Abstract
Background
Cancer is a serious public health problem worldwide, and difficulty in early diagnosis has been the chief obstacle to improve the prognosis of patients. Recently, microRNAs (miRNAs) were widely studied to be potential biomarkers for cancer detection. miR-16 is a prevalent but sophisticated one. In the current study, we aimed to assess the diagnostic value of serum miR-16 for cancer detection.
Methods
A total of 1458 cancer patients, containing ten types of cancers, and 1457 non-cancer controls were recruited in this study. qRT-PCR was used for the amplification of miRNAs. In addition, a meta-analysis of reported studies was performed to confirm our findings systematically.
Results
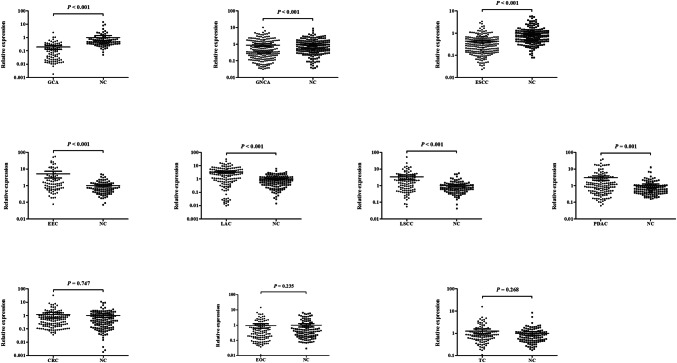
Consequently, miR-16 was down-regulated in ESCC, GCA and GNCA patients compared with NCs (all P < 0.001), while up-regulated in PDAC patients (P = 0.001), LAC, LSCC and EEC patients (all P < 0.001). But no significant differences were observed in CRC, EOC and TC patients when compared to NCs (P = 0.747, 0.235 and 0.268, respectively). The areas under the receiver operating characteristic (ROC) curve of miR-16 in GCA, ESCC, LAC, LSCC, GNCA, PDAC and EEC were 0.881, 0.780, 0.757, 0.693, 0.602, 0.614 and 0.681, respectively. Results of meta-analysis showed that miR-16 achieved an overall pooled sensitivity of 0.72, specificity of 0.79, and AUC of 0.85, suggesting that miR-16 was a promising biomarker in cancer detection.
Conclusions
We provided a comprehensive view of the diagnostic value of serum miR-16 in cancer diagnosis, and confirmed that circulating miR-16 could play an important role in cancer detection.
Electronic supplementary material
The online version of this article (10.1007/s00432-019-02849-8) contains supplementary material, which is available to authorized users.
Keywords: Cancer, Circulation, MicroRNA-16, Diagnosis, Meta-analysis
Introduction
Cancer remains an enormous burden on society in both developed and developing countries. According to the latest research, an estimated 14.1 million new cancer cases and 8.2 million cancer deaths occurred in 2012 worldwide (Torre et al. 2015). Risk factors for cancer include the adoption of lifestyle behaviors and reproductive changes, and deteriorating environment is also to blame. Among various cancers, lung and breast cancer are the most common malignancies and the leading causes of cancer death in men and women, respectively. Liver, stomach, colorectum and cervix uteri cancer are also frequently diagnosed cancers worldwide. What is more, most cancers are diagnosed in advanced-stage because of non-specific symptoms, and there is currently a lack of effective treatments for patients with advanced-stage cancer. Therefore, it is imperative to seek suitable diagnostic tools in early diagnosis of tumor. Currently, histological diagnosis of biopsy is still the gold standard for cancer detection. In spite of its relatively high sensitivity and specificity, this cancer screening is inconvenient and invasive (Wittmann and Jack 2010). Moreover, the circulating biomarkers, such as carcinoembryonic antigen (CEA), carbohydrate antigen 19-9 (CA19-9) and prostate-specific antigen (PSA), are limited in clinical practice for insufficient diagnostic accuracy (Hanash et al. 2011). Thus, reliable non-invasive markers are urgently needed for the early diagnosis of various cancers.
