Abstract
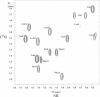
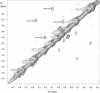
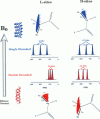
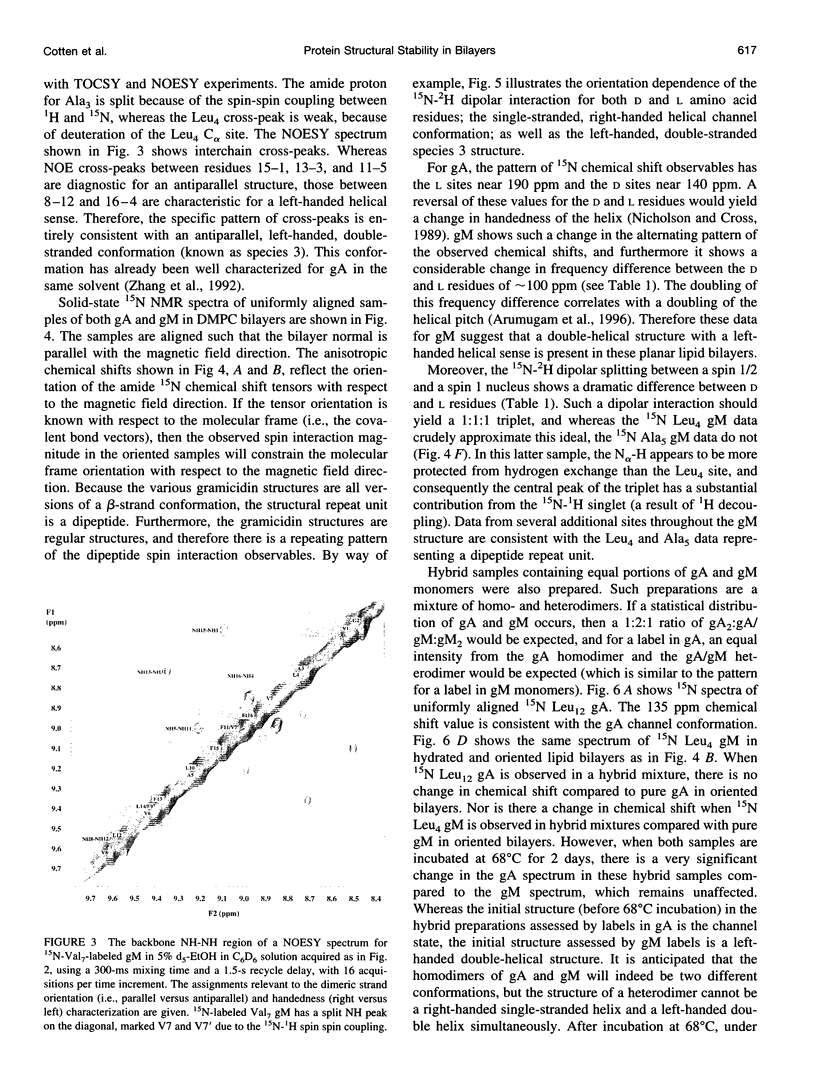
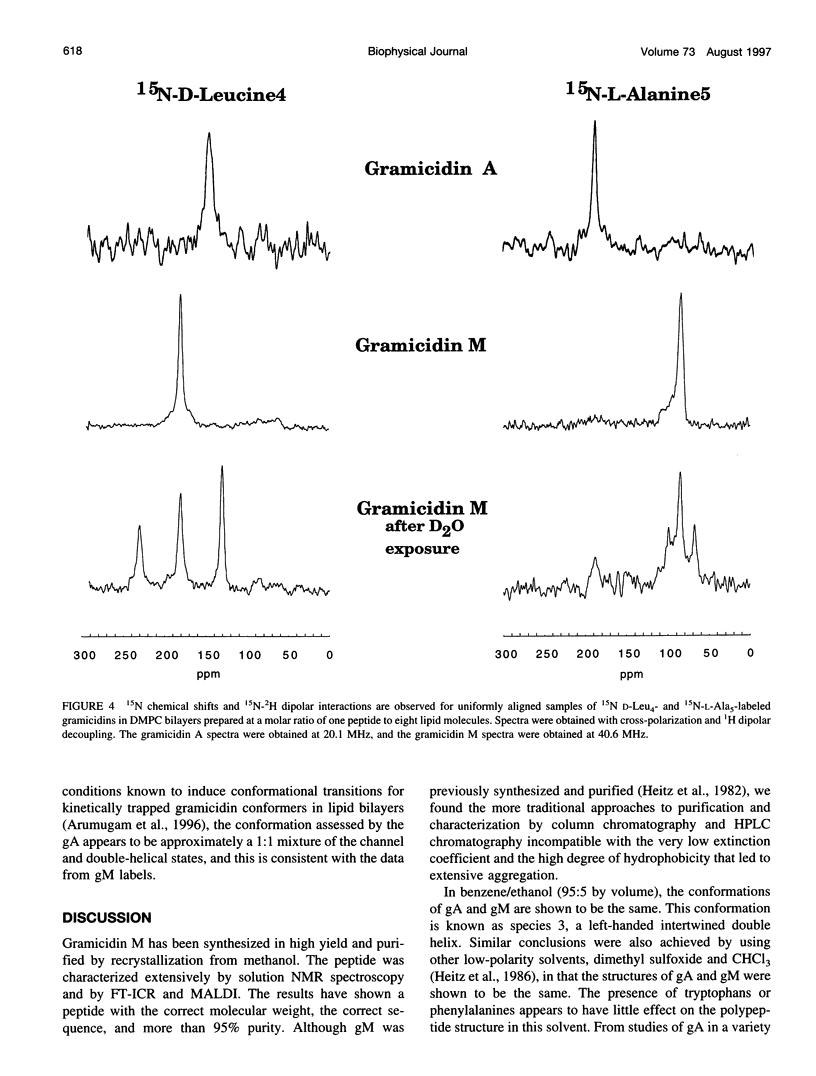
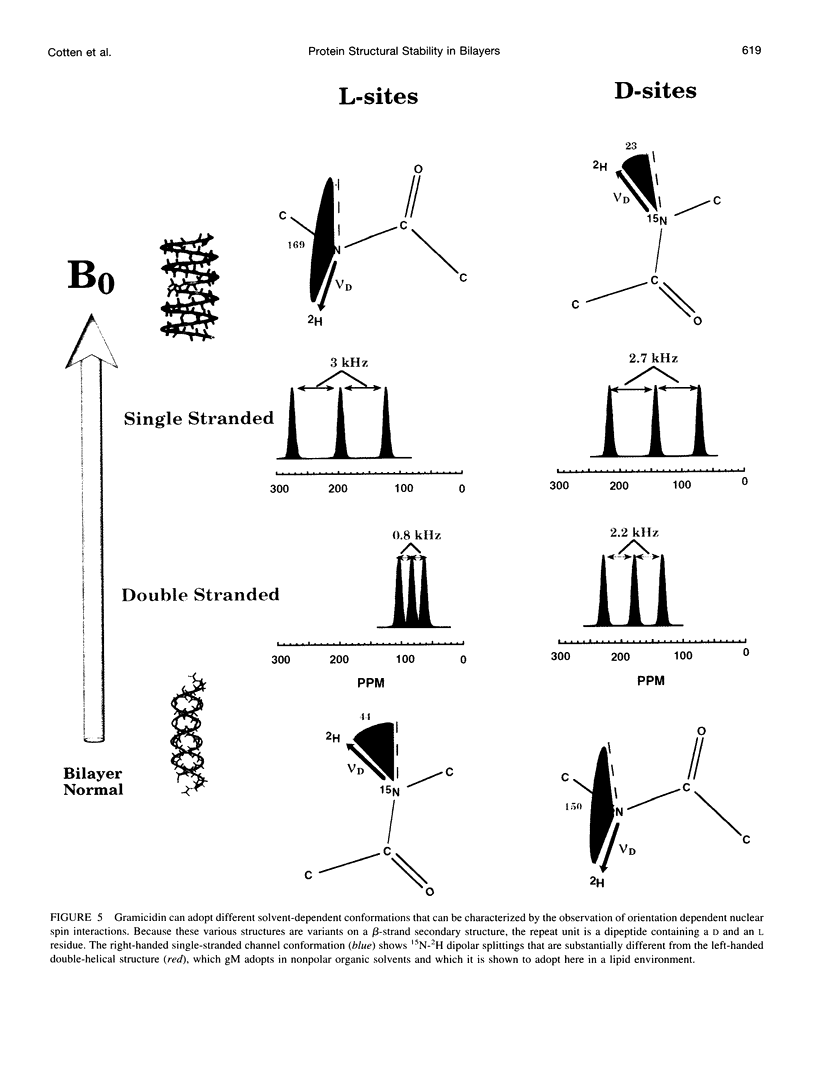
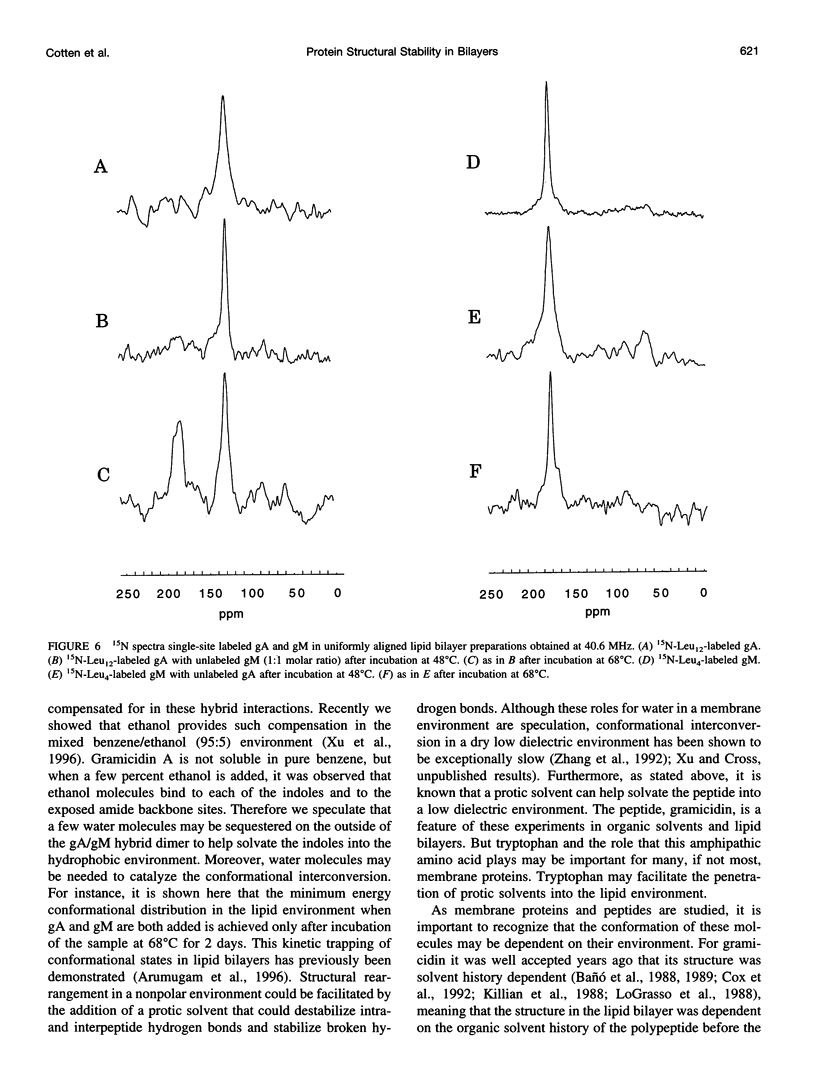
The replacement of four tryptophans in gramicidin A by four phenylalanines (gramicidin M) causes no change in the molecular fold of this dimeric peptide in a low dielectric isotropic organic solvent, but the molecular folds are dramatically different in a lipid bilayer environment. The indoles of gramicidin A interact with the anisotropic bilayer environment to induce a change in the molecular fold. The double-helical fold of gramicidin M, as opposed to the single-stranded structure of gramicidin A, is not compatible with ion conductance. Gramicidin A/gramicidin M hybrid structures have also been prepared, and like gramicidin M homodimers, these dimeric hybrids appear to have a double-helical fold, suggesting that a couple of indoles are being buried in the bilayer interstices. To achieve this equilibrium structure (i.e., minimum energy conformation), incubation at 68 degrees C for 2 days is required. Kinetically trapped metastable structures may be more common in lipid bilayers than in an aqueous isotropic environment. Structural characterizations in the bilayers were achieved with solid-state NMR-derived orientational constraints from uniformly aligned lipid bilayer samples, and characterizations in organic solvents were accomplished by solution NMR.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abdul-Manan N., Hinton J. F. Conformation states of gramicidin A along the pathway to the formation of channels in model membranes determined by 2D NMR and circular dichroism spectroscopy. Biochemistry. 1994 Jun 7;33(22):6773–6783. doi: 10.1021/bi00188a005. [DOI] [PubMed] [Google Scholar]
- Andersen O. S., Koeppe R. E., 2nd Molecular determinants of channel function. Physiol Rev. 1992 Oct;72(4 Suppl):S89–158. doi: 10.1152/physrev.1992.72.suppl_4.S89. [DOI] [PubMed] [Google Scholar]
