Abstract
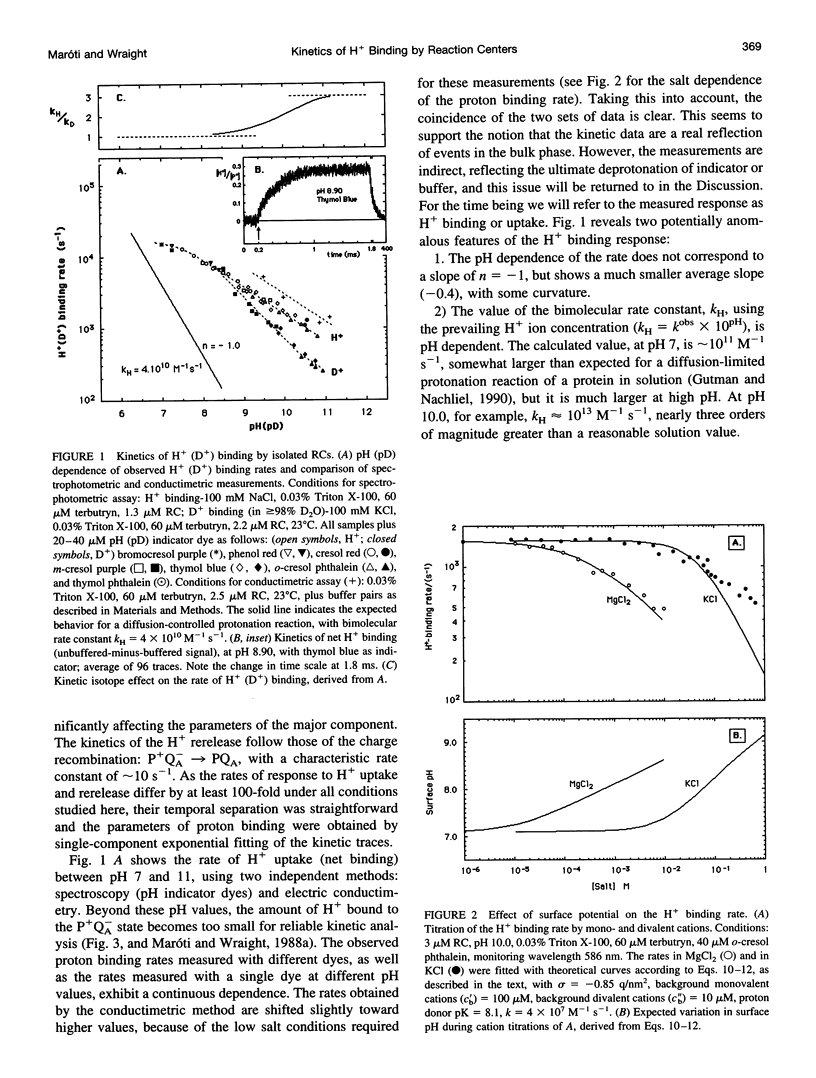
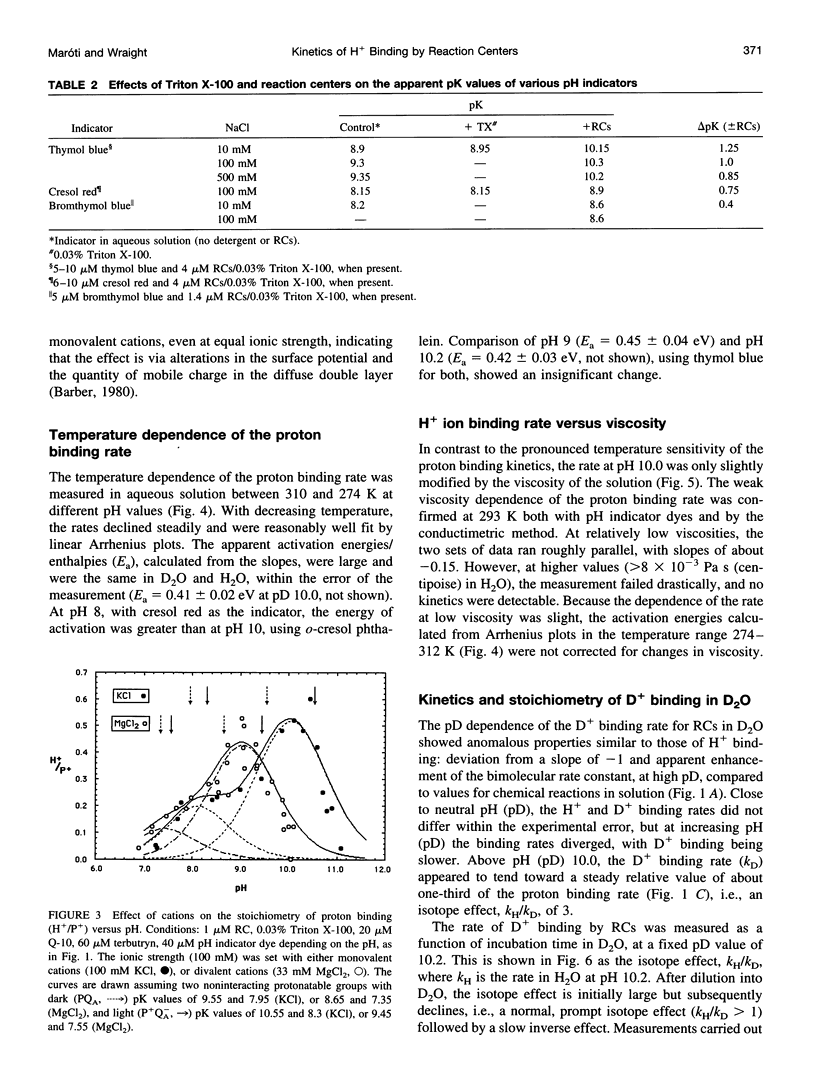
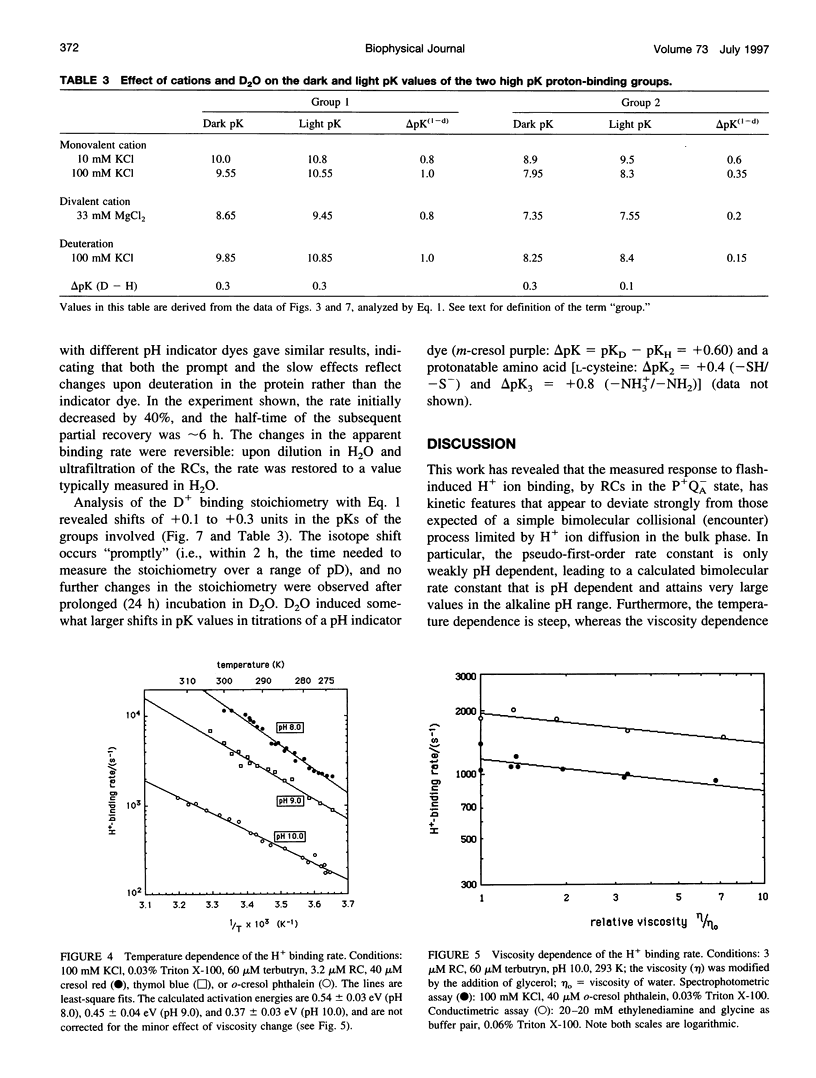
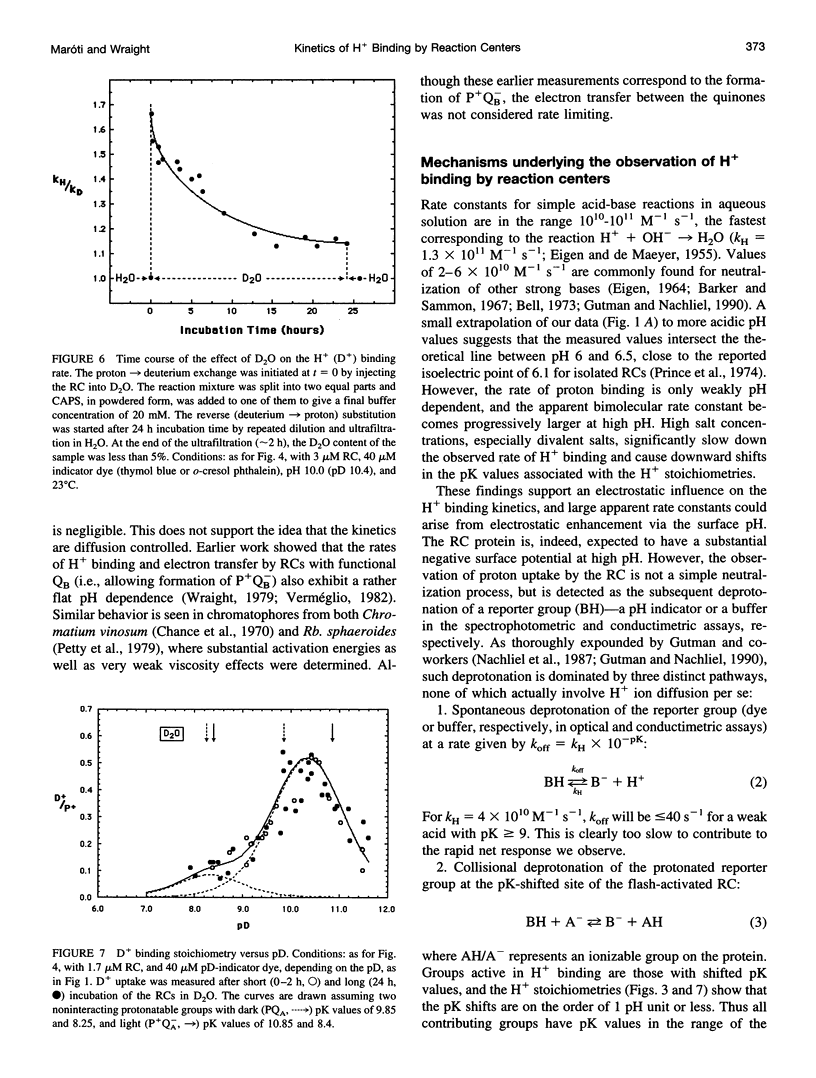
The kinetics of flash-induced H+ ion binding by isolated reaction centers (RCs) of Rhodobacter sphaeroides, strain R-26, were measured, using pH indicators and conductimetry, in the presence of terbutryn to block electron transfer between the primary and secondary quinones (QA and QB), and in the absence of exogenous electron donors to the oxidized primary donor, P+, i.e., the P+QA-state. Under these conditions, proton binding by RCs is to the protein rather than to any of the cofactors. After light activation to form P+QA-, the kinetics of proton binding were monoexponential at all pH values studied. At neutral pH, the apparent bimolecular rate constant was close to the diffusional limit for proton transfer in aqueous solution (approximately 10(11) M-1 s-1), but increased significantly in the alkaline pH range (e.g., 2 x 10(13) M-1 s-1 at pH 10). The average slope of the pH dependence was -0.4 instead of -1.0, as might be expected for a H+ diffusion-controlled process. High activation energy (0.54 eV at pH 8.0) and weak viscosity dependence showed that H+ ion uptake by RCs is not limited by diffusion. The salt dependence of the H+ ion binding rate and the pK values of the protonatable amino acid residues of the reaction center implicated surface charge influences, and Gouy-Chapman theory provided a workable description of the ionic effects as arising from modulation of the pH at the surface of the RC. Incubation in D2O caused small increases in the pKs of the protonatable groups and a small, pH (pD)-dependent slowing of the binding rate. The salt, pH, temperature, viscosity, and D2O dependences of the proton uptake by RCs in the P+QA- state were accounted for by three considerations: 1) parallel pathways of H+ delivery to the RC, contributing to the observed (net) H+ disappearance; 2) rate limitation of the protonation of target groups within the protein by conformational dynamics; and 3) electrostatic influences of charged groups in the protein, via the surface pH.