Recently, microRNAs (miRNAs) were widely studied in cancer biology. MiRNAs are endogenous 20–22 nt non-coding RNAs that post-transcriptionally regulate gene expression and function as oncogenes or tumor suppressors by degrading target mRNAs or blocking their translation (Bartel 2004; Calin and Croce 2006). Accumulating evidence has shown that miRNAs could be stably detected in circulating plasma or serum, and circulating miRNAs have emerged as reliable biomarkers for the early diagnosis of cancer (Mitchell et al. 2008). A series of studies have proved the diagnostic value of circulating miRNAs in various cancers (Johansen et al. 2016; Yamada et al. 2015; Zhou et al. 2015). Among numerous miRNAs, miR-16 is one of the most representative miRNAs, which has been extensively studied in many tumors. Generally, miR-16 was used as a reference gene for the quantification of circulating microRNAs in cancer patients (Song et al. 2012). Nevertheless, many studies have shown that circulating miR-16 could act as a diagnostic biomarker to discriminate cancer patients from non-cancer controls, and yet the results of diagnostic accuracy were inconsistent.
In view of the inconformity above, we measured the expression level of miR-16 using qRT-PCR in serum samples from non-cancer controls (NCs) and ten types of common cancers, including colorectal cancer (CRC), endometrioid endometrial cancer (EEC), epithelial ovarian cancer (EOC), esophageal squamous cell carcinoma (ESCC), gastric non-cardia adenocarcinoma (GNCA), gastric cardia adenocarcinoma (GCA), lung adenocarcinoma (LAC), lung squamous cell carcinoma (LSCC), pancreatic ductal adenocarcinoma (PDAC) and thyroid cancer (TC), to assess the diagnostic performance of miR-16 in various cancers. In addition, we performed a meta-analysis of previous reported studies to evaluate the pooled accuracy of circulating miR-16 in cancer detection.
Materials and methods
Clinical samples
A total of 2915 participants, which consist of 190 CRC patients vs. 197 NCs, 82 EEC patients vs. 90 NCs, 128 EOC patients vs. 128 NCs, 208 ESCC patients vs. 208 NCs, 219 GNCA patients vs. 212 NCs, 91 GCA patients vs. 100 NCs, 159 LAC patients vs. 159 NCs, 110 LSCC patients vs. 110 NCs, 155 PDAC patients vs. 137 NCs and 116 TC patients vs. 116 NCs, were recruited from the First Affiliated Hospital of Nanjing Medical University and the Affiliated Hospital of Jiangnan University between 2013 and 2016. All the cancer patients were histopathologically conformed by histopathological examination or biopsy, and all participants were without previous history of cancer and previous history of receiving chemotherapy or radiotherapy. And a group of 1457 normal controls (NCs) collected from 2013 to 2016 were recruited from a large pool of people seeking a routine health check-up at Hospital. People who showed no evidence of disease (including cancer and precancerous lesion) were selected as NCs. All the procedures were approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University and the Affiliated Hospital of Jiangnan University in compliance with the Declaration of Helsinki, and the written informed consent was obtained from each participant.
Sample preparation and RNA extraction
Five millilitres of peripheral blood sample was collected from each participant before initial treatment. Blood specimens were separated into serum and cellular fractions by centrifugation at 3000 rpm for 10 min and 12,000 rpm for 2 min within 12 h after collection. Serum samples were stored at − 80 °C for further processing.
Total RNA was extracted from 200 µl serum sample using the mirVana PARIS Kit (Ambion, Austin, TX, USA) in accordance with the manufacturer’s protocol. After denaturising, 5 µl of synthetic C. elegans miR-39 (cel-miR-39) (5 nM/L, RiboBio, Guangzhou, China) was spiked into each sample for controlling variability in RNA extraction and/or purification procedures. The ultraviolet spectrophotometer was used to evaluate the concentration and purity of the extracted total RNA.
