Abstract
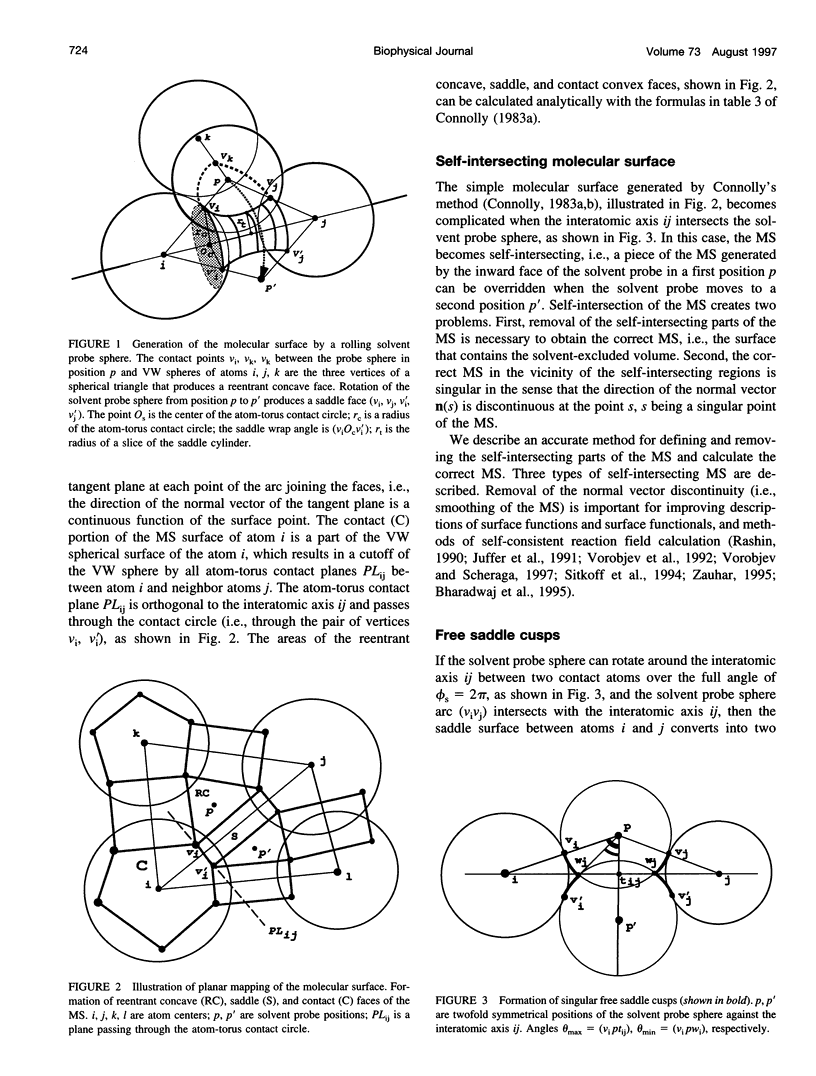
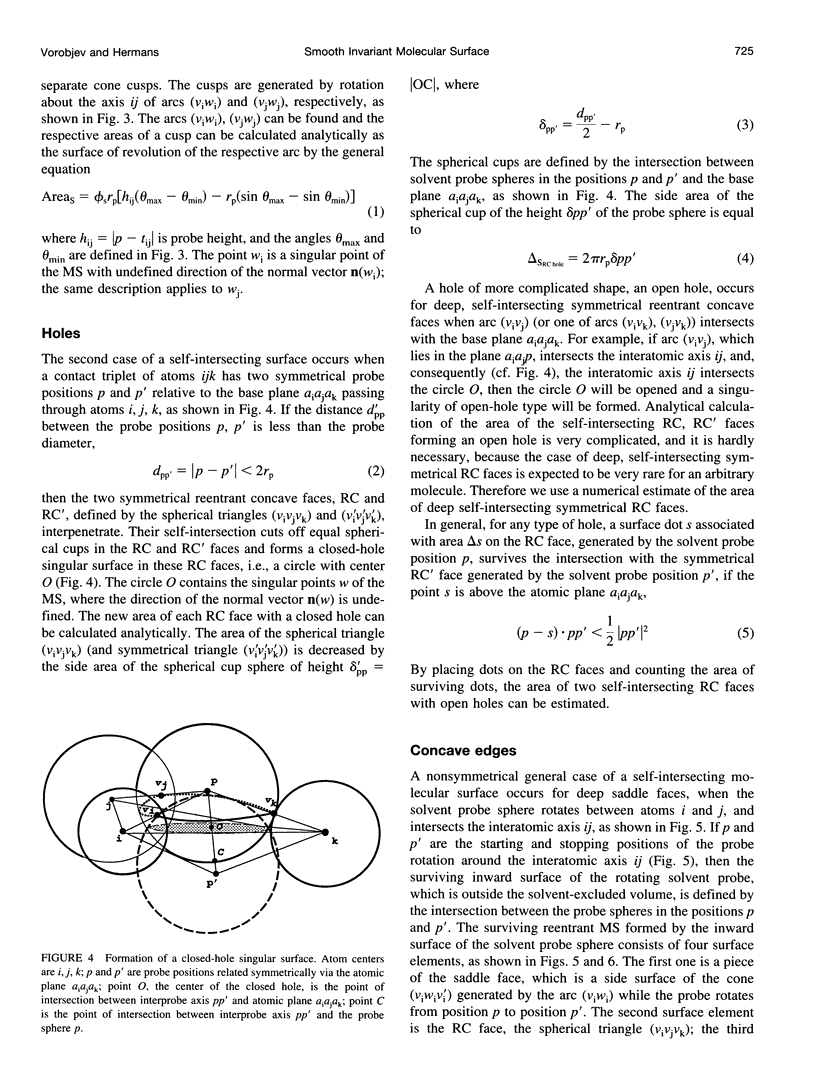
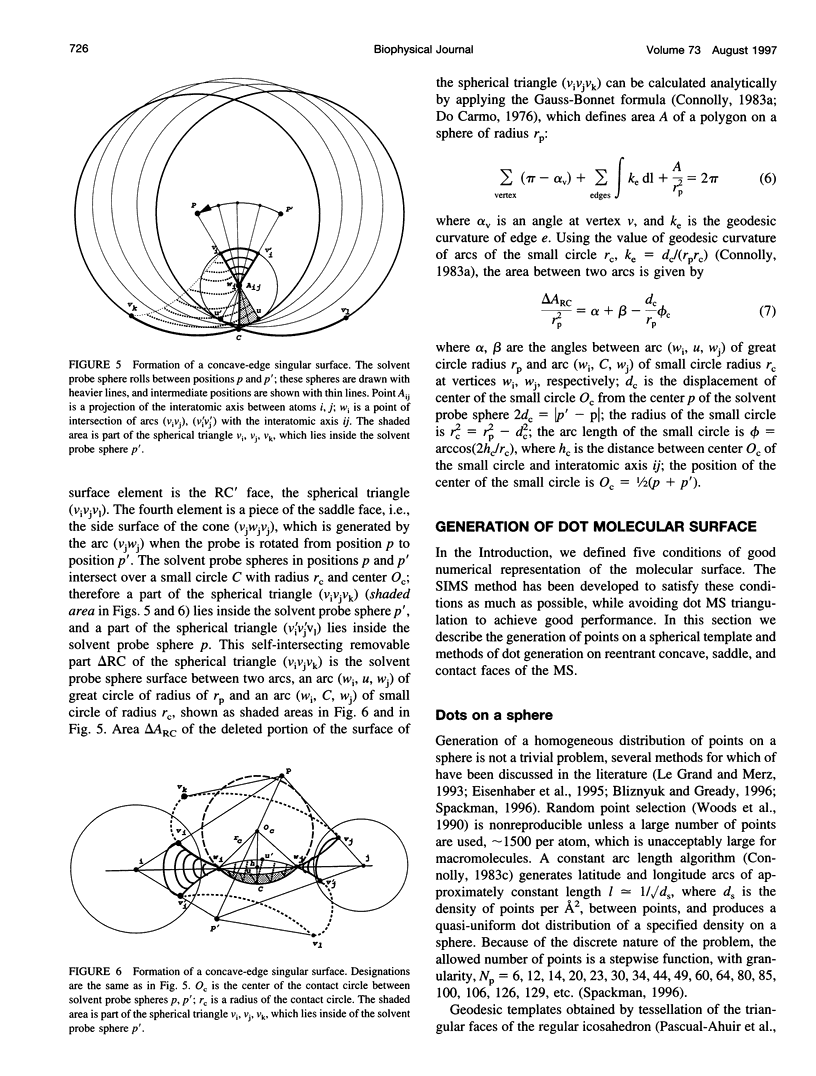
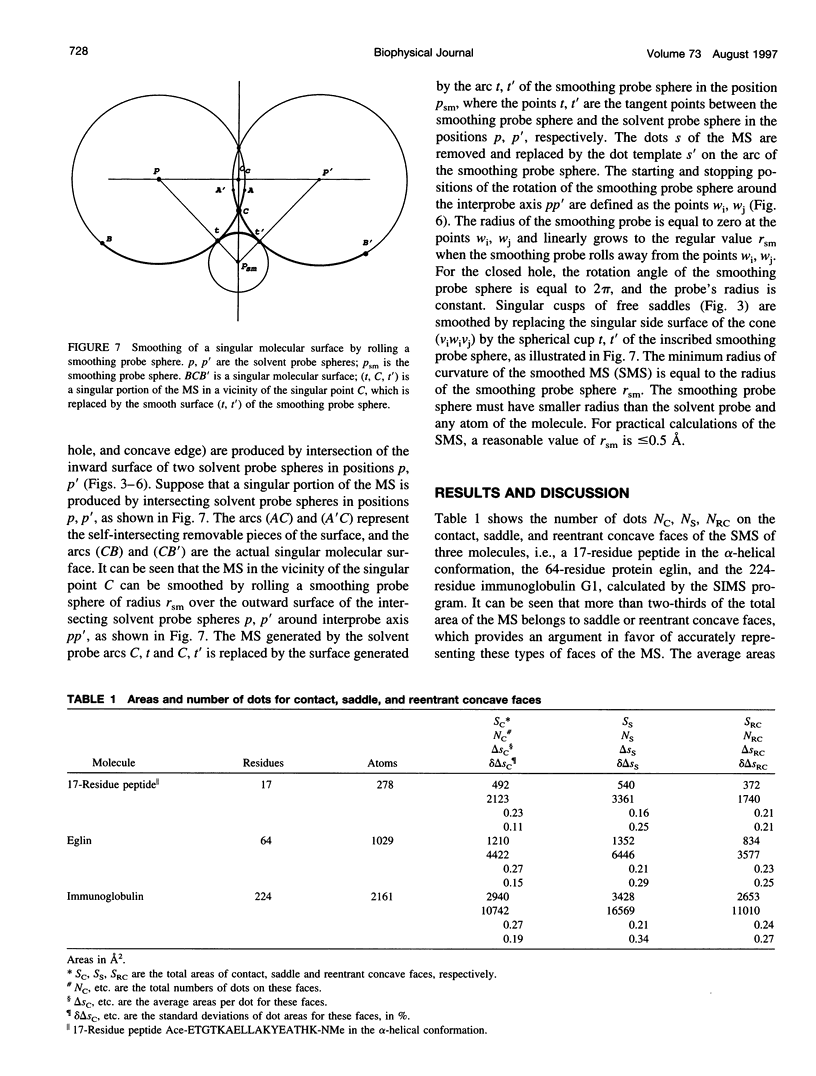
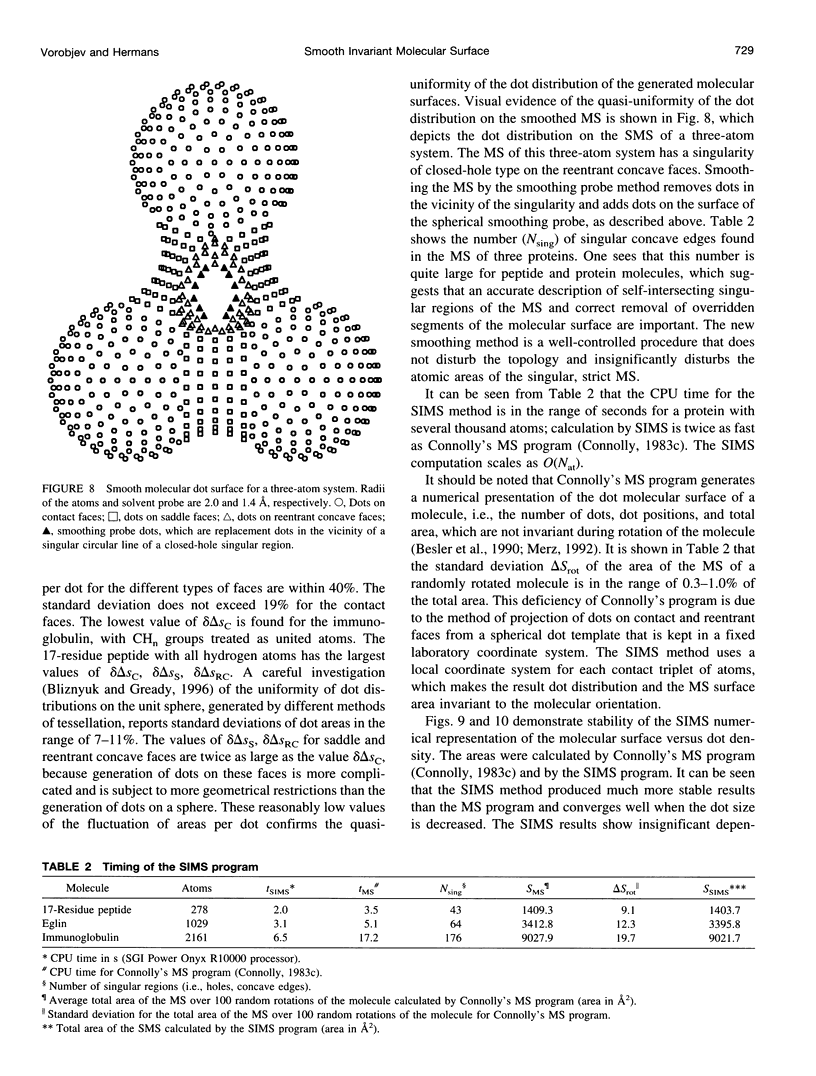
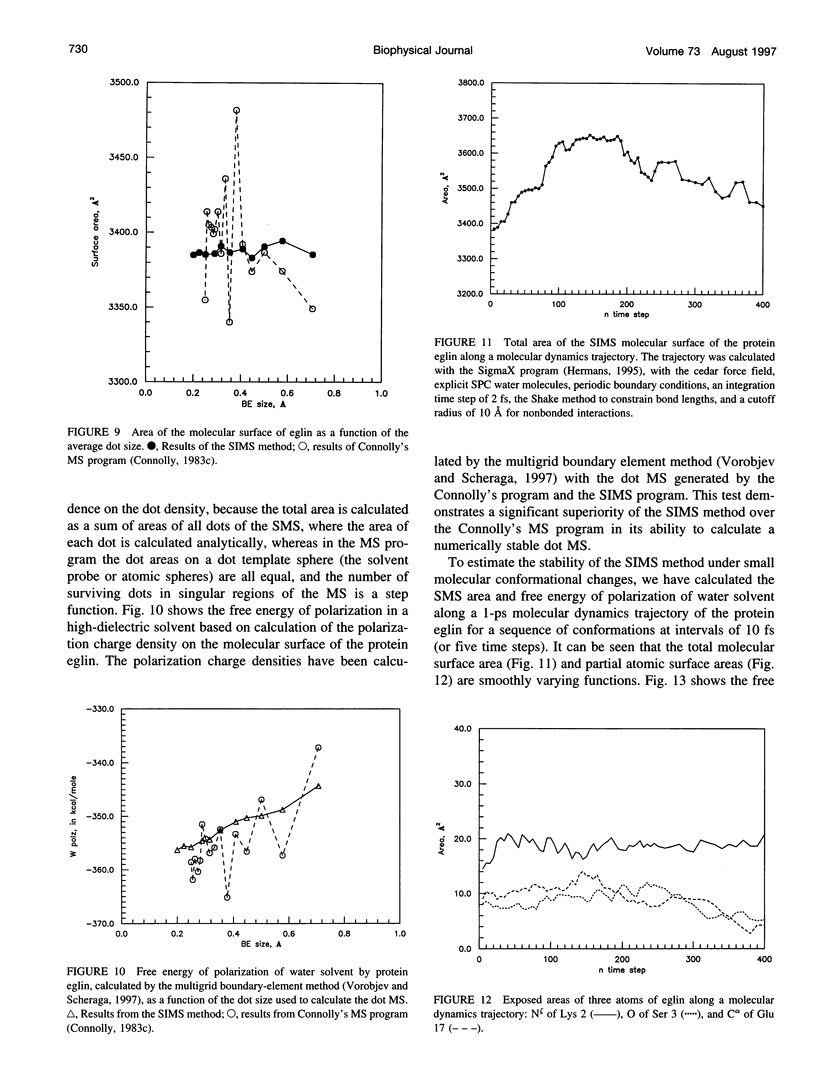
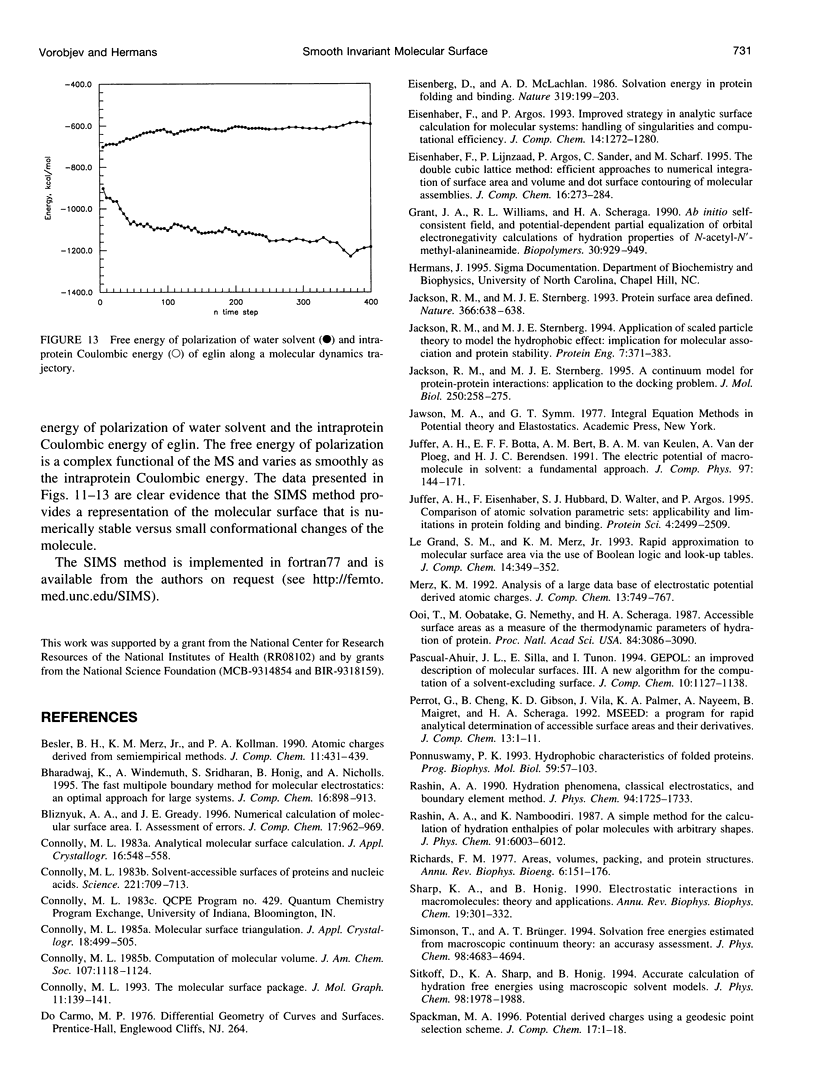
SIMS, a new method of calculating a smooth invariant molecular dot surface, is presented. The SIMS method generates the smooth molecular surface by rolling two probe spheres. A solvent probe sphere is rolled over the molecule and produces a Richards-Connolly molecular surface (MS), which envelops the solvent-excluded volume of the molecule. In deep crevices, Connolly's method of calculating the MS has two deficiencies. First, it produces self-intersecting parts of the