Abstract
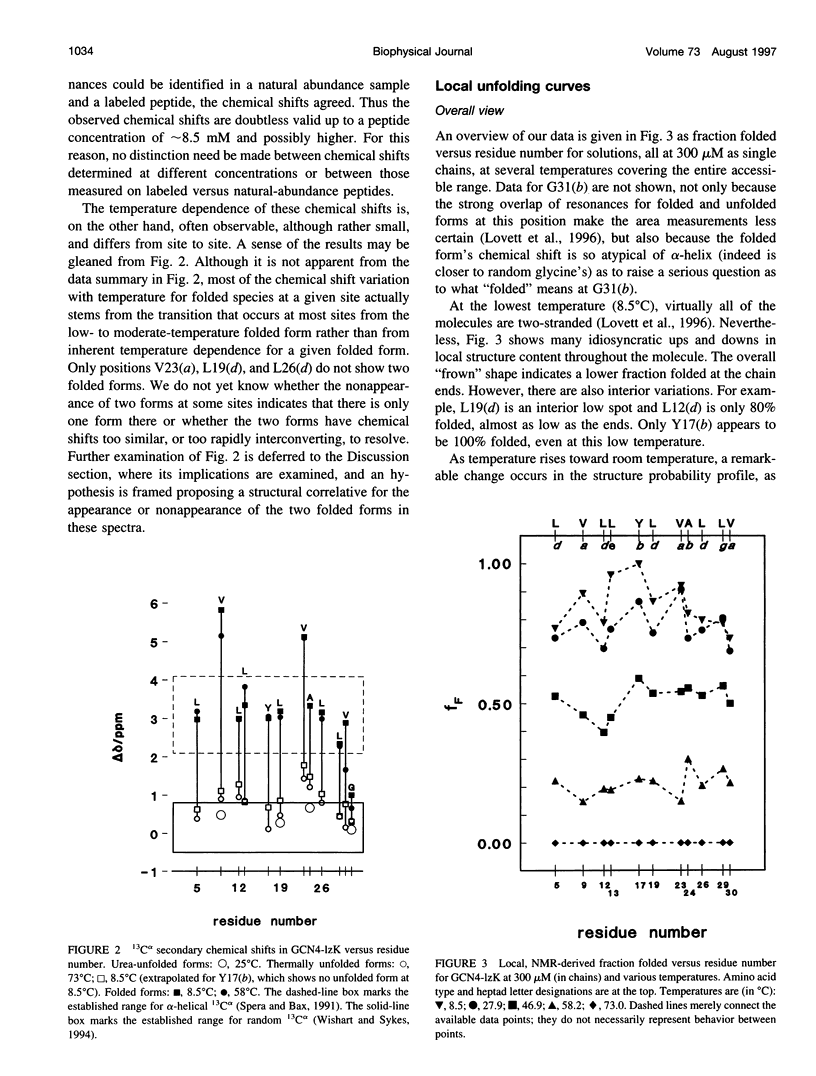
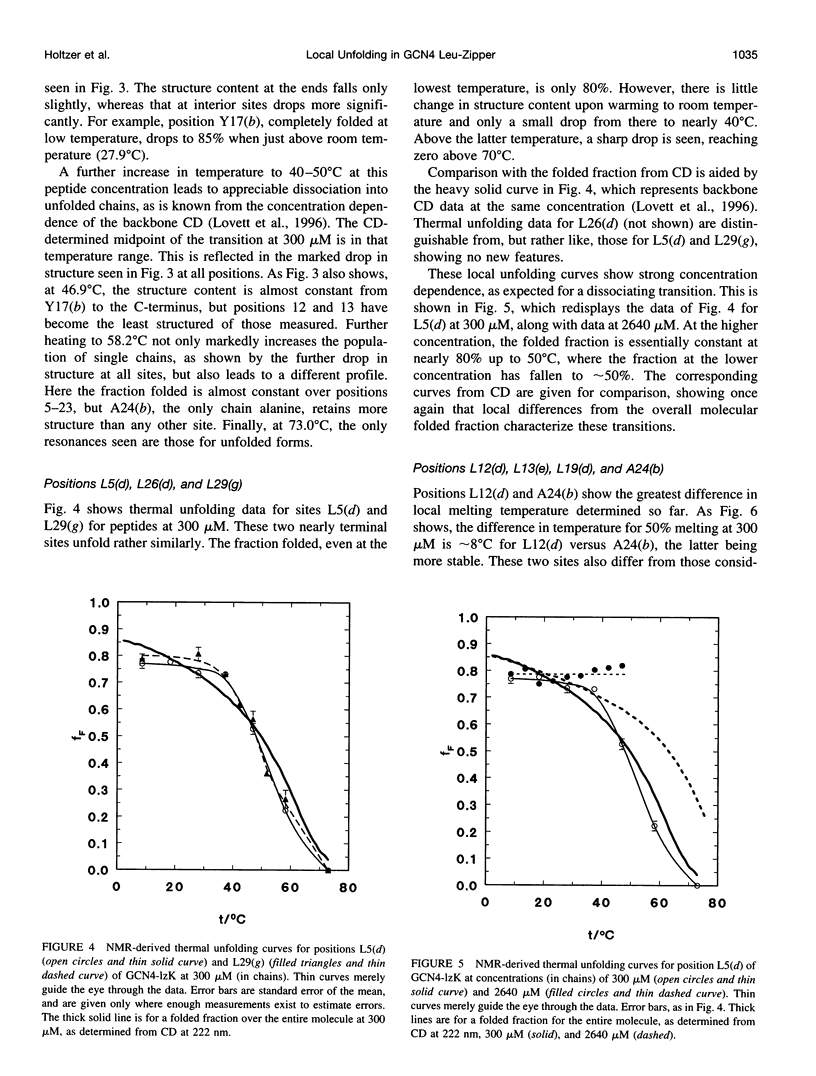
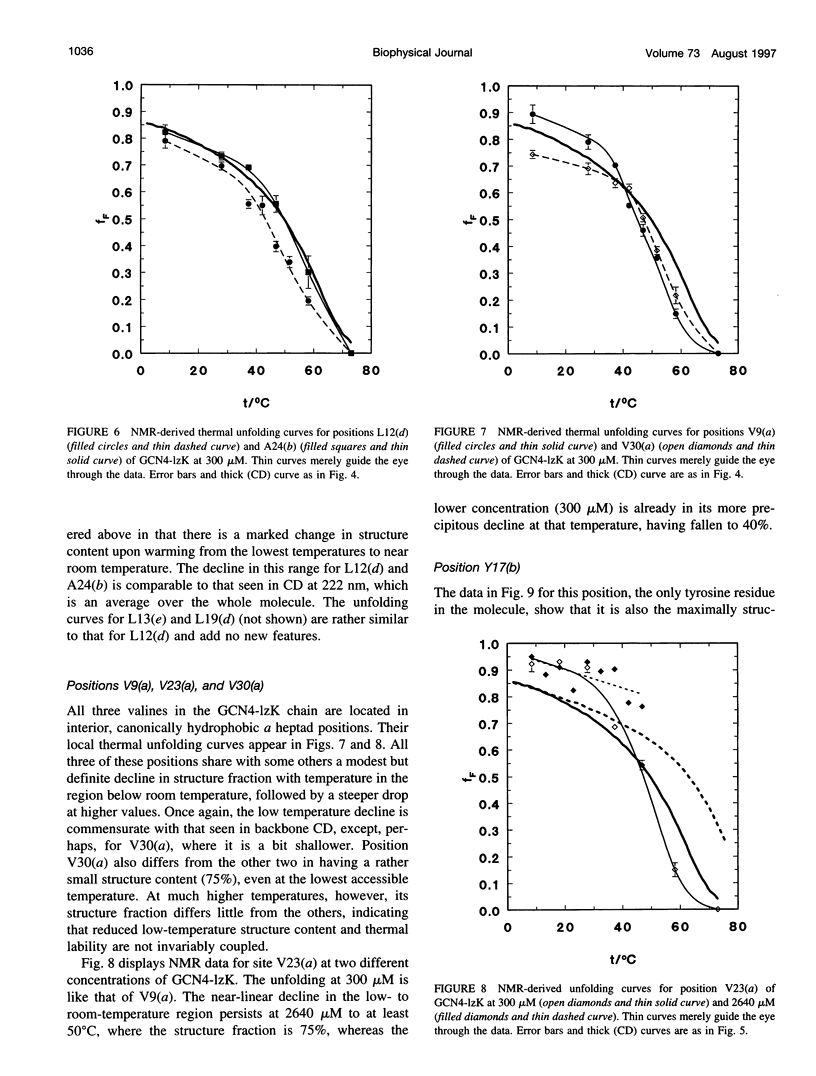
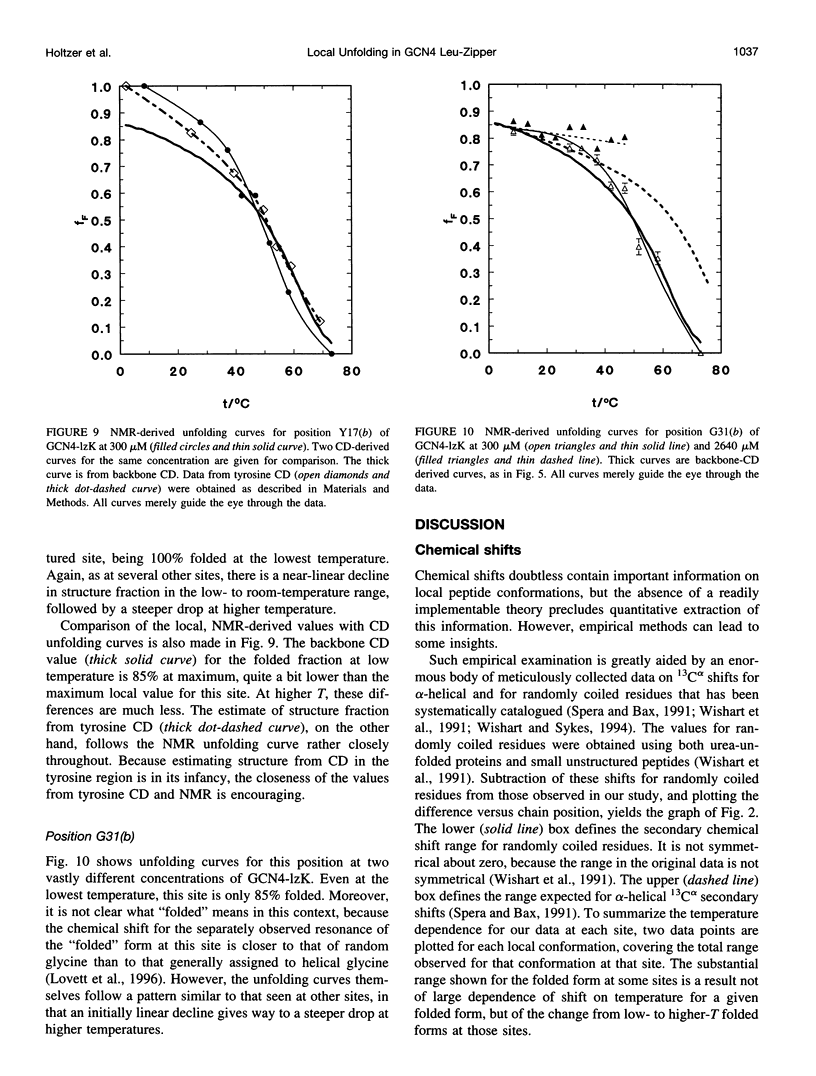
13C alpha chemical shifts and site-specific unfolding curves are reported for 12 sites on a 33-residue, GCN4-like leucine zipper peptide (GCN4-lzK), ranging over most of the chain and sampling most heptad positions. Data were derived from NMR spectra of nine synthetic, isosequential peptides bearing 99% 13C alpha at sites selected to avoid spectral overlap in each peptide. At each site, separate resonances appear for unfolded and folded forms, and most sites show resonances for two folded forms near room temperature. The observed chemical shifts suggest that 1) urea-unfolded GCN4-lzK chains are randomly coiled; 2) thermally unfolded chains include significant transient structure, except at the ends; 3) the coiled-coli structure in the folded chains is atypical near the C-terminus; 4) only those interior sites surrounded by canonical interchain salt bridges fail to show two folded forms. Local unfolding