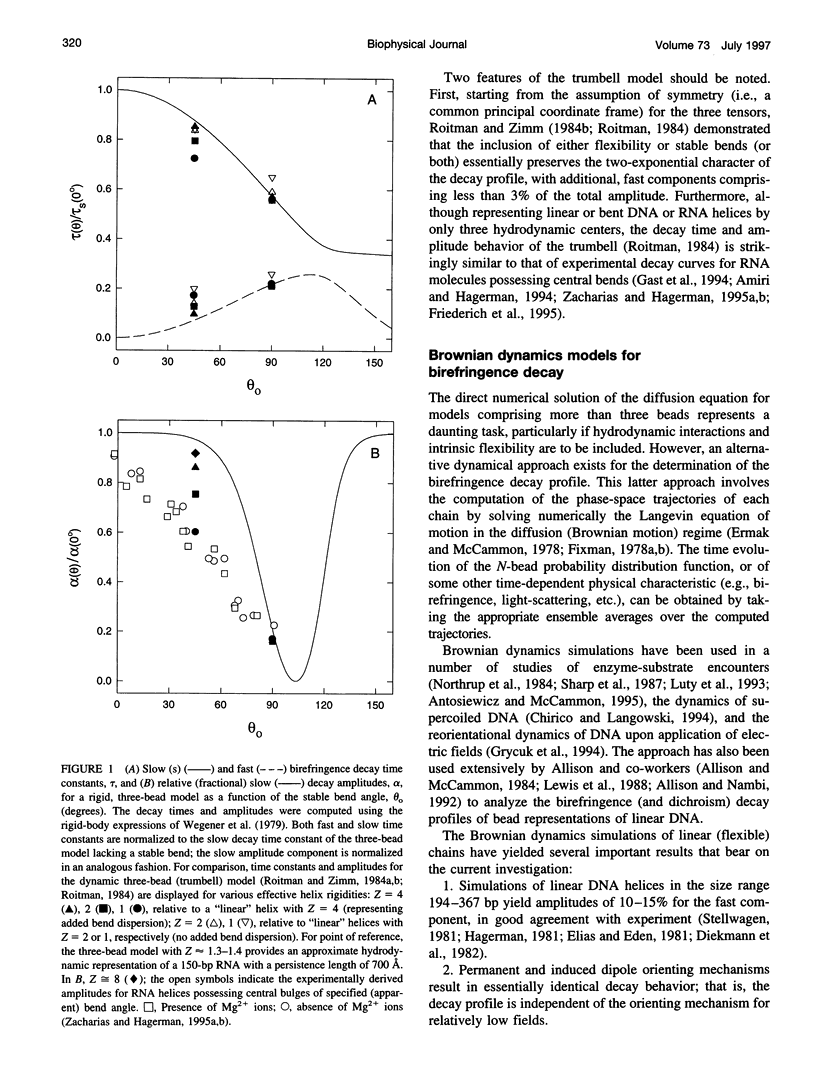
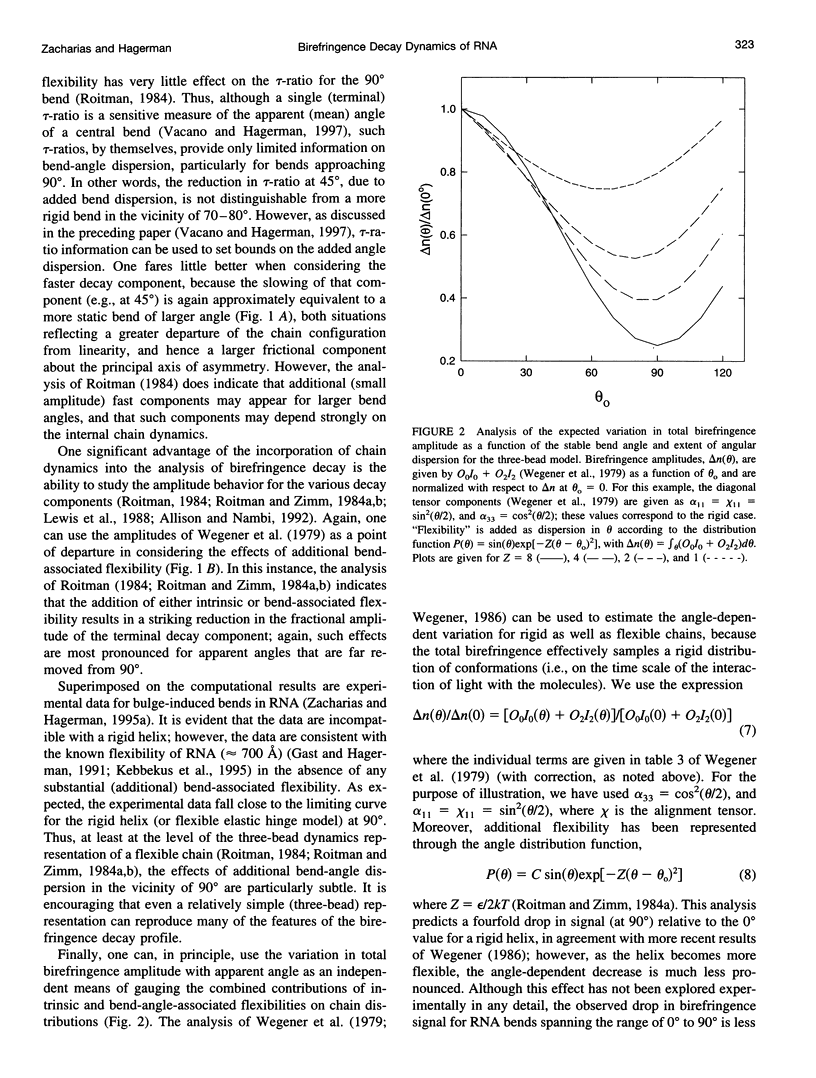
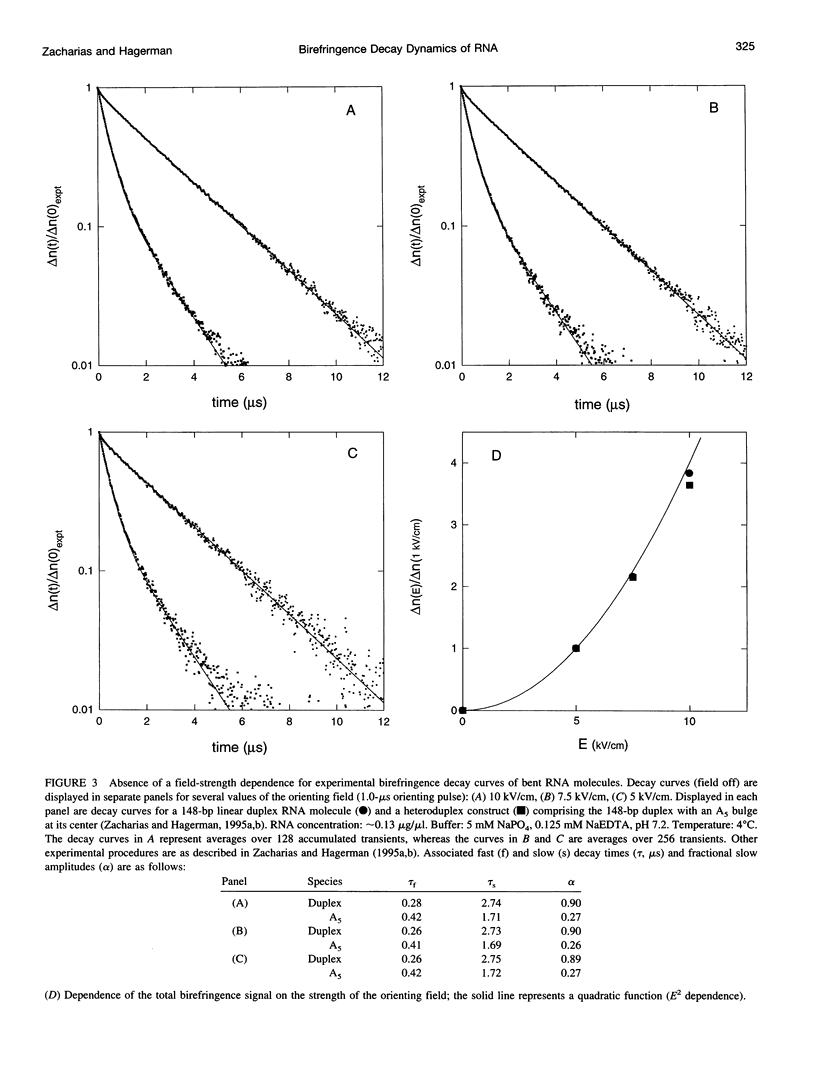
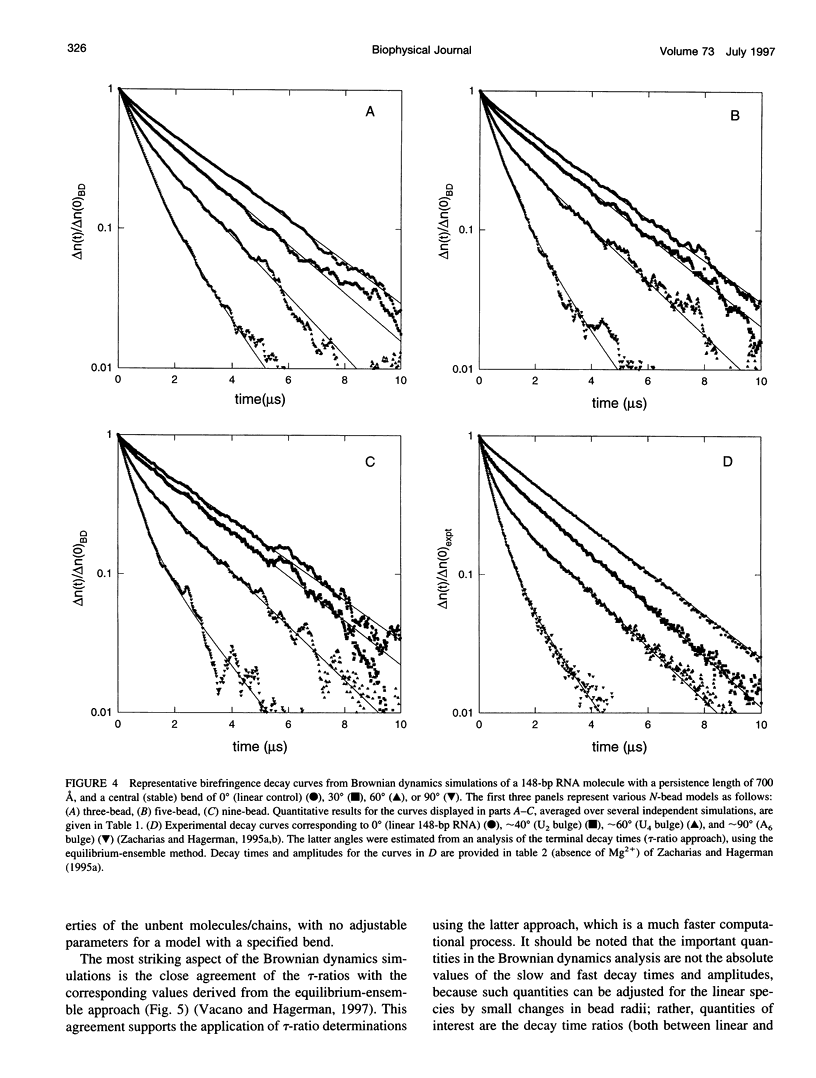
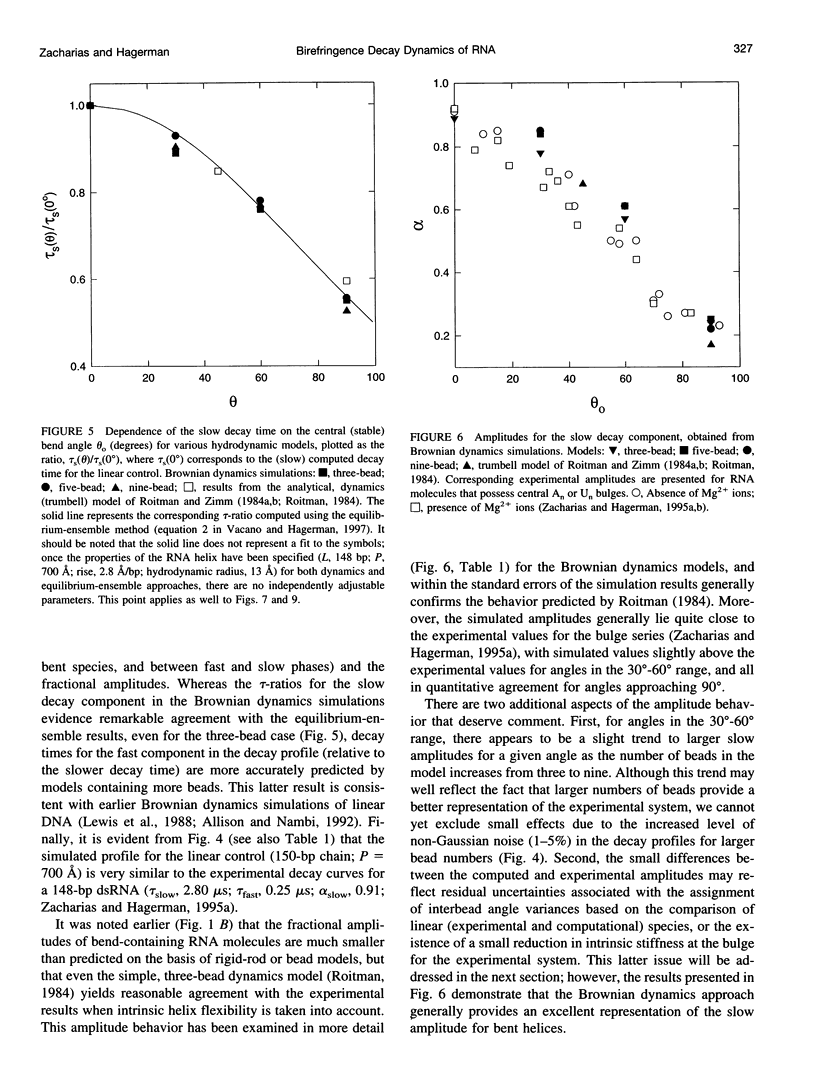
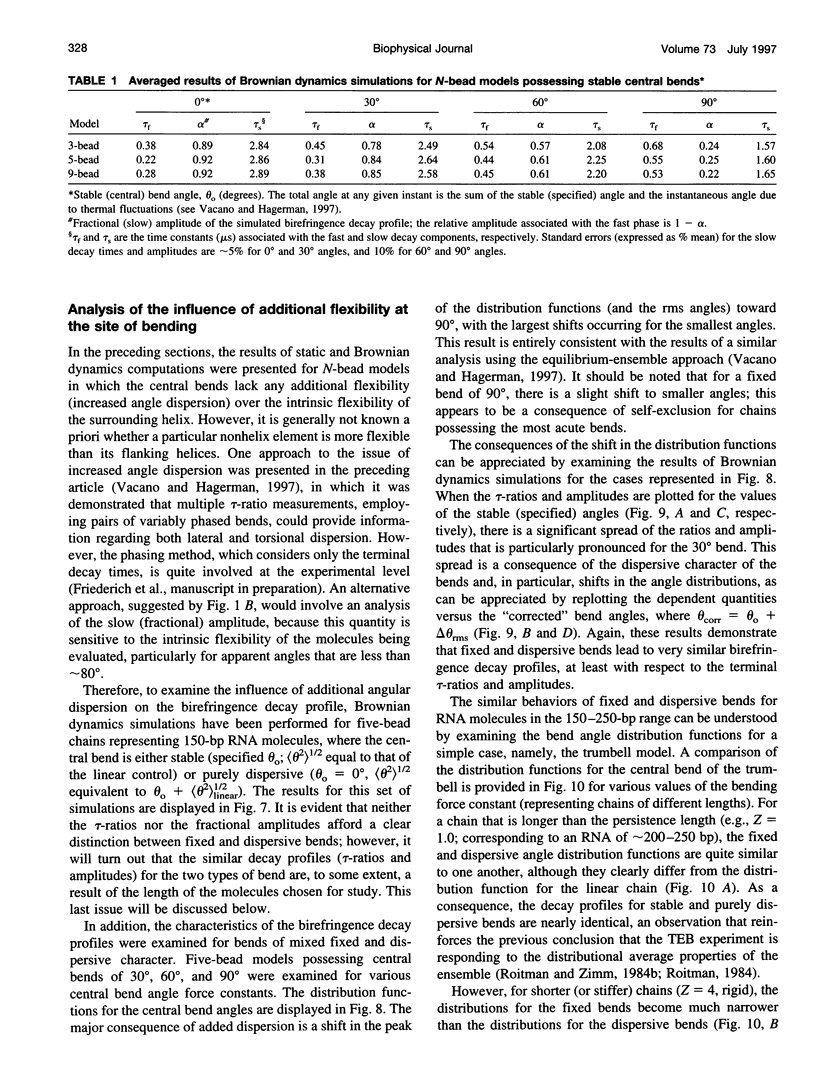
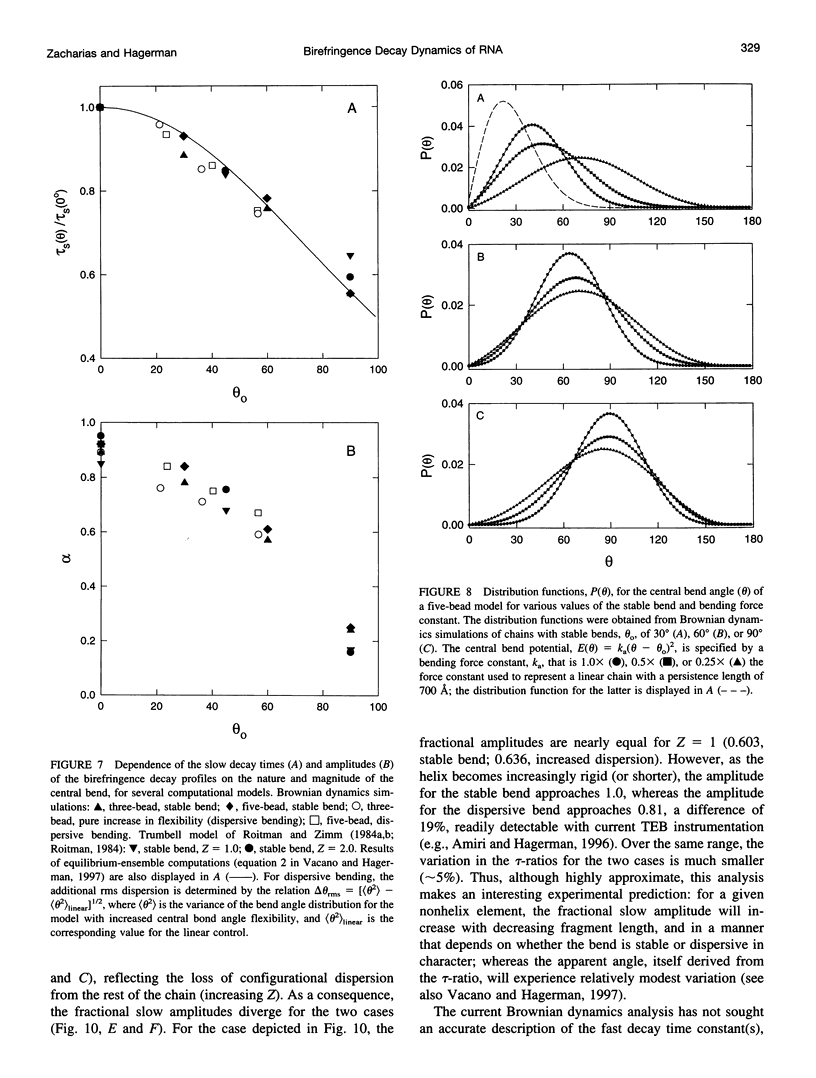
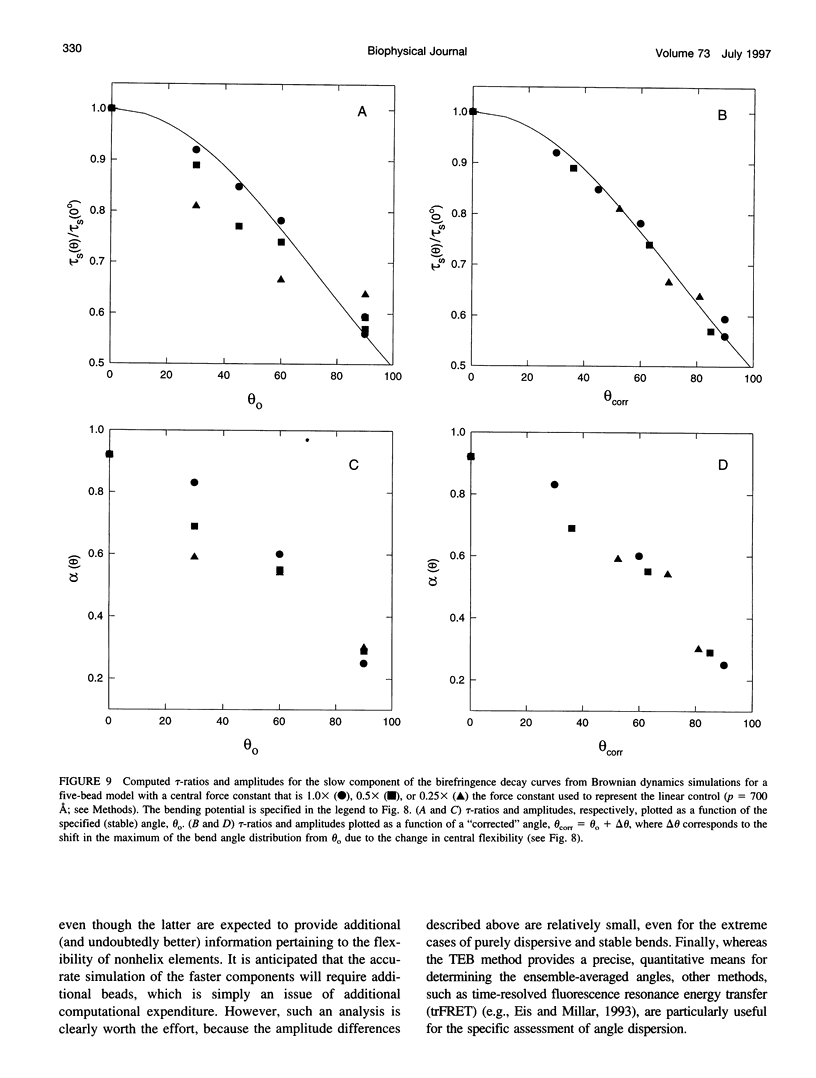
Bends in nucleic acid helices can be quantified in a transient electric birefringence (TEB) experiment from the ratio of the terminal decay times of the bent molecule and its fully duplex counterpart (tau-ratio method). The apparent bend angles can be extracted from the experimental tau-ratios through the application of static (equilibrium-ensemble) hydrodynamic models; however, such models do not properly address the faster component(s) of the birefringence decay profile, which can represent up to 80% of the total birefringence signal for large band angles. To address this latter issue, the relative amplitudes