Quantitative RT-PCR and data normalization
The amplification of miRNAs was conducted using the specific primers of reverse transcription (RT) and polymerase chain reaction (PCR) from Bulge-Loop™ miRNA qRT-PCR Primer Set (RiboBio, Guangzhou, China). The quantification of PCR product was evaluated by the level of fluorescence in emitted by SYBR Green (SYBR® Premix Ex Taq™ II, TaKaRa). RT reactions were carried out at 42 °C for 60 min followed by 70 °C for 10 min. The qRT-PCR was conducted on LightCycler® 480 Real-Time PCR System (Roche Diagnostics, Mannheim, Germany) in 384-well plates at 95 °C for 20 s, followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s and then 70 °C for 10 s. The specificity of PCR products was evaluated by the melting curve analysis. All reactions were performed in triplicate. The relative expression levels of miR-16 were determined using the 2−△△Ct method relative to the control miRNA (cel-miR-39), ΔCt = CtmiR− 16 − Ctcel−miR− 39.
Statistical analysis
Mann–Whitney test was used to analyze differential miRNAs expression between cancer patients and NCs. Clinical characteristics among different groups and their associations with miRNA were evaluated with one-way ANOVA or χ2 test. Receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) were used to estimate the diagnostic value of miR-16 for various cancers. All the statistical analyses were performed using SPSS software (version 20.0, IBM, USA). A P value < 0.05 was defined statistically significant.
Meta-analysis
A comprehensive literature search in PubMed, Embase, Web of Science and the Cochrane Library until August 16, 2016, was conducted using the following search terms: (neoplasms or cancer or tumor or malignancy) and (microRNA-16 or miRNA-16 or miR-16) and (diagnosis or sensitivity or specificity). We manually searched the references of articles to explore potentially additional studies. And publication language was limited to English.
Studies were eligible if they met the following criteria: (1) expression of circulating (blood, plasma, and serum) miR-16 was assessed in any type of cancer; (2) cancer patients should be confirmed by a golden standard test; and (3) the association between miR-16 expression levels and cancer diagnosis was investigated. Articles were excluded if they met the exclusion criteria: (1) reviews or comments or letters; and (2) studies were identified as duplicates or lack of key data.
Two investigators carefully and independently reviewed eligible articles and extracted the following data: first author, publication year, country of origin, ethnicity, total number of participants, cancer types, source of control, specimen, sensitivity, specificity and other relevant data for meta-analysis. Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) was used to assess the quality of selected studies (Whiting et al. 2011).
All statistical analyses were performed using Meta-Disc 1.4 and Stata 12.0 software. Numbers of true-positive (TP), false-positive (FP), false-negative (FN), and true-negative (TN) were analyzed and pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) were calculated using the bivariate meta-analysis model. The summary receiver operator characteristic (SROC) curve was also plotted to evaluate the diagnostic accuracy of miR-16 in cancer. Deek’s funnel plot was performed to assess the publication bias.
Results
Characteristics of study subjects
A total of 2915 participants were recruited in this study. Ten types of cancers, including CRC, EEC, EOC, ESCC, GNCA, GCA, LAC, LSCC, PDAC and TC. The clinical features of patients and NCs are listed in Table 1 and Table S1–S10. For each type of cancer, there was no significant difference in the distribution of gender or age between patients and NCs.
Table 1.