curves, obtained from integrated resonance intensities, show that 1) sites differ in structure content and in melting temperature, so the equilibrium population must comprise more than two molecular conformations; 2) there is significant end-fraying, even at the lowest temperatures, but thermal unfolding is not a progressive unwinding from the ends; 3) residues 9-16 are in the lowest melting region; 4) heptad position does not dictate stability; 5) significant unfolding occurs below room temperature, so the shallow, linear decline in backbone CD seen there has conformational significance. It seems that only a relatively complex array of conformational states could underlie these findings.
Full text
PDF



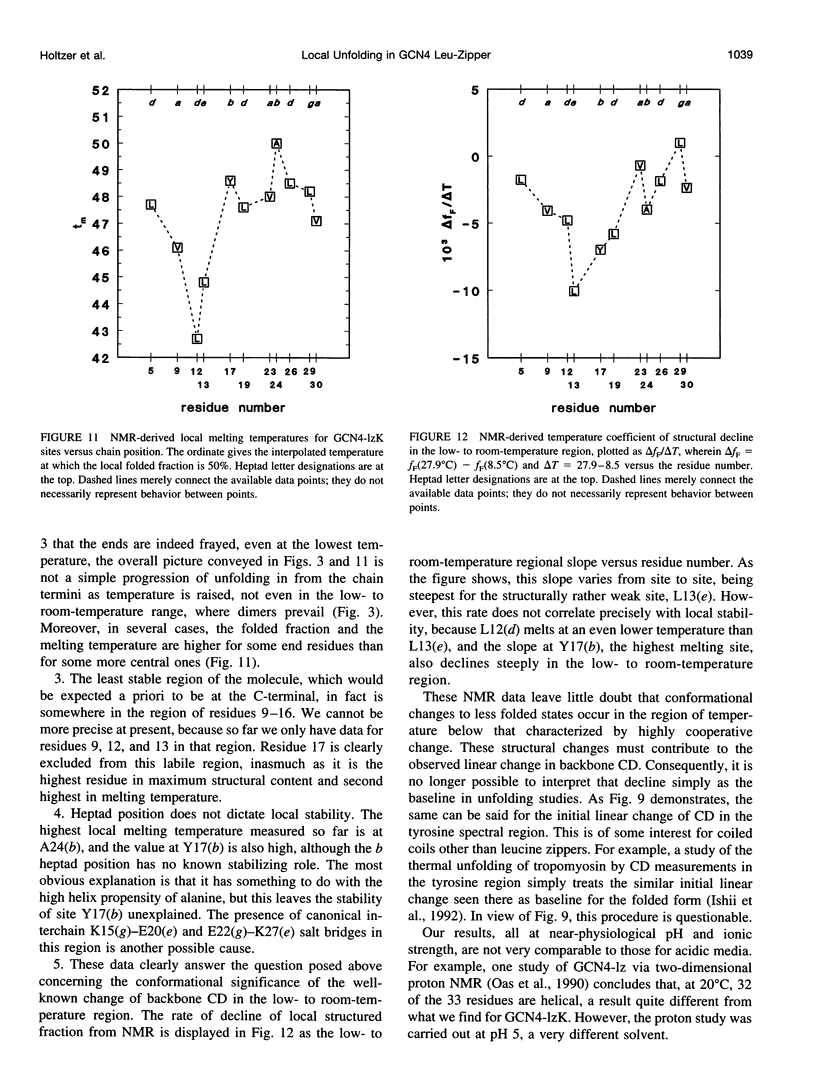

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aurora R., Srinivasan R., Rose G. D. Rules for alpha-helix termination by glycine. Science. 1994 May 20;264(5162):1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974 Jul 30;13(16):3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Esposito G., Dhanapal B., Dumy P., Varma V., Mutter M., Bodenhausen G. Lysine as helix C-capping residue in a synthetic peptide. Biopolymers. 