- Arumugam S., Pascal S., North C. L., Hu W., Lee K. C., Cotten M., Ketchem R. R., Xu F., Brenneman M., Kovacs F. Conformational trapping in a membrane environment: a regulatory mechanism for protein activity? Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5872–5876. doi: 10.1073/pnas.93.12.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker D., Agard D. A. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994 Jun 21;33(24):7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- Baumgärtner A. Insertion and hairpin formation of membrane proteins: a Monte Carlo study. Biophys J. 1996 Sep;71(3):1248–1255. doi: 10.1016/S0006-3495(96)79324-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañ M. C., Braco L., Abad C. Conformational transitions of gramicidin A in phospholipid model membranes. A high-performance liquid chromatography assessment. Biochemistry. 1991 Jan 29;30(4):886–894. doi: 10.1021/bi00218a002. [DOI] [PubMed] [Google Scholar]
- Bañ M. C., Braco L., Abad C. HPLC study on the 'history' dependence of gramicidin A conformation in phospholipid model membranes. FEBS Lett. 1989 Jun 19;250(1):67–71. doi: 10.1016/0014-5793(89)80686-6. [DOI] [PubMed] [Google Scholar]
- Bañ M. C., Braco L., Abad C. New high-performance liquid chromatography-based methodology for monitoring the conformational transitions of self-associating hydrophobic peptides, incorporated into liposomes. J Chromatogr. 1988 Dec 23;458:105–116. doi: 10.1016/s0021-9673(00)90557-0. [DOI] [PubMed] [Google Scholar]
- Becker M. D., Greathouse D. V., Koeppe R. E., 2nd, Andersen O. S. Amino acid sequence modulation of gramicidin channel function: effects of tryptophan-to-phenylalanine substitutions on the single-channel conductance and duration. Biochemistry. 1991 Sep 10;30(36):8830–8839. doi: 10.1021/bi00100a015. [DOI] [PubMed] [Google Scholar]