of the components in the birefringence decay profile have been analyzed through a series of Brownian dynamics (BD) simulations. Decay profiles have been simulated for three-, five-, and nine-bead models representing RNA molecules with central bends of 30 degrees, 60 degrees, and 90 degrees, and with various degrees of associated angle dispersion. The BD simulations are in close agreement with experimental results for the fractional amplitudes, suggesting that both amplitudes and terminal tau-ratios can be used as a measure of the magnitudes of bends in the helix axis. Although the current results indicate that it is generally not possible to distinguish between relatively fixed and highly flexible bends from single tau-ratio measurements, because they can lead to similar reductions in terminal decay time and amplitude, measurements of the dependence of the fractional amplitudes on helix length may afford such a distinction.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amiri K. M., Hagerman P. J. Global conformation of a self-cleaving hammerhead RNA. Biochemistry. 1994 Nov 15;33(45):13172–13177. doi: 10.1021/bi00249a003. [DOI] [PubMed] [Google Scholar]
- Amiri K. M., Hagerman P. J. The global conformation of an active hammerhead RNA during the process of self-cleavage. J Mol Biol. 1996 Aug 16;261(2):125–134. doi: 10.1006/jmbi.1996.0446. [DOI] [PubMed] [Google Scholar]
- Antosiewicz J., McCammon J. A. Electrostatic and hydrodynamic orientational steering effects in enzyme-substrate association. Biophys J. 1995 Jul;69(1):57–65. doi: 10.1016/S0006-3495(95)79874-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Hillen W., Morgeneyer B., Wells R. D., Pörschke D. Orientation relaxation of DNA restriction fragments and the internal mobility of the double helix. Biophys Chem. 1982 Jul;15(4):263–270. doi: 10.1016/0301-4622(82)80009-4. [DOI] [PubMed] [Google Scholar]