Clinical characteristics of cancer patients and non-cancer controls
| Cancer type | Number | Age (mean ± SD) | Gender | |
|---|---|---|---|---|
| Male | Female | |||
| CRC | 190 | 63.72 ± 10.29 | 123 (64.7%) | 67 (35.3%) |
| EEC | 82 | 61.02 ± 15.83 | 0 | 82 |
| EOC | 128 | 57.73 ± 9.23 | 0 | 128 |
| ESCC | 208 | 65.04 ± 11.55 | 122 (58.7%) | 86 (42.3%) |
| GNCA | 219 | 60.78 ± 10.83 | 123 (56.2%) | 96 (43.8%) |
| GCA | 91 | 66.13 ± 12.46 | 68 (74.7%) | 23 (25.3%) |
| LAC | 159 | 61.38 ± 11.21 | 102 (64.2%) | 57 (35.8%) |
| LSCC | 110 | 62.57 ± 13.37 | 87 (79.1%) | 23 (20.9%) |
| PC | 155 | 62.38 ± 12.59 | 89 (57.4%) | 66 (42.6%) |
| TC | 116 | 44.78 ± 8.06 | 30 (25.9%) | 86 (74.1%) |
Expression patterns of serum miR-16
The expression level of serum miR-16 was evaluated in all ten common types of cancers and in the ten independent NCs. As shown in Fig. 1, the scatter dot plot demonstrated the relative expression of miR-16 in cancers and NCs. However, the results of different cancers varied. In digestive system cancers, miR-16 was down-regulated in ESCC, GCA and GNCA patients compared with NCs (all P < 0.001), while up-regulated in PDAC patients (P = 0.001). But there was no significant difference of miR-16 expression was observed in CRC patients when compared to NCs (P = 0.747). Unlike esophagus and gastric cancers but similar to pancreatic cancer, miR-16 was significantly overexpressed in NSCLC (non-small-cell lung cancer), including LAC and LSCC (both P < 0.001). In female genital tumors, such as EEC and EOC, and in TC which generally occurred in women, we only observed up-regulation of miR-16 in EEC (P < 0.001), but there was no significant difference in EOC and TC (P = 0.235 and 0.268, respectively).
Fig. 1.
Expression level of miR-16 in the serum of ten cancer patients and NCs. Horizontal line: mean with 95% CI
Diagnostic value of serum miR-16
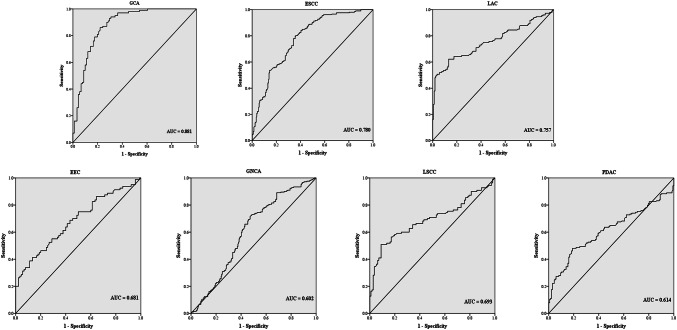
ROC curve analysis was applied to evaluate the diagnostic value of serum miR-16 in the groups between which statistically significant expression level of miR-16 was observed. As shown in Fig. 2, the diagnostic accuracy of miR-16 was relatively good in GCA against NCs (AUC = 0.881, 95% CI 0.831–0.930, sensitivity = 86.0%, specificity = 78.0%). And the diagnostic performance of miR-16 in discriminating ESCC from NCs (AUC = 0.780, 95% CI 0.736–0.824, sensitivity = 80.2%, specificity = 64.0%) and LAC from NCs (AUC = 0.757, 95% CI 0.701–0.813, sensitivity = 62.2%, specificity = 82.8%) was also meaningful. However, miR-16 exhibited relatively low AUC values of 0.693 (95% CI 0.622–0.765, sensitivity = 50.9%, specificity = 91.0%) for LSCC, 0.602 (95% CI 0.548–0.656, sensitivity = 73.0%, specificity = 51.4%) for GNCA, 0.614 (95% CI 0.548–0.679, sensitivity = 48.1%, specificity = 81.0%) for PDAC and 0.681 (95% CI 0.601–0.762, sensitivity = 41.3%, specificity = 86.7%) for EEC in the discrimination of cancer patients from NCs.
Fig. 2.