- Cox K. J., Ho C., Lombardi J. V., Stubbs C. D. Gramicidin conformational studies with mixed-chain unsaturated phospholipid bilayer systems. Biochemistry. 1992 Feb 4;31(4):1112–1117. doi: 10.1021/bi00119a020. [DOI] [PubMed] [Google Scholar]
- Deber C. M., Goto N. K. Folding proteins into membranes. Nat Struct Biol. 1996 Oct;3(10):815–818. doi: 10.1038/nsb1096-815. [DOI] [PubMed] [Google Scholar]
- Durkin J. T., Providence L. L., Koeppe R. E., 2nd, Andersen O. S. Formation of non-beta 6.3-helical gramicidin channels between sequence-substituted gramicidin analogues. Biophys J. 1992 Apr;62(1):145–159. doi: 10.1016/S0006-3495(92)81801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Fields C. G., Fields G. B., Noble R. L., Cross T. A. Solid phase peptide synthesis of 15N-gramicidins A, B, and C and high performance liquid chromatographic purification. Int J Pept Protein Res. 1989 Apr;33(4):298–303. doi: 10.1111/j.1399-3011.1989.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Fields G. B., Fields C. G., Petefish J., Van Wart H. E., Cross T. A. Solid-phase peptide synthesis and solid-state NMR spectroscopy of [Ala3-15N][Val1]gramicidin A. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1384–1388. doi: 10.1073/pnas.85.5.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein A., Andersen O. S. The gramicidin A channel: a review of its permeability characteristics with special reference to the single-file aspect of transport. J Membr Biol. 1981 Apr 30;59(3):155–171. doi: 10.1007/BF01875422. [DOI] [PubMed] [Google Scholar]
- Fonseca V., Daumas P., Ranjalahy-Rasoloarijao L., Heitz F., Lazaro R., Trudelle Y., Andersen O. S. Gramicidin channels that have no tryptophan residues. Biochemistry. 1992 Jun 16;31(23):5340–5350. doi: 10.1021/bi00138a014. [DOI] [PubMed] [Google Scholar]
- Heitz F., Gavach C., Spach G., Trudelle Y. Analysis of the ion transfer through the channel of 9,11,13,15-phenylalanylgramicidin A. Biophys Chem. 1986 Jul;24(2):143–148. doi: 10.1016/0301-4622(86)80007-2. [DOI] [PubMed] [Google Scholar]
