Abstract
Purpose
Breast cancer is one of the most common malignancies among females, and its prognosis is affected by a complex network of gene interactions. Weighted gene co-expression network analysis was used to construct free-scale gene co-expression networks and to identify potential biomarkers for breast cancer progression.
Methods
The gene expression profiles of GSE42568 were downloaded from the Gene Expression Omnibus database. RNA-sequencing data and clinical information of breast cancer from TCGA were used for validation.
Results
A total of ten modules were established by the average linkage hierarchical clustering. We identified 58 network hub genes in the significant module (R2 = 0.44) and 6 hub genes (AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK), which were significantly correlated with prognosis. Receiver-operating characteristic curve validated that the mRNA levels of these six genes exhibited excellent diagnostic efficiency in the test data set of GSE42568. RNA-sequencing data from TCGA showed that the expression levels of these six genes were higher in triple-negative tumors. One-way ANOVA suggested that these six genes were upregulated at more advanced stages. The results of independent sample t test indicated that MCM10 and TTK were associated with tumor size, and that AGO2, CDC20, CDCA5, MCM10, and MYBL2 were overexpressed in lymph-node positive breast cancer.
Conclusions
AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK were identified as candidate biomarkers for further basic and clinical research on breast cancer based on co-expression analysis.
Electronic supplementary material
The online version of this article (10.1007/s00432-019-02974-4) contains supplementary material, which is available to authorized users.
Keywords: Breast cancer, Weighted gene co-expression network analysis, Prognosis, Gene Expression Omnibus, Biomarker
Purpose
Breast cancer is a popular diagnosed malignancy around the world. It accounts for 24% cancer cases and 15% cancer deaths of females, as the leading cause of female cancer death. According to the estimates from the Global Cancer Statistics 2018, there was nearly 2.1 million breast cancers diagnosed in the world, with approximately 62,000 deaths (Bray et al. 2018). The incident rates of breast cancer rose in most developing countries during last decades, resulting from a combination of social and economic factors, including the postponement of childbearing, increased obesity and physical inactivity (Bray et al. 2018). The incidence of breast cancer was also markedly high in developed countries, in which nearly 60% death-causing cases were diagnosed at advanced stages, despite the developments of diagnosis and treatment strategies. Among the females diagnosed at early stages, 30% developed metastatic lesions months or even years after surgical removal of the primary tumors (O’Shaughnessy 2005; Redig and McAllister 2013).
In breast cancer initiation and progression, inheritance plays important roles. It was reported that 5–10% breast cancer cases resulted from hereditary and genetic factors, such as inherited mutations and family history (Bray et al. 2018). BRCA mutations are observed in 20% triple-negative breast cancer patients, while, in the general population, the mutations of BRCA are substantially less. BRCA1/2 mutations are currently detected to assess the risk for inherited breast cancer (Trainer et al. 2011). The risks of breast cancer associated with BRCA1/2 mutations were 87% and 84% in cancer-prone families, while, in population-based studies, such risk decreased to 65% and 45% (Ford et al. 1994; Gonzalez-Angulo et al. 2011). Human epidermal growth factor receptor 2 (HER2) is another important prognostic biomarker, up to 30% breast cancers overexpressing HER2 (King et al. 1985). Overexpression of HER2 is associated with higher risk of local recurrence and worse overall survival (Paterson et al. 1991; Press et al. 1997). These results suggested HER2 as a therapeutic target and a predictive marker. The monoclonal antibody, trastuzumab, and the dual tyrosine dual kinase inhibitor, lapatinib, were approved to treat HER2-positive breast cancer. Overexpression of HER2 was used to predict the response to trastuzumab and lapatinib (Jung et al. 2018; Rimawi et al. 2018; Sagara et al. 2018; Takada et al. 2018; von Minckwitz et al. 2018). Expression of the hormone receptors (ER/PR) was also used to predict the response to endocrine therapies; thus, patients with positive hormone receptors were often predicted to have a favorable prognosis. Women with ER-positive breast cancer treated with tamoxifen were reported to have a significant decrease of recurrence and death (Fisher et al. 2001; Slamon et al. 1987). Epithelial growth factor receptor was also reported to be overexpressed in 50% inflammatory and triple-negative breast cancers (Khalil et al. 2003). Detection of these biomarkers alone or in combination assisted early diagnosis, therapeutic strategy determination, and prognosis predication after treatment. Limited knowledge on precise molecular targets for breast cancer limits advanced disease treatment. Therefore, it is crucial to identify novel candidate genes.
The high-throughput platforms for genomic analysis provided promising tools in medical oncology with great clinical applications. Co-expression analysis is increasingly used to analyze these high-dimensional data. To find candidate biomarkers and to describe the correlation patterns among genes, we constructed co-expression networks using weighted gene co-expression network analysis (WGCNA). This method has been used to explore candidate prognostic genes and therapeutic targets (Langfelder and Horvath 2008). In the presented study, we used WGCNA algorithm to explore candidate predictive tumor-associated genes.