molecular surface, which must be removed to obtain the correct MS. Second, the correct MS is not smooth, i.e., the direction of the normal vector of the MS is not continuous, and some points of the MS are singular. We present an exact method for removing self-intersecting parts and smoothing the singular regions of the MS. The singular MS is smoothed by rolling a smoothing probe sphere over the inward side of the singular MS. The MS in the vicinity of singularities is replaced with the reentrant surface of the smoothing probe sphere. The smoothing method does not disturb the topology of a singular MS, and the smooth MS is a better approximation of the dielectric border between high dielectric solvent and the low dielectric molecular interior. The SIMS method generates a smooth molecular dot surface, which has a quasi-uniform dot distribution in two orthogonal directions on the molecular surface, which is invariant with molecular rotation and stable under changes in the molecular conformation, and which can be used in a variety of implicit methods of modeling solvent effects. The SIMS program is faster than the Connolly MS program, and in a matter of seconds generates a smooth dot MS of a 200-residue protein. The program is available from the authors on request (see http:@femto.med.unc.edu/SIMS).
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. The molecular surface package. J Mol Graph. 1993 Jun;11(2):139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., McLachlan A. D. Solvation energy in protein folding and binding. Nature. 1986 Jan 16;319(6050):199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- Grant J. A., Williams R. L., Scheraga H. A. Ab initio self-consistent field and potential-dependent partial equalization of orbital electronegativity calculations of hydration properties of N-acetyl-N'-methyl-alanineamide. Biopolymers. 1990;30(9-10):929–949. doi: 10.1002/bip.360300908. [DOI] [PubMed] [Google Scholar]
- Jackson R. M., Sternberg M. J. A continuum model for protein-protein interactions: application to the docking problem. J Mol Biol. 1995 Jul 7;250(2):258–275. doi: 10.1006/jmbi.1995.0375. [DOI] [PubMed] [Google Scholar]
- Jackson R. M., Sternberg M. J. Application of scaled particle theory to model the hydrophobic effect: implications for molecular