- Eis P. S., Millar D. P. Conformational distributions of a four-way DNA junction revealed by time-resolved fluorescence resonance energy transfer. Biochemistry. 1993 Dec 21;32(50):13852–13860. doi: 10.1021/bi00213a014. [DOI] [PubMed] [Google Scholar]
- Friederich M. W., Gast F. U., Vacano E., Hagerman P. J. Determination of the angle between the anticodon and aminoacyl acceptor stems of yeast phenylalanyl tRNA in solution. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4803–4807. doi: 10.1073/pnas.92.11.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gast F. U., Amiri K. M., Hagerman P. J. Interhelix geometry of stems I and II of a self-cleaving hammerhead RNA. Biochemistry. 1994 Feb 22;33(7):1788–1796. doi: 10.1021/bi00173a023. [DOI] [PubMed] [Google Scholar]
- Gast F. U., Hagerman P. J. Electrophoretic and hydrodynamic properties of duplex ribonucleic acid molecules transcribed in vitro: evidence that A-tracts do not generate curvature in RNA. Biochemistry. 1991 Apr 30;30(17):4268–4277. doi: 10.1021/bi00231a024. [DOI] [PubMed] [Google Scholar]
- Hagerman K. R., Hagerman P. J. Helix rigidity of DNA: the meroduplex as an experimental paradigm. J Mol Biol. 1996 Jul 12;260(2):207–223. doi: 10.1006/jmbi.1996.0393. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Investigation of the flexibility of DNA using transient electric birefringence. Biopolymers. 1981 Jul;20(7):1503–1535. doi: 10.1002/bip.1981.360200710. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sometimes a great motion: the application of transient electric birefringence to the study of macromolecular structure. Curr Opin Struct Biol. 1996 Oct;6(5):643–649. doi: 10.1016/s0959-440x(96)80031-5. [DOI] [PubMed] [Google Scholar]
- Kebbekus P., Draper D. E., Hagerman P. Persistence length of RNA. Biochemistry. 1995 Apr 4;34(13):4354–4357. doi: 10.1021/bi00013a026. [DOI] [PubMed] [Google Scholar]