Receiver-operating characteristic (ROC) curves for serum miR-16 to discriminate EEC, ESCC, GCA, GNCA, LAC, LSCC and PDAC patients from NCs, respectively. AUC: areas under the curve
Meta-analysis of circulating miR-16 in cancer detection
After carefully and strictly filtering, a total of 13 eligible articles were included in the final meta-analysis (El-Abd et al. 2015; Ell et al. 2013; Fan et al. 2016; Fang et al. 2012; Gao et al. 2014; Guo et al. 2016; Liu et al. 2013; Maclellan et al. 2012; Mahn et al. 2011; Qu et al. 2011; Wang et al. 2014; Zhang et al. 2015; Zhu et al. 2014). The main characteristics of the 13 included studies are listed in Table 2. The publication year of the 13 articles range from 2011 to 2015. A total of 2120 subjects including Asians, Caucasians and a few Africans were enrolled in our meta-analysis. Serum or plasma miR-16 was evaluated in various cancers including gastric cancer, NSCLC, hepatocellular carcinoma, pancreatic cancer, prostate cancer, oral cancer, melanoma, NPC (nasopharyngeal carcinoma) and DLBCL (diffuse large B cell lymphoma). The QUADAS-2 tool was used to assess the quality of articles, and most studies turned out to be relatively high quality (Figure S1).
Table 2.
Main characteristics of all studies included in the meta-analysis
| First author | Year | Country | Ethnicity | Sample number | Cancer type | Sample | Test method | Normalizer | Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Controls | ||||||||||
| Mahn 12 | 2011 | Germany | Caucasian | 37 | 18BPH + 20HI | Prostate Cancer | Serum | qRT-PCR (SYBR Green) | cel-miR-39 | 54.1 | 68.4 |
| Qu 13 | 2011 | USA | Caucasian | 105 | 107CLD | HCC | Serum | qRT-PCR (TaqMan) | U6 snRNA | 72.1 | 88.8 |
| MacLellan 14 | 2012 | Canada | Caucasian | 30 | 26 | HRL | Serum | qRT-PCR (SYBR Green) | ROX reference dye | 61.5 | 93.3 |
| Fang 15 | 2012 | China | Asian | 75 | 77 | DLBCL | Serum | qRT-PCR (SYBR Green) | cel-miR-39 | 94.0 | 51.0 |
| Ell 16 | 2013 | USA | Caucasian | 54 | 42 | BM | Serum | qRT-PCR (TaqMan) | RNU6B | 95.0 | 82.1 |
| Wang 17 | 2014 | China | Asian | 50 | 47 | Gastric cancer | Serum | qRT-PCR (TaqMan) | U6 snRNA | 79.0 | 78.0 |
| EL-Abd 18 | 2015 | Egyptian | African | 40 | 40 chronic HCV | HCC | Serum | qRT-PCR (TaqMan) | RNU48 | 57.5 | 70.0 |
| Fan 19 | 2015 | China | Asian | 94 | 58 | NSCLC | Serum | qRT-PCR (TaqMan) | Standard curve | 88.0 | 86.0 |
| Guo 20 | 2016 | China | Asian | 120 | 120 | Melanoma | Serum | qRT-PCR (SYBR Green) | cel-miR-39 | 80.0 | 71.7 |
| Liu 21 | 2013 | China | Asian | 217 | 73 | NPC | Plamsa | qRT-PCR (SYBR Green) | U6 snRNA | 68.0 | 50.7 |
| Zhu 22 | 2014 | China | Asian | 160 | 124 | GNCA | Plamsa | qRT-PCR (TaqMan) | cel-miR-39 | 75.0 | 92.3 |