association and protein stability. Protein Eng. 1994 Mar;7(3):371–383. doi: 10.1093/protein/7.3.371. [DOI] [PubMed] [Google Scholar]
- Juffer A. H., Eisenhaber F., Hubbard S. J., Walther D., Argos P. Comparison of atomic solvation parametric sets: applicability and limitations in protein folding and binding. Protein Sci. 1995 Dec;4(12):2499–2509. doi: 10.1002/pro.5560041206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi T., Oobatake M., Némethy G., Scheraga H. A. Accessible surface areas as a measure of the thermodynamic parameters of hydration of peptides. Proc Natl Acad Sci U S A. 1987 May;84(10):3086–3090. doi: 10.1073/pnas.84.10.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnuswamy P. K. Hydrophobic characteristics of folded proteins. Prog Biophys Mol Biol. 1993;59(1):57–103. doi: 10.1016/0079-6107(93)90007-7. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Vila J., Williams R. L., Vásquez M., Scheraga H. A. Empirical solvation models can be used to differentiate native from near-native conformations of bovine pancreatic trypsin inhibitor. Proteins. 1991;10(3):199–218. doi: 10.1002/prot.340100305. [DOI] [PubMed] [Google Scholar]
- Wilson I. Plastids better red than dead. Nature. 1993 Dec 16;366(6456):638–638. doi: 10.1038/366638a0. [DOI] [PubMed] [Google Scholar]
- Zauhar R. J. SMART: a solvent-accessible triangulated surface generator for molecular graphics and boundary element applications. J Comput Aided Mol Des. 1995 Apr;9(2):149–159. doi: 10.1007/BF00124405. [DOI] [PubMed] [Google Scholar]