Materials and methods
Data processing and differentially expressed genes (DEGs) screening
The gene expression profiles of GSE42568 were downloaded from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). The GSE42568 was based on GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array). This data set included 17 normal breast tissues and 104 breast cancer tissues. Probes were annotated by the annotation files. The downloaded raw data were preprocessed with the Robust Multichip Average method in R software including background adjustment, quintile normalization, and summarization. Using linear models for microarray data package in R, the expression value of each gene was compared between cancer samples and normal controls to identify DEGs. The false discovery rate method was used to adjust the p value. |log2 fold-change (FC)| > 1 and adjusted p value < 0.05 were set as the cut-off criteria to select genes for further network construction.
Co-expression network construction
The DEG expression data were first tested to evaluate their usability. WGCNA package in R was then used to construct gene co-expression network. The adjacency matrix Amn was defined as follows: Amn = |smn |β, Aij encoded the adjacency between gene m and gene n, and Smn represented the Pearson’s correlation between gene m and gene n. The soft-thresholding parameter β = 6 (scale free R2 = 0.88) was selected to emphasize strong correlations between genes and penalize weak correlations. The adjacent matrix was then transformed into topological overlap matrix (TOM) to counter the effects of spurious or missing connections between network nodes. We conducted average linkage hierarchical clustering to classify genes with high absolute correlations into gene modules according to the TOM-based dissimilarity measure with a minimum size of 30.
Identification of clinically significant modules
To identify modules related to clinical information of breast cancer. We calculated the correlation between module eigengenes and clinical trait. A module eigengene is the first principal component of the gene module, and considered as a representative of the gene expression profiles in a module. In addition, we measured the module significance of each module, as the average gene significance for all the genes in a module. The higher absolute value of module significance represented more biologically significant of a given module. In general, the module significance tended to be highly associated with correlation between module eigengenes and clinical trait.
Gene enrichment analysis
After screening out the clinically significant module, we used the database for annotation, visualization, and integrated discovery (http://david.abcc.ncifcrf.gov/) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of candidate DEGs (da Huang et al. 2009a, b). The ontology contains three hierarchies: biological process, cellular component, and molecular function. Adjusted p value < 0.05 was set as the cut-off criterion to identify enriched GO terms and KEGG pathways.
Identification and validation of hub genes