1997 Jan;41(1):27–35. doi: 10.1002/(SICI)1097-0282(199701)41:1<27::AID-BIP3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
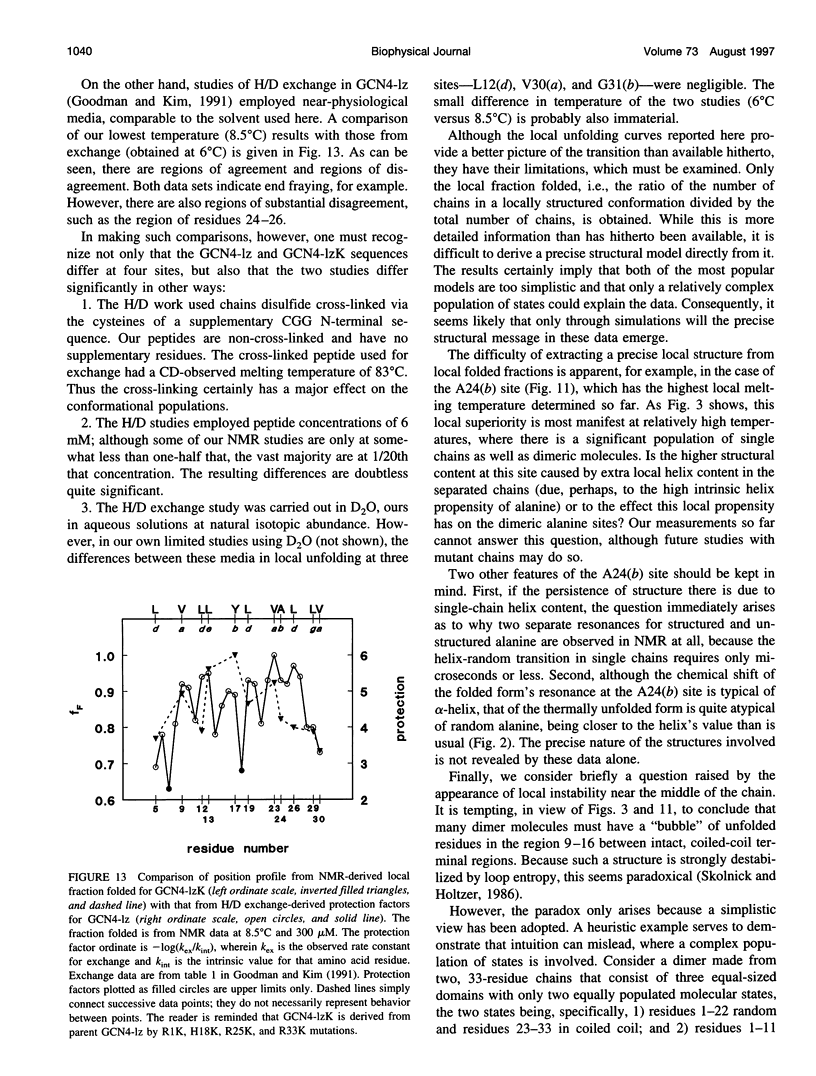
- Goodman E. M., Kim P. S. Periodicity of amide proton exchange rates in a coiled-coil leucine zipper peptide. Biochemistry. 1991 Dec 17;30(50):11615–11620. doi: 10.1021/bi00114a002. [DOI] [PubMed] [Google Scholar]
- Holtzer M. E., Adams K., Lovett E. G., Holtzer A. Tyrosines in two-stranded coiled coils are CD active near 280 nm even in the absence of interhelix tyrosine-tyrosine interactions. Biopolymers. 1996 May;38(5):669–671. doi: 10.1002/(SICI)1097-0282(199605)38:5%3C669::AID-BIP11%3E3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Holtzer M. E., Crimmins D. L., Holtzer A. Structural stability of short subsequences of the tropomyosin chain. Biopolymers. 1995 Jan;35(1):125–136. doi: 10.1002/bip.360350113. [DOI] [PubMed] [Google Scholar]