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barber J. Membrane surface charges and potentials in relation to photosynthesis. Biochim Biophys Acta. 1980 Dec;594(4):253–308. doi: 10.1016/0304-4173(80)90003-8. [DOI] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Electrostatic calculations of amino acid titration and electron transfer, Q-AQB-->QAQ-B, in the reaction center. Biophys J. 1995 Jun;68(6):2233–2250. doi: 10.1016/S0006-3495(95)80406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Protonation of interacting residues in a protein by a Monte Carlo method: application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B., Crofts A. R., Nishimura M., Price B. Fast membrane H+ binding in the light-activated state of Chromatium chromatophores. Eur J Biochem. 1970 Apr;13(2):364–374. doi: 10.1111/j.1432-1033.1970.tb00938.x. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Gekko K., Timasheff S. N. Thermodynamic and kinetic examination of protein stabilization by glycerol. Biochemistry. 1981 Aug 4;20(16):4677–4686. doi: 10.1021/bi00519a024. [DOI] [PubMed] [Google Scholar]
- Gutman M., Huppert D., Pines E., Nachliel E. Probing the micelle/water interface by a rapid laser-induced proton pulse. Biochim Biophys Acta. 1981 Mar 20;642(1):15–26. doi: 10.1016/0005-2736(81)90133-4. [DOI] [PubMed] [Google Scholar]
- Gutman M., Nachliel E., Gershon E., Giniger R. Kinetic analysis of the protonation of a surface group of a macromolecule. Eur J Biochem. 1983 Jul 15;134(1):63–69. doi: 10.1111/j.1432-1033.1983.tb07531.x. [DOI] [PubMed] [Google Scholar]