| Gao 23 | 2014 | China | Asian | 70 | 50 | PC | Plamsa | qRT-PCR (TaqMan) | cel-miR-39 | 73.7 | 86.4 |
| Zhang 24 | 2015 | China | Asian | 155 | 111 | Gastric cancer | Plamsa | qRT-PCR (TaqMan) | cel-miR-39 | 49.0 | 91.0 |
BPH benign prostate hyperplasia, HI healthy volunteers, HCC hepatocellular carcinoma, CLD chronic liver disease, HRL oral cancer or carcinoma in situ, DLBCL diffuse large B cell lymphoma, BM bone metastasis, HCV hepatitis C virus, NSCLC non-small cell lung cancer, NPC nasopharyngeal carcinoma, GNCA gastric non-cardia adenocarcinoma, PC pancreatic cancer
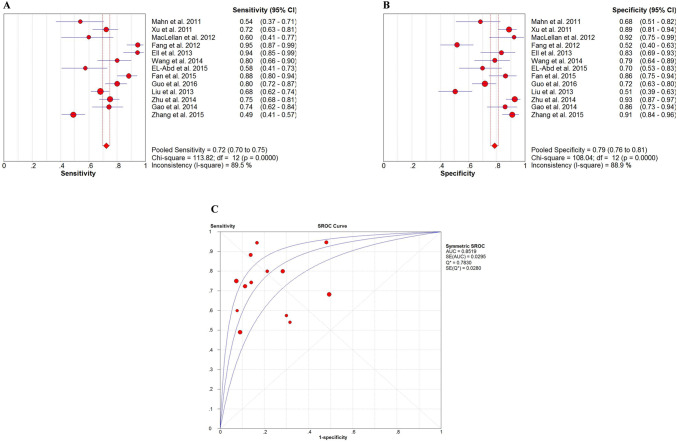
The forest plots of sensitivity and specificity for circulating miR-16 in detecting cancers are shown in Fig. 3. The pooled results for sensitivity and specificity were 0.72 (95% CI 0.70–0.75) and 0.79 (95% CI 0.76–0.81), respectively. And the pooled PLR, NLR and DOR were 3.70 (95% CI 2.51–5.45), 0.33 (95% CI 0.24–0.44) and 12.84 (95% CI 6.88–23.98), respectively (Figure S2). The SROC curve with an overall AUC of 0.8519 indicated a relatively high diagnostic accuracy of circulating miR-16 in cancers.
Fig. 3.
Meta-analysis of included studies in differentiating cancer from NCs. a The forest plots of sensitivity for miR-16 in cancer diagnosis. b The forest plots of specificity for miR-16 in cancer diagnosis. c Summary receiver operating characteristic (SROC) curve for miR-16 in cancer diagnosis
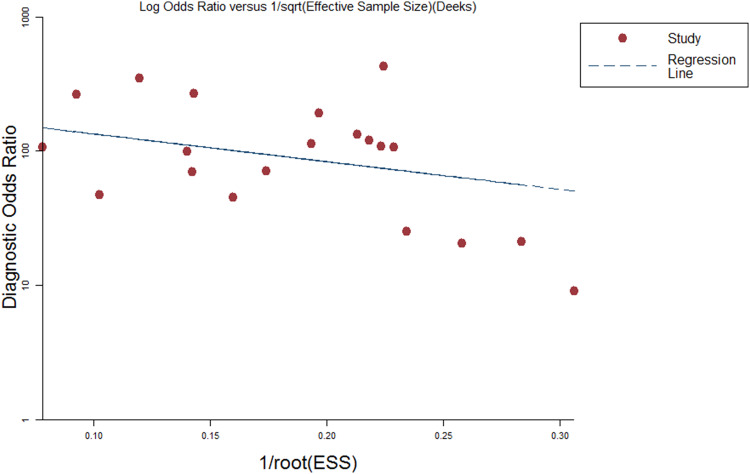
Publication bias of the included studies was checked by Deeks’ funnel plot asymmetry test. The slope coefficient was associated with a P value of 0.144 suggesting an existing low likelihood of publication bias (Fig. 4).
Fig. 4.
Deeks et al.’s funnel plots asymmetry test with regression line to explore publication bias
Discussion
Despite the improvements in diagnostic and therapeutic approaches, malignant tumors still remain one of the leading causes of death all over the world. Thus, novel biomarkers are urgently needed for cancer early detection. Over the past decade, emerging evidence has shown that circulating miRNAs played an important role in the diagnosis of various cancers. Among the various miRNAs, miR-16 is a relatively special one. Although the researches on miR-16 develop fast, the role of miR-16 in cancer is dubious. Interestingly, circulating miR-16 was found as a suppressor gene in many researches but an oncogene in some other studies. It was also common to apply miR-16 as the housekeeping gene for PCR data normalization in some cancers. We were very curious about the discrepancy between different studies and cancer types. In view of this, in the present study, we evaluated the expression level of serum miR-16 in various cancers and non-cancer controls.