- Mellado P., Iniesta A., Diaz F. G., García de la Torre J. Diffusion coefficients of segmentally flexible macromolecules with two subunits: a study of broken rods. Biopolymers. 1988 Nov;27(11):1771–1786. doi: 10.1002/bip.360271107. [DOI] [PubMed] [Google Scholar]
- Sharp K., Fine R., Honig B. Computer simulations of the diffusion of a substrate to an active site of an enzyme. Science. 1987 Jun 12;236(4807):1460–1463. doi: 10.1126/science.3589666. [DOI] [PubMed] [Google Scholar]
- Shen Z., Hagerman P. J. Conformation of the central, three-helix junction of the 5 S ribosomal RNA of Sulfolobus acidocaldarius. J Mol Biol. 1994 Aug 19;241(3):415–430. doi: 10.1006/jmbi.1994.1517. [DOI] [PubMed] [Google Scholar]
- Stellwagen N. C. Electric birefringence of restriction enzyme fragments of DNA: optical factor and electric polarizability as a function of molecular weight. Biopolymers. 1981 Mar;20(3):399–434. doi: 10.1002/bip.1981.360200302. [DOI] [PubMed] [Google Scholar]
- Vacano E., Hagerman P. J. Analysis of birefringence decay profiles for nucleic acid helices possessing bends: the tau-ratio approach. Biophys J. 1997 Jul;73(1):306–317. doi: 10.1016/S0006-3495(97)78071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P. J. Bulge-induced bends in RNA: quantification by transient electric birefringence. J Mol Biol. 1995 Mar 31;247(3):486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P. J. The bend in RNA created by the trans-activation response element bulge of human immunodeficiency virus is straightened by arginine and by Tat-derived peptide. Proc Natl Acad Sci U S A. 1995 Jun 20;92(13):6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Hagerman P. J. The influence of symmetric internal loops on the flexibility of RNA. J Mol Biol. 1996 Mar 29;257(2):276–289. doi: 10.1006/jmbi.1996.0162. [DOI] [PubMed] [Google Scholar]