- Heberle J., Dencher N. A. Surface-bound optical probes monitor protein translocation and surface potential changes during the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5996–6000. doi: 10.1073/pnas.89.13.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D., Okamura M. Y., Feher G. Electron transfer in reaction centers of Rhodopseudomonas sphaeroides. I. Determination of the charge recombination pathway of D+QAQ(-)B and free energy and kinetic relations between Q(-)AQB and QAQ(-)B. Biochim Biophys Acta. 1984 Jul 27;766(1):126–140. doi: 10.1016/0005-2728(84)90224-x. [DOI] [PubMed] [Google Scholar]
- Klinman J. P. Kinetic isotope effects in enzymology. Adv Enzymol Relat Areas Mol Biol. 1978;46:415–494. doi: 10.1002/9780470122914.ch7. [DOI] [PubMed] [Google Scholar]
- Lancaster C. R., Michel H., Honig B., Gunner M. R. Calculated coupling of electron and proton transfer in the photosynthetic reaction center of Rhodopseudomonas viridis. Biophys J. 1996 Jun;70(6):2469–2492. doi: 10.1016/S0006-3495(96)79820-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maróti P., Hanson D. K., Schiffer M., Sebban P. Long-range electrostatic interaction in the bacterial photosynthetic reaction centre. Nat Struct Biol. 1995 Dec;2(12):1057–1059. doi: 10.1038/nsb1295-1057. [DOI] [PubMed] [Google Scholar]
- McLaughlin S. The electrostatic properties of membranes. Annu Rev Biophys Biophys Chem. 1989;18:113–136. doi: 10.1146/annurev.bb.18.060189.000553. [DOI] [PubMed] [Google Scholar]
- Miksovska J., Maróti P., Tandori J., Schiffer M., Hanson D. K., Sebban P. Distant electrostatic interactions modulate the free energy level of QA- in the photosynthetic reaction center. Biochemistry. 1996 Dec 3;35(48):15411–15417. doi: 10.1021/bi961299u. [DOI] [PubMed] [Google Scholar]
- Nachliel E., Gutman M., Kiryati S., Dencher N. A. Protonation dynamics of the extracellular and cytoplasmic surface of bacteriorhodopsin in the purple membrane. Proc Natl Acad Sci U S A. 1996 Oct 1;93(20):10747–10752. doi: 10.1073/pnas.93.20.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M. Y., Feher G. Isotope effect on electron transfer in reaction centers from Rhodopseudomonas sphaeroides. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8152–8156. doi: 10.1073/pnas.83.21.8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M. Y., Feher G. Proton transfer in reaction centers from photosynthetic bacteria. Annu Rev Biochem. 1992;61:861–896. doi: 10.1146/annurev.bi.61.070192.004241. [DOI] [PubMed] [Google Scholar]