Hub genes are often considered functionally significant and highly connected with other nodes in the module. After relating modules to clinical traits, we calculated module connectivity of each gene, which was measured by absolute value of the module membership (MM). MM represented the Pearson’s correlation between a gene and the module eigengene. Hub genes tended to be highly connected and to have high MM. In addition, we measured the absolute value of gene significance (GS) which represented the Pearson’s correlation between a given gene and the clinical trait. The biologically significant genes often had higher GS absolute values. In this study, hub genes were identified using the cut-off criteria of absolute MM > 0.7 and absolute GS > 0.4. TCGA database was used to validate the expression levels of hub genes. Human Protein Atlas (http://www.proteinatlas.org) was also applied to validate the immunohistochemistry of candidate hub genes (Uhlen et al. 2010, 2015). Kaplan–Meier-plotter (www.kmplot.com) was used for survival analysis (Gyorffy et al. 2010).
Results
Construction of weighted co-expression network and identification of key modules
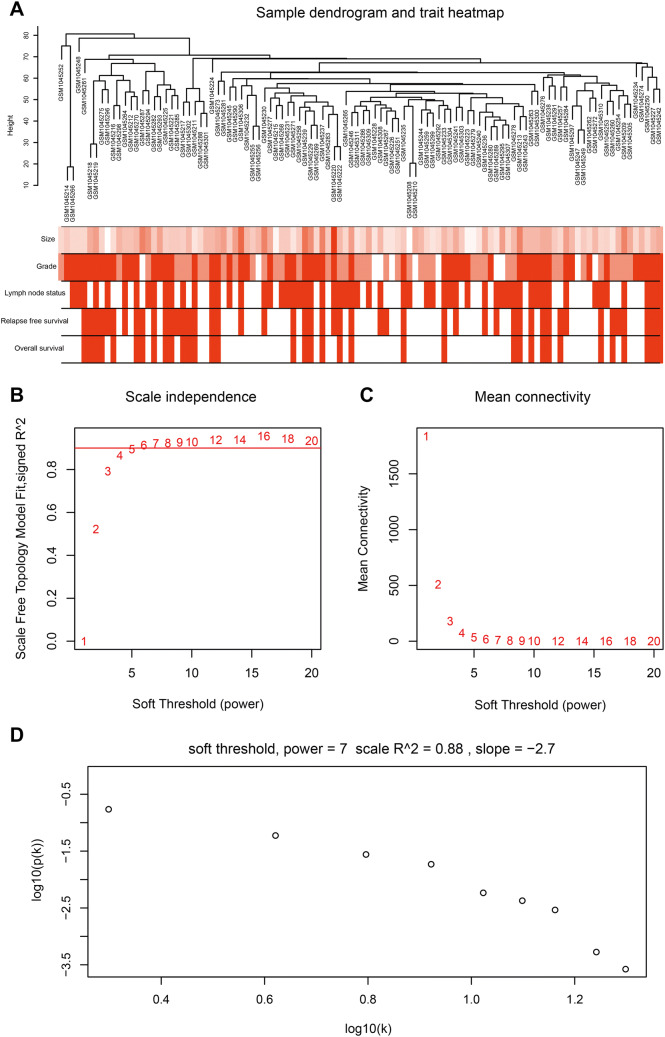
The workflow of current study is shown in Fig. S1. After data preprocessing and quality assessment, the expression matrices were obtained from the 104 samples in the data set GSE42568. The data set details are represented in Table S1. Based on the threshold of adjusted p value < 0.05 and |log2 fold-change (FC)| > 1, a total of 3771 DEGs (3188 upregulated and 583 downregulated) were selected for subsequent WGCNA. To assess the microarray quality and exclude outlier samples, sample cluster of GSE42568 was performed in Pearson’s correlation matrices and with average linkage method (Fig. 1a). To ensure a scale-free network, the power of β = 6 (scale free R2 = 0.88) was selected as the soft-thresholding in this study (Fig. 1b–d). Based on the average linkage hierarchical clustering, a total of ten modules were established. Turquoise module had the highest correlation with pathological grade (Fig. 2), which was selected as the clinically significant module for further analysis.
Fig. 1.
Clustering dendrogram and determination of soft-thresholding power in the WGCNA. a Clustering dendrogram of 104 samples. b Analysis of the scale-free fit index for various soft-thresholding powers (β). c Analysis of the mean connectivity for various soft-thresholding powers. d Checking the scale-free topology when β = 6
Fig. 2.
Identification of modules associated with the clinical traits of breast cancer. a Dendrogram of all differentially expressed genes clustered based on a dissimilarity measure (1-TOM). b Heatmap of the correlation between module eigengenes and clinical traits of breast cancer. c Distribution of average gene significance and errors in the modules associated with tumor grades of breast cancer
Gene enrichment analysis