- Heitz F., Spach G., Trudelle Y. Single channels of 9, 11, 13, 15-destryptophyl-phenylalanyl-gramicidin A. Biophys J. 1982 Oct;40(1):87–89. doi: 10.1016/S0006-3495(82)84462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Cross T. A. Tryptophan hydrogen bonding and electric dipole moments: functional roles in the gramicidin channel and implications for membrane proteins. Biochemistry. 1995 Oct 31;34(43):14147–14155. doi: 10.1021/bi00043a020. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Killian J. A. Gramicidin and gramicidin-lipid interactions. Biochim Biophys Acta. 1992 Dec 11;1113(3-4):391–425. doi: 10.1016/0304-4157(92)90008-x. [DOI] [PubMed] [Google Scholar]
- Killian J. A., Prasad K. U., Hains D., Urry D. W. The membrane as an environment of minimal interconversion. A circular dichroism study on the solvent dependence of the conformational behavior of gramicidin in diacylphosphatidylcholine model membranes. Biochemistry. 1988 Jun 28;27(13):4848–4855. doi: 10.1021/bi00413a040. [DOI] [PubMed] [Google Scholar]
- Koeppe R. E., 2nd, Anderson O. S. Engineering the gramicidin channel. Annu Rev Biophys Biomol Struct. 1996;25:231–258. doi: 10.1146/annurev.bb.25.060196.001311. [DOI] [PubMed] [Google Scholar]
- Langs D. A., Smith G. D., Courseille C., Précigoux G., Hospital M. Monoclinic uncomplexed double-stranded, antiparallel, left-handed beta 5.6-helix (increases decreases beta 5.6) structure of gramicidin A: alternate patterns of helical association and deformation. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5345–5349. doi: 10.1073/pnas.88.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langs D. A. Three-dimensional structure at 0.86 A of the uncomplexed form of the transmembrane ion channel peptide gramicidin A. Science. 1988 Jul 8;241(4862):188–191. doi: 10.1126/science.2455345. [DOI] [PubMed] [Google Scholar]
- LoGrasso P. V., Moll F., 3rd, Cross T. A. Solvent history dependence of gramicidin A conformations in hydrated lipid bilayers. Biophys J. 1988 Aug;54(2):259–267. doi: 10.1016/S0006-3495(88)82955-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L. K., Cross T. A. Gramicidin cation channel: an experimental determination of the right-handed helix sense and verification of beta-type hydrogen bonding. Biochemistry. 1989 Nov 28;28(24):9379–9385. doi: 10.1021/bi00450a019. [DOI] [PubMed] [Google Scholar]