- Holtzer M. E., Holtzer A. Alpha-helix to random coil transitions: interpretation of the CD in the region of linear temperature dependence. Biopolymers. 1992 Nov;32(11):1589–1591. doi: 10.1002/bip.360321116. [DOI] [PubMed] [Google Scholar]
- Hvidt S., Rodgers M. E., Harrington W. F. Temperature-dependent optical rotatory dispersion properties of helical muscle proteins and homopolymers. Biopolymers. 1985 Sep;24(9):1647–1662. doi: 10.1002/bip.360240902. [DOI] [PubMed] [Google Scholar]
- Ishii Y., Hitchcock-DeGregori S., Mabuchi K., Lehrer S. S. Unfolding domains of recombinant fusion alpha alpha-tropomyosin. Protein Sci. 1992 Oct;1(10):1319–1325. doi: 10.1002/pro.5560011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenar K. T., García-Moreno B., Freire E. A calorimetric characterization of the salt dependence of the stability of the GCN4 leucine zipper. Protein Sci. 1995 Sep;4(9):1934–1938. doi: 10.1002/pro.5560040929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S. S., Stafford W. F., 3rd Preferential assembly of the tropomyosin heterodimer: equilibrium studies. Biochemistry. 1991 Jun 11;30(23):5682–5688. doi: 10.1021/bi00237a007. [DOI] [PubMed] [Google Scholar]