We categorized the genes of turquoise module into three GO groups: biological process, cellular component, and molecular function. In biological process analysis, the genes were significantly enriched in cell division, mitotic nuclear division, sister chromatid cohesion, DNA replication, G1/S transition of mitotic cell cycle, and chromosome segregation (Fig. 3a). In molecular function group, the genes were mainly enriched in protein binding, ATP binding, chromatin binding, microtubule binding, and single-stranded DNA-dependent ATPase activity (Fig. 3b). For cellular component, the genes were enriched in nucleoplasm, nucleus, condensed chromosome kinetochore, chromosome, centromeric region, and midbody (Fig. 3c). According to KEGG pathway analysis, our results indicated that these genes were significantly enriched in cell cycle, DNA replication, pyrimidine metabolism, p53 signaling pathway, and progesterone-mediated oocyte maturation (Fig. 3d). Our enrichment analysis of the clinically significant module demonstrated that mitotic cell cycle process played important roles in the tumor progression.
Fig. 3.
GO and pathway enrichment analysis of blue module genes. a Biological process analysis. b Cellular component analysis. c Molecular function analysis. d KEGG pathway analysis
Identification and validation of hub genes
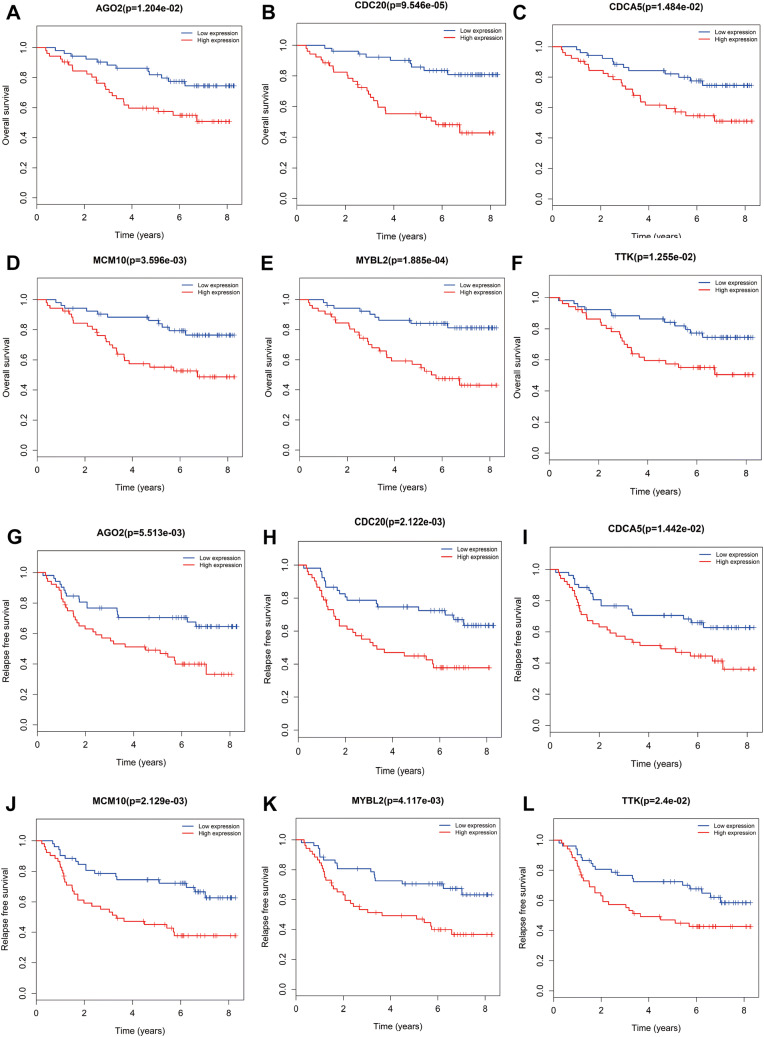
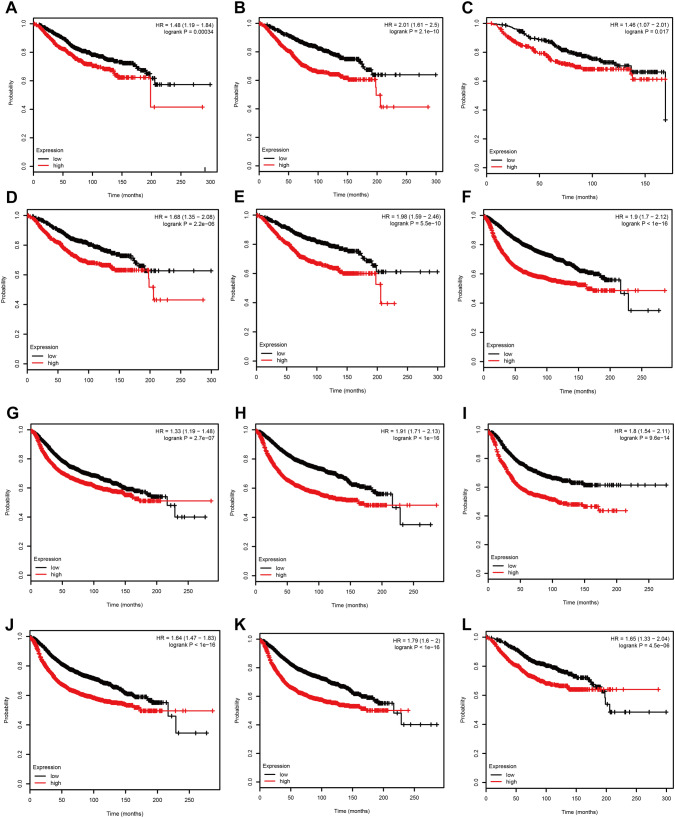
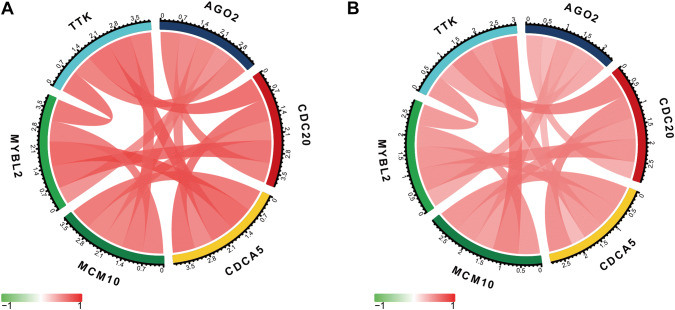
Based the cut-off criteria (absolute MM > 0.7 and absolute GS > 0.4), we selected a total of 58 genes as hub genes which had high functional significance in the clinically significant module. Among them, AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK were negatively associated with the prognosis of breast cancer patients using Kaplan–Meier survival curves by log-rank test (Fig. 4). The same results were found based on Kaplan–Meier-plotter (www.kmplot.com) (Fig. 5). Therefore, these six genes were chosen for further analysis. In the test data set of GSE42568, receiver-operating characteristic (ROC) curve validated that their expression could distinguish normal samples from breast cancer. The hierarchical cluster analysis of the data demonstrated that they were highly expressed in more aggressive tumors (Fig. 6). In addition, based on RNA-sequencing data from TCGA, ROC curve indicated that CDC20, CDCA5, MCM10, MYBL2, and TTK exhibited prominently diagnostic roles in tumor tissues. The expression levels of these six genes were higher in triple-negative tumors. One-way ANOVA suggested that these six genes were upregulated at more advanced stages. The results of independent sample t test demonstrated that MCM10 and TTK were associated with tumor size; AGO2, CDC20, CDCA5, MCM10, and MYBL2 were overexpressed in lymph-node positive breast cancer (Fig. 7). Protein levels of CDC20, CDCA5, MCM10, MYBL2, and TTK were significantly higher in tumor tissues compared with normal tissues based on the IHC data from database of Human Protein Atlas (Fig. S2). Since GO2, CDC20, CDCA5, MCM10, and MYBL2 were hub genes in turquoise module, we evaluated the correlation among these genes. Our results demonstrated that their expression levels were highly correlated (Fig. 8).