The results were pretty interesting. The expression levels of serum miR-16 varied in different tumors. Compared with NCs, miR-16 exhibited lower differential expression levels in ESCC, GCA and GNCA patients, which implied that miR-16 might function as a tumor suppressor in esophageal and stomach tumors. In a previous study, plasma miR-16 was validated as a down-regulated biomarker in GC (Zhang et al. 2015). A classic paper confirmed that miR-16 was located at 13q14 and was down-regulated in chronic lymphocytic lymphoma (Calin et al. 2002). This research team further revealed that miR-16 negatively regulated B-cell lymphoma 2 (Bcl2) at the post-transcriptional level to induce apoptosis (Cimmino et al. 2005). The similar mechanism was also observed in gastric cancer (Xia et al. 2008). Meanwhile, HGF/c-Met pathway might be a potential target of miR-16 in GC (Li et al. 2016). However, in contrast, miR-16 was proved to have the oncogenic function in GC and ESCC in some other studies (Ren et al. 2016; Wang et al. 2014). This oncogenic role of miR-16 in cancers was embodied in our study, which showed the overexpression of miR-16 in PDAC, NSCLC and EEC. Liu et al. reported that plasma miR-16 was significantly aberrantly up-regulated in the pancreatic cancer compared with NC (Liu et al. 2012). Besides in circulation, miR-16 was overexpressed in pancreatic cancer tissues. Functional studies illustrated that up-regulation of miR-16 could result in the reduction of dendritic cells and thus diminish immune responses, which led to immune escape and limitless proliferation of cancer cells (Min et al. 2013). In addition, miR-16 might target transcriptional corepressor, the silencing mediator for retinoid and thyroid hormone receptor (SMRT) and modulate NF-kappaB-regulated transactivation of interleukin-8 gene, which was a key gene in tumorigenesis and metastasis of cancers (Zhou et al. 2012). In contradiction to our results, Fan et al. found that serum miR-16 was significantly down-regulated in NSCLC (Fan et al. 2016). This may be because miR-16 directly targeted hepatoma-derived growth factor (HDGF) to inhibit growth, colony formation, migration and invasion in NSCLC (Ke et al. 2013). There are no data assessing the diagnostic role of circulating miR-16 in EEC patients. In the present study, the expression of miR-16 in CRC, EOC, TC and NCs was no difference. The diagnostic potential of miR-16 in ovarian cancer patients has been studied in previous studies, which also identified that the expression level of miR-16 was similar between the patients and healthy controls (Meng et al. 2015). That might be why miR-16 has been chosen as an internal reference gene for data normalization in some cancers. Results of the present study suggested that miR-16 could act as a tumor suppressor in one scenario, but an oncogenic miRNA (oncomiR) in another, or even served as a suitable reference gene in the circulation for cancers. The duplicity of miR-16 might be because of the imperfect complementarity of the interactions between miRNAs and their target genes. It was known that one miRNA can target hundreds of mRNAs, and one mRNA can be regulated by a few miRNAs. In particular, specific miRNAs, such as miR-16, can simultaneously produce competing oncogenic and tumor suppressive effects by suppressing both tumor suppressive mRNAs and oncogenic mRNAs, respectively. Moreover, miRNAs can modulate tumor-modifying extrinsic factors to reach a dynamic balance, which determines whether miRNAs produce oncogenic or tumor suppressive effects (Svoronos et al. 2016). More evidence needs to be dug to further understand these mechanisms from a holistic standpoint.