- O'Connell A. M., Koeppe R. E., 2nd, Andersen O. S. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 1990 Nov 30;250(4985):1256–1259. doi: 10.1126/science.1700867. [DOI] [PubMed] [Google Scholar]
- Pascal S. M., Cross T. A. High-resolution structure and dynamic implications for a double-helical gramicidin A conformer. J Biomol NMR. 1993 Sep;3(5):495–513. doi: 10.1007/BF00174606. [DOI] [PubMed] [Google Scholar]
- Pascal S. M., Cross T. A. Structure of an isolated gramicidin A double helical species by high-resolution nuclear magnetic resonance. J Mol Biol. 1992 Aug 20;226(4):1101–1109. doi: 10.1016/0022-2836(92)91055-t. [DOI] [PubMed] [Google Scholar]
- Popot J. L., Engelman D. M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990 May 1;29(17):4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- Salom D., Bañ M. C., Braco L., Abad C. HPLC demonstration that an all Trp-->Phe replacement in gramicidin A results in a conformational rearrangement from beta-helical monomer to double-stranded dimer in model membranes. Biochem Biophys Res Commun. 1995 Apr 17;209(2):466–473. doi: 10.1006/bbrc.1995.1525. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Chang C. H., Stevens F. J. The functions of tryptophan residues in membrane proteins. Protein Eng. 1992 Apr;5(3):213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- Smart O. S., Goodfellow J. M., Wallace B. A. The pore dimensions of gramicidin A. Biophys J. 1993 Dec;65(6):2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solouki T., Marto J. A., White F. M., Guan S., Marshall A. G. Attomole biomolecule mass analysis by matrix-assisted laser desorption/ionization Fourier transform ion cyclotron resonance. Anal Chem. 1995 Nov 15;67(22):4139–4144. doi: 10.1021/ac00118a017. [DOI] [PubMed] [Google Scholar]