Fig. 4.
Overall survival (OS) and relapse-free survival (RFS) of the six hub genes in breast cancer based on the data set GSE42568. The patients were stratified into high-level group and low-level group according to median expression. a OS of AGO2. b OS of CDC20. c OS of CDCA5. d OS of MCM10. e OS of MYBL2. f OS of TTK. g RFS of AGO2. h RFS of CDC20. i RFS of CDCA5. j RFS of MCM10. k RFS of MYBL2. l RFS of TTK
Fig. 5.
OS and RFS of the six hub genes in breast cancer based on Kaplan–Meier-plotter. The patients were stratified into high-level group and low-level group according to median expression. a OS of AGO2. b OS of CDC20. c OS of CDCA5. d OS of MCM10. e OS of MYBL2. f OS of TTK. g RFS of AGO2. h RFS of CDC20. i RFS of CDCA5. j RFS of MCM10. k RFS of MYBL2. l RFS of TTK
Fig. 6.
Validation of AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK. a ROC curve of AGO2. b ROC curve of CDC20. c ROC curve of CDCA5. d ROC curve of MCM10. e ROC curve of MYBL2. f ROC curve of TTK. g Heatmap of the expression of hub genes in different stages of breast cancer
Fig. 7.
Validation of AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK based on TCGA data set. a ROC curve of AGO2. b ROC curve of CDC20. c ROC curve of CDCA5. d ROC curve of MCM10. e ROC curve of MYBL2. f ROC curve of TTK. g AGO2 expression at different stages of breast cancer. h CDC20 expression at different stages of breast cancer. i CDCA5 expression at different stages of breast cancer. j MCM10 expression at different stages of breast cancer. k MYBL2 expression at different stages of breast cancer. l TTK expression at different stages of breast cancer. m MCM10 expression and tumor size. n TTK expression and tumor size. o AGO2 expression and lymph-node metastasis. p CDC20 expression and lymph-node metastasis. q CDCA5 expression and lymph-node metastasis. r MCM10 expression and lymph-node metastasis. s MYBL2 expression and lymph-node metastasis. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
Fig. 8.
Correlation among AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK. a GSE42568 and b TCGA
Discussion
Breast cancer is the leading cause of cancer death in females and easy to recur. The high-throughput platforms for genomic analysis provided promising tools in medical oncology with great clinical applications, while it is difficult to use such a large number of genes for clinical application. Various genetic changes were found to regulate breast cancer initiation and progression. Currently, many biomarkers have been identified for the diagnosis and treatment of breast cancer. However, novel biomarkers are to be investigated for better understanding the mechanisms of tumor progression and prediction of prognosis. In the present study, we performed WGCNA to identify candidate biomarkers associated with the tumor grades of breast cancer. A total of 3771 DEGs were screened out and ten modules were identified via co-expression network analysis. Turquoise module had the highest association with tumor grade. We screened 58 genes with high functional significance from this module. Using Kaplan–Meier survival curves and log-rank test, AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK were negatively associated with the prognosis of breast cancer patients.
The protein encoded by AGO2 modulates RNA interference and is associated with tumor progression. AGO2 was upregulated in ERɑ-negative breast cancer cell lines compared with ERɑ-positive ones, and overexpression of AGO2 was a response to the epithelial growth factor receptor/mitogen-activated protein kinase signaling pathway. In addition, ERɑ-positive MCF7 cells transfected with AGO2 displayed enhanced proliferation, reduced cell–cell adhesion, and increased migratory ability (Adams et al. 2009). A recent study demonstrated that acetylation of AGO2 enhanced the maturation of miR-19b by recruiting pre-miR-19b form the miRNA precursor deposit complex. Immunohistochemistry staining of lung cancer tissues and xenograft mouse models confirmed that AGO2 acetylation levels were associated with poor prognosis in lung cancer patients (Zhang et al. 2018a). In the data set GSE42568, AGO2 was highly expressed in breast cancer tissues compared with normal breast tissues, and ROC curve indicated that AGO2 could excellently distinguish normal tissues from tumor tissues. However, the diagnostic efficiency was not observed in TCGA data set. One-way ANOVA test demonstrated that AGO2 was upregulated in ERɑ-negative, especially triple-negative tumors. Survival analysis revealed that high expression of AGO2 was correlated with poor survival. AGO2 is a potential prognostic biomarker.