- Lovett E. G., D'Avignon D. A., Holtzer M. E., Braswell E. H., Zhu D., Holtzer A. Observation via one-dimensional 13Calpha NMR of local conformational substates in thermal unfolding equilibria of a synthetic analog of the GCN4 leucine zipper. Proc Natl Acad Sci U S A. 1996 Mar 5;93(5):1781–1785. doi: 10.1073/pnas.93.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb K. J., Carr C. M., Kim P. S. Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry. 1994 Jun 14;33(23):7361–7367. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Stewart M. Tropomyosin coiled-coil interactions: evidence for an unstaggered structure. J Mol Biol. 1975 Oct 25;98(2):293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- Miller D. W., Dill K. A. A statistical mechanical model for hydrogen exchange in globular proteins. Protein Sci. 1995 Sep;4(9):1860–1873. doi: 10.1002/pro.5560040921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Oas T. G., McIntosh L. P., O'Shea E. K., Dahlquist F. W., Kim P. S. Secondary structure of a leucine zipper determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1990 Mar 27;29(12):2891–2894. doi: 10.1021/bi00464a001. [DOI] [PubMed] [Google Scholar]
- Privalov P. L. Stability of proteins. Proteins which do not present a single cooperative system. Adv Protein Chem. 1982;35:1–104. [PubMed] [Google Scholar]
- Skolnick J., Holtzer A. Alpha-helix-to-random-coil transitions of two-chain, coiled coils: a theoretical model for the "pretransition" in cysteine-190-cross-linked tropomyosin. Biochemistry. 1986 Oct 7;25(20):6192–6202. doi: 10.1021/bi00368a054. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Jackson S., Wilk R. R., Englander S. W., DeGrado W. F. The role of helix formation in the folding of a fully alpha-helical coiled coil. Proteins. 1996 Apr;24(4):427–432. doi: 10.1002/(SICI)1097-0134(199604)24:4<427::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Sykes B. D. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Sykes B. D., Richards F. M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991 Nov 20;222(2):311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]