- Paddock M. L., McPherson P. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of serine-L223 by alanine inhibits electron and proton transfers associated with reduction of quinone to dihydroquinone. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6803–6807. doi: 10.1073/pnas.87.17.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock M. L., Rongey S. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of glutamic acid 212 in the L subunit by glutamine inhibits quinone (secondary acceptor) turnover. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6602–6606. doi: 10.1073/pnas.86.17.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty K. M., Dutton P. L. Properties of the flash-induced proton binding encountered in membranes of Rhodopseudomonas sphaeroides: a functional pK on the ubisemiquinone? Arch Biochem Biophys. 1976 Feb;172(2):335–345. doi: 10.1016/0003-9861(76)90085-0. [DOI] [PubMed] [Google Scholar]
- Petty K., Jackson J. B., Dutton P. L. Factors controlling the binding of two protons per electron transferred through the ubiquinone and cytochrome b/c2 segment of Rhodopseudomonas sphaeroides chromatophores. Biochim Biophys Acta. 1979 Apr 11;546(1):17–42. doi: 10.1016/0005-2728(79)90167-1. [DOI] [PubMed] [Google Scholar]
- Prince R. C., Cogdell R. J., Crofts A. R. The photo-oxidation of horse heart cytochrome c and native cytochrome c2 by reaction centres from Rhodopseudomonas spheroides R26. Biochim Biophys Acta. 1974 Apr 23;347(1):1–13. doi: 10.1016/0005-2728(74)90194-7. [DOI] [PubMed] [Google Scholar]
- Schowen K. B., Schowen R. L. Solvent isotope effects of enzyme systems. Methods Enzymol. 1982;87:551–606. [PubMed] [Google Scholar]