CDC20 regulates cell cycle and interacts with several other proteins. CDC20 was recognized as an oncogene, and its overexpression was reported in various malignancies (Gao et al. 2018). CDC20 promoted proteasomal degradation of the tumor suppressor SMAR1, thus enhancing cell migration and invasion in breast cancer (Paul et al. 2017). In glioma, CDC20 knockdown enhanced the drug sensitivity of glioma cells to temozolomide, suggesting that CDC20 inactivation contributed to human cancer control (Wang et al. 2017). In our current analysis, CDC20 was highly expressed in breast cancer tissues compared with normal ones. ROC curve indicated that CDC20 exhibited excellent diagnostic efficiency. One-way ANOVA test demonstrated that CDC20 was highly expressed in triple-negative tumor and associated with tumor stages. Human Protein Atlas database with the immunohistochemistry staining of CDC20 was used to further investigate its translational levels. Our results indicated that CDC20 protein levels were significantly upregulated in breast cancer tissues compared with normal ones. Survival analysis revealed that high expression levels of CDC20 were associated with poor prognosis.
CDCA5 was overexpressed in various types of tumors and associated with the tumor progression (Chang et al. 2015; Chen et al. 2017; Zhang et al. 2018b). It was required for stable cohesion of chromatids during S and G2/M cell cycle phases. In gastric cancer, CDCA5 silence downregulated Cyclin E1 expression and suppressed proliferation of gastric cancer cells by inducing G1-phase arrest (Zhang et al. 2018b). CDCA5 knockdown also reduced viability and induced cycle arrest of hepatic cell carcinoma cells via downregulating CCNB1 and CDK1 (Shen et al. 2018). In addition, CDCA5 silencing in hepatic cell carcinoma was attributed to inactivation of the ERK/AKT pathway (Wang et al. 2018). The results of our study demonstrated that CDCA5 was overexpressed in breast cancer, especially in triple-negative tumor. Overexpression of CDCA5 was correlated with lymph-node metastasis and negatively correlated with the prognosis.
MCM10 encodes a protein modulating the initiation of eukaryotic genome replication. MCM10 was reported to be overexpressed in pancreatic, cervical, esophageal, urothelial, and breast cancers, and mutated in early gastric cancer specimens (Kang et al. 2013; Li et al. 2016; Lu et al. 2014; Peng et al. 2016). In breast cancer, highly MCM10 expression levels were associated with shorter survival time. MCM10 deficiency decreased cell proliferation and migration of breast cancer cells MCF7 (Mahadevappa et al. 2018). In both the data sets GSE42568 and TCGA, ROC curve indicated that MCM10 could effectively distinguish normal tissues from tumor ones. MCM10 overexpression was positively correlated with tumor size and lymph-node metastasis, and negatively with the survival possibilities.
MYBL2 is a transcription of the MYB family. It is a crucial regulator of cell proliferation, differentiation, and apoptosis, and involved in tumor progression (Fan et al. 2018; Sala 2005). MYBL2 was upregulated in various types of cancers and associated with poor patient outcome (Guan et al. 2018; Musa et al. 2017; Ren et al. 2015; Thorner et al. 2009), and previous studies revealed that MYBL2 overcome p53-induced G1 checkpoint arrest and DNA damage-induced G2 checkpoint arrest in p53 mutant cells, thus enhancing cancer cell cycle progression and survival (Lin et al. 1994; Mannefeld et al. 2009). In breast cancer cells, MYBL2 knockdown restored E-Cadherin, and suppressed cell invasion, growth and tumor formation. Conversely, MYBL2 overexpression increased expression of mesenchymal markers but downregulated E-cadherin. It was proposed that MYBL2 promoted tumor invasion via inducing epithelial–mesenchymal transition (Tao et al. 2015). In our study, MYBL2 was significantly upregulated in breast cancer. MYBL2 overexpression was associated with lymph-node metastasis and poor survival.
TTK is a critical mitotic checkpoint protein and essential for chromosome alignment at the centromere during mitosis, thus required for centrosome duplication. TTK mRNA levels were elevated in lung, anaplastic thyroid, and breast cancer (Landi et al. 2008; Yuan et al. 2006). Decreased TTK protein levels were associated with suppressed cell proliferation, migration, and invasion, suggesting the tumorigenic role of TTK (Chen et al. 2018; Thu et al. 2018). In the current study, ROC curve demonstrated that TTK could efficiently distinguish normal from tumor tissues. TTK mRNA expression was positively correlated with tumor size and negatively with the patient survival.
Conclusions
In the present study, a gene co-expression network was constructed using co-expression analysis, and a clinically significant module was identified. Functional enrichment analysis indicated that this clinically significant module regulated mitotic cell cycle process. In addition, we identified 58 hub genes closely correlated with the tumor grades, and 6 hub genes (AGO2, CDC20, CDCA5, MCM10, MYBL2, and TTK) significantly correlated with the survival of breast cancer patients. RNA-sequencing data from TCGA and immunohistochemical data from the Human Protein Atlas database were used to validate the credibility of co-expression results. Our studies enlightened further in vivo and in vitro studies to investigate the underlying molecular mechanisms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The excellent technical assistance of Jianing Tang is gratefully acknowledged.