- Solouki T., Pasa-Tolić L., Jackson G. S., Guan S., Marshall A. G. High-resolution multistage MS, MS2, and MS3 matrix-assisted laser desorption/ionization FT-ICR mass spectra of peptides from a single laser shot. Anal Chem. 1996 Nov 1;68(21):3718–3725. doi: 10.1021/ac960312d. [DOI] [PubMed] [Google Scholar]
- Steffen M. A., Lao K., Boxer S. G. Dielectric asymmetry in the photosynthetic reaction center. Science. 1994 May 6;264(5160):810–816. doi: 10.1126/science.264.5160.810. [DOI] [PubMed] [Google Scholar]
- Urry D. W., Long M. M., Jacobs M., Harris R. D. Conformation and molecular mechanisms of carriers and channels. Ann N Y Acad Sci. 1975 Dec 30;264:203–220. doi: 10.1111/j.1749-6632.1975.tb31484.x. [DOI] [PubMed] [Google Scholar]
- Veatch W. R., Fossel E. T., Blout E. R. The conformation of gramicidin A. Biochemistry. 1974 Dec 17;13(26):5249–5256. doi: 10.1021/bi00723a001. [DOI] [PubMed] [Google Scholar]
- Wallace B. A., Ravikumar K. The gramicidin pore: crystal structure of a cesium complex. Science. 1988 Jul 8;241(4862):182–187. doi: 10.1126/science.2455344. [DOI] [PubMed] [Google Scholar]
- Wiener M. C., White S. H. Structure of a fluid dioleoylphosphatidylcholine bilayer determined by joint refinement of x-ray and neutron diffraction data. III. Complete structure. Biophys J. 1992 Feb;61(2):434–447. doi: 10.1016/S0006-3495(92)81849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley W. C., White S. H. Membrane partitioning: distinguishing bilayer effects from the hydrophobic effect. Biochemistry. 1993 Jun 29;32(25):6307–6312. doi: 10.1021/bi00076a001. [DOI] [PubMed] [Google Scholar]
- Zaks A., Klibanov A. M. The effect of water on enzyme action in organic media. J Biol Chem. 1988 Jun 15;263(17):8017–8021. [PubMed] [Google Scholar]
- Zhang Z., Pascal S. M., Cross T. A. A conformational rearrangement in gramicidin A: from a double-stranded left-handed to a single-stranded right-handed helix. Biochemistry. 1992 Sep 22;31(37):8822–8828. doi: 10.1021/bi00152a019. [DOI] [PubMed] [Google Scholar]