- Sebban P., Maróti P., Hanson D. K. Electron and proton transfer to the quinones in bacterial photosynthetic reaction centers: insight from combined approaches of molecular genetics and biophysics. Biochimie. 1995;77(7-8):677–694. doi: 10.1016/0300-9084(96)88183-1. [DOI] [PubMed] [Google Scholar]
- Shinkarev V. P., Wraight C. A. The interaction of quinone and detergent with reaction centers of purple bacteria. I. Slow quinone exchange between reaction center micelles and pure detergent micelles. Biophys J. 1997 May;72(5):2304–2319. doi: 10.1016/S0006-3495(97)78875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R. R., Castellvi A. L., Bogacz J. P., Wraight C. A. Herbicide-quinone competition in the acceptor complex of photosynthetic reaction centers from Rhodopseudomonas sphaeroides: a bacterial model for PS-II-herbicide activity in plants. J Cell Biochem. 1984;24(3):243–259. doi: 10.1002/jcb.240240306. [DOI] [PubMed] [Google Scholar]
- Stowell M. H., McPhillips T. M., Rees D. C., Soltis S. M., Abresch E., Feher G. Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer. Science. 1997 May 2;276(5313):812–816. doi: 10.1126/science.276.5313.812. [DOI] [PubMed] [Google Scholar]
- Takahashi E., Wraight C. A. Proton and electron transfer in the acceptor quinone complex of Rhodobacter sphaeroides reaction centers: characterization of site-directed mutants of the two ionizable residues, GluL212 and AspL213, in the QB binding site. Biochemistry. 1992 Jan 28;31(3):855–866. doi: 10.1021/bi00118a031. [DOI] [PubMed] [Google Scholar]
- Wraight C. A. Electron acceptors of bacterial photosynthetic reaction centers. II. H+ binding coupled to secondary electron transfer in the quinone acceptor complex. Biochim Biophys Acta. 1979 Nov 8;548(2):309–327. doi: 10.1016/0005-2728(79)90138-5. [DOI] [PubMed] [Google Scholar]
- Zhu Z. Y., Karlin S. Clusters of charged residues in protein three-dimensional structures. Proc Natl Acad Sci U S A. 1996 Aug 6;93(16):8350–8355. doi: 10.1073/pnas.93.16.8350. [DOI] [PMC free article] [PubMed] [Google Scholar]