Funding
This study was supported by National Natural Science Foundation of China (Grant No. 81800429), the Fundamental Research Funds for the Central Universities (Grant No. 2042018kf0065), Health Commission of Hubei Province Scientific Research Project (Grant No. WJ2019Q047), and Zhongnan Hospital of Wuhan University Science, Technology and Innovation Seed Fund (Grant Nos. znpy2017001 and znpy2018028).
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qiang Zhou and Jiangbo Ren have contributed equally to this work.
Contributor Information
Yu Xiao, Email: yu.xiao@whu.edu.cn.
Yan Gong, Email: yan.gong@whu.edu.cn.
References
- Adams BD, Claffey KP, White BA (2009) Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology 150:14–23. 10.1210/en.2008-0984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Chang IW et al (2015) CDCA5 overexpression is an indicator of poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladder. Am J Transl Res 7:710–722 [PMC free article] [PubMed] [Google Scholar]
- Chen T et al (2017) Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol Rep 38:1822–1832. 10.3892/or.2017.5846 [DOI] [PubMed] [Google Scholar]
- Chen X et al (2018) A novel USP9X substrate TTK contributes to tumorigenesis in non-small-cell lung cancer. Theranostics 8:2348–2360. 10.7150/thno.22901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Huang W, Sherman BT, Lempicki RA (2009a) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Huang W, Sherman BT, Lempicki RA (2009b) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Fan X et al (2018) B-Myb mediates proliferation and migration of non-small-cell lung cancer via suppressing IGFBP3. Int J Mol Sci. 10.3390/ijms19051479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B, Land S, Mamounas E, Dignam J, Fisher ER, Wolmark N (2001) Prevention of invasive breast cancer in women with ductal carcinoma in situ: an update of the National Surgical Adjuvant Breast and Bowel Project experience. Semin Oncol 28:400–418 [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet (Lond, Engl) 343:692–695 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang B, Wang Y, Shang G (2018) Cdc20 inhibitor apcin inhibits the growth and invasion of osteosarcoma cells. Oncol Rep 40:841–848. 10.3892/or.2018.6467 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM et al (2011) Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res 17:1082–1089. 10.1158/1078-0432.Ccr-10-2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Cheng W, Huang D, Wei A (2018) High MYBL2 expression and transcription regulatory activity is associated with poor overall survival in patients with hepatocellular carcinoma. Curr Res Transl Med 66:27–32. 10.1016/j.retram.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res Treat 123:725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Jung KH et al (2018) Adjuvant subcutaneous trastuzumab for HER2-positive early breast cancer: subgroup analyses of safety and active medical conditions by body weight in the SafeHer phase III study. The oncologist 23:1137–1143. 10.1634/theoncologist.2018-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang G et al (2013) Exome sequencing identifies early gastric carcinoma as an early stage of advanced gastric cancer. PloS One 8:e82770. 10.1371/journal.pone.0082770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil MY, Grandis JR, Shin DM (2003) Targeting epidermal growth factor receptor: novel therapeutics in the management of cancer. Expert Rev Anticancer Ther 3:367–380. 10.1586/14737140.3.3.367 [DOI] [PubMed] [Google Scholar]
- King CR, Kraus MH, Aaronson SA (1985) Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science (New York) 229:974–976 [DOI] [PubMed] [Google Scholar]
- Landi MT et al (2008) Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PloS One 3:e1651. 10.1371/journal.pone.0001651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WM et al (2016) MCM10 overexpression implicates adverse prognosis in urothelial carcinoma. Oncotarget 7:77777–77792. 10.18632/oncotarget.12795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D et al (1994) Constitutive expression of B-myb can bypass p53-induced Waf1/Cip1-mediated G1 arrest. Proc Natl Acad Sci USA 91:10079–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P et al (2014) Genome-wide gene expression profile analyses identify CTTN as a potential prognostic marker in esophageal cancer. PloS One 9:e88918. 10.1371/journal.pone.0088918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevappa R et al (2018) DNA replication licensing protein MCM10 promotes tumor progression and is a novel prognostic biomarker and potential therapeutic target in breast cancer. Cancers. 10.3390/cancers10090282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannefeld M, Klassen E, Gaubatz S (2009) B-MYB is required for recovery from the DNA damage-induced G2 checkpoint in p53 mutant cells. Cancer Res 69:4073–4080. 10.1158/0008-5472.can-08-4156 [DOI] [PubMed] [Google Scholar]
- Musa J, Aynaud MM, Mirabeau O, Delattre O, Grunewald TG (2017) MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 8:e2895. 10.1038/cddis.2017.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy J (2005) Extending survival with chemotherapy in metastatic breast cancer. Oncologist 10(Suppl 3):20–29. 10.1634/theoncologist.10-90003-20 [DOI] [PubMed] [Google Scholar]
- Paterson MC et al (1991) Correlation between c-erbB-2 amplification and risk of recurrent disease in node-negative breast cancer. Cancer Res 51:556–567 [PubMed] [Google Scholar]
- Paul D, Ghorai S, Dinesh US, Shetty P, Chattopadhyay S, Santra MK (2017) Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis 8:e2882. 10.1038/cddis.2017.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YP et al (2016) The expression and prognostic roles of MCMs in pancreatic cancer. PloS One 11:e0164150. 10.1371/journal.pone.0164150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MF et al (1997) HER-2/neu gene amplification characterized by fluorescence in situ hybridization: poor prognosis in node-negative breast carcinomas. J Clin Oncol 15:2894–2904. 10.1200/jco.1997.15.8.2894 [DOI] [PubMed] [Google Scholar]
- Redig AJ, McAllister SS (2013) Breast cancer as a systemic disease: a view of metastasis. J Intern Med 274:113–126. 10.1111/joim.12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F et al (2015) MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am J Cancer Res 5:1542–1552 [PMC free article] [PubMed] [Google Scholar]
- Rimawi M et al (2018) First-line trastuzumab plus an aromatase inhibitor, with or without pertuzumab, in human epidermal growth factor receptor 2-positive and hormone receptor-positive metastatic or locally advanced breast cancer (PERTAIN): a randomized open-label phase II trial. J Clin Oncol 36:2826–2835. 10.1200/jco.2017.76.7863 [DOI] [PubMed] [Google Scholar]
- Sagara Y et al (2018) Effectiveness of neo-adjuvant systemic therapy with trastuzumab for basal HER2 type breast cancer: results from retrospective cohort study of Japan Breast Cancer Research Group (JBCRG)-C03. Breast Cancer Res Treat 171:675–683. 10.1007/s10549-018-4873-0 [DOI] [PubMed] [Google Scholar]
- Sala A (2005) B-MYB, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur J Cancer (Oxford, England: 1990) 41:2479–2484. 10.1016/j.ejca.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Shen Z et al (2018) CDCA5 regulates proliferation in hepatocellular carcinoma and has potential as a negative prognostic marker. OncoTargets Ther 11:891–901. 10.2147/ott.s154754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL (1987) Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science (New York) 235:177–182 [DOI] [PubMed] [Google Scholar]
- Takada M et al (2018) Prediction of postoperative disease-free survival and brain metastasis for HER2-positive breast cancer patients treated with neoadjuvant chemotherapy plus trastuzumab using a machine learning algorithm. Breast Cancer Res Treat. 10.1007/s10549-018-4958-9 [DOI] [PubMed] [Google Scholar]
- Tao D, Pan Y, Jiang G, Lu H, Zheng S, Lin H, Cao F (2015) B-Myb regulates snail expression to promote epithelial-to-mesenchymal transition and invasion of breast cancer cell. Med Oncol (Northwood, London, England) 32:412. 10.1007/s12032-014-0412-y [DOI] [PubMed] [Google Scholar]
- Thorner AR, Hoadley KA, Parker JS, Winkel S, Millikan RC, Perou CM (2009) In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene 28:742–751. 10.1038/onc.2008.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thu KL et al (2018) Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc Natl Acad Sci USA 115:E1570–e1577. 10.1073/pnas.1719577115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainer AH, Thompson E, James PA (2011) BRCA and beyond: a genome-first approach to familial breast cancer risk assessment. Discov Med 12:433–443 [PubMed] [Google Scholar]
- Uhlen M et al (2010) Towards a knowledge-based Human Protein Atlas. Nat Biotechnol 28:1248–1250. 10.1038/nbt1210-1248 [DOI] [PubMed] [Google Scholar]
- Uhlen M et al (2015) Proteomics. Tissue-based map of the human proteome. Science (New York) 347:1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- von Minckwitz G et al (2018) Efficacy and safety of ABP 980 compared with reference trastuzumab in women with HER2-positive early breast cancer (LILAC study): a randomised, double-blind, phase 3 trial. Lancet Oncol 19:987–998. 10.1016/s1470-2045(18)30241-9 [DOI] [PubMed] [Google Scholar]
- Wang J et al (2017) Cdc20 overexpression is involved in temozolomide-resistant glioma cells with epithelial-mesenchymal transition. Cell Cycle (Georget) 16:2355–2365. 10.1080/15384101.2017.1388972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J et al (2018) Silencing of CDCA5 inhibits cancer progression and serves as a prognostic biomarker for hepatocellular carcinoma. Oncol Rep 40:1875–1884. 10.3892/or.2018.6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B et al (2006) Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res 12:405–410. 10.1158/1078-0432.Ccr-05-0903 [DOI] [PubMed] [Google Scholar]
- Zhang H et al (2018a) Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene. 10.1038/s41388-018-0530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shen M, Zhou G (2018b) Upregulation of CDCA5 promotes gastric cancer malignant progression via influencing cyclin E1. Biochem Biophys Res Commun 496:482–489. 10.1016/j.bbrc.2018.01.046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.