To examine the reported diagnostic accuracy of circulating miR-16 in cancer detection, we further performed a meta-analysis on 13 previous eligible studies. As it turned out, circulating miR-16 achieved an overall pooled sensitivity of 0.72, specificity of 0.79, and AUC of 0.85, suggesting that miR-16 was a promising biomarker in circulation for discriminating cancer and non-cancer. The DOR, an important indicator for evaluation of diagnostic accuracy (Glas et al. 2003), was presented as 12.84, indicating a moderate diagnostic accuracy. Meanwhile, the pooled PLR and NLR were 3.70 and 0.33, respectively. Both the PLR and NLR were not ideal enough, but they demonstrated a more particular knowledge of diagnostic accuracy of miR-16. Obviously, there were still several limitations. First, the number of eligible previous articles was relatively small. Even so, no significant publication bias was observed. Second, the different reference for RT-PCR or inconsistent cut-off values across studies could affect the final outcome of experiments, which may also be the potential source of heterogeneity in the meta-analysis. Besides that, only one article was conducted on the African populations and provided limited data. Despite of the limitations, we performed the first meta-analysis to investigate the diagnostic value of circulating miR-16 for cancer detection.
Conclusions
Taken together, the present study provided a more comprehensive view of the diagnostic value of serum miR-16 in cancer detection based on a large-scale population. To our knowledge, this is the first study to investigate the effectiveness of serum miR-16 for the diagnosis of various cancers, and our findings suggest that serum miR-16 is a promising diagnostic biomarker. Furthermore, meta-analysis of previous studies further confirmed that circulating miR-16 could serve as a potential biomarker for cancer diagnosis. We are conducting the similar experiment of plasma miR-16, and further, more in-depth research is warranted.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1 Risk of bias and applicability concerns graph a review of authors’ judgments about each domain presented as percentages across included studies.
Figure S2 Forest plots of pooled A: positive likelihood ratio (PLR); B: negative likelihood ratio (NLR); C: diagnostic odds ratio (DOR) for miR-16 in cancer diagnosis.
Acknowledgements
We thank Ping Liu and Wei Zhu (Department of Oncology, the First Affiliated Hospital of Nanjing Medical University) for constructive suggestion.
Author contributions
HZB, CWJ and DYP performed experiments; HZB conducted the meta-analysis; GQ and SD reviewed eligible articles; WXH and MY provided the serum samples of patients and normal controls; HZB and ZX drafted the manuscript; ZX and HD designed the study, critically interpreted results; All authors read and approved the final manuscript.
Funding
Supported by grants: (1) Wuxi Health and Family Planning Commission Project; Grant number: Q201724. (2) Kunshan Social Development Science and Technology Project; Grant number: KS1643.
Compliance with ethical standards
Conflict of interest
All authors declare that they have no conflict of interest.
Ethics approval
All the procedures were approved by the Ethics Committee of the First Affiliated Hospital of Nanjing Medical University and the Affiliated Hospital of Jiangnan University in compliance with the Declaration of Helsinki, and the written informed consent was obtained from each participant.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zebo Huang, Wenjiao Chen and Yiping Du contributed equally to this work.
Contributor Information
Xin Zhou, Email: ivorchou1989@126.com.
Dong Hua, Email: huanglixin1123@163.com.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11):857–866 [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99(24):15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102(39):13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Abd NE, Fawzy NA, El-Sheikh SM, Soliman ME (2015) Circulating miRNA-122, miRNA-199a, and miRNA-16 as biomarkers for early detection of hepatocellular carcinoma in egyptian patients with chronic hepatitis C virus Infection. Mol Diagn Therapy 19(4):213–220 [DOI] [PubMed] [Google Scholar]
- Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y (2013) Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer cell 24(4):542–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Qi H, Teng J, Su B, Chen H, Wang C, Xia Q (2016) Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol 37(6):7777–7784 [DOI] [PubMed] [Google Scholar]
- Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, Fan L, Miao KR, Liu P, Xu W, Li JY (2012) Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol 91(4):553–559 [DOI] [PubMed] [Google Scholar]
- Gao L, He SB, Li DC (2014) Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clin Lab 60(1):73–77 [DOI] [PubMed] [Google Scholar]
- Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PMM (2003) The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 56(11):1129–1135 [DOI] [PubMed] [Google Scholar]
- Guo S, Guo W, Li S, Dai W, Zhang N, Zhao T, Wang H, Ma J, Yi X, Ge R, Wang G, Gao T, Li C (2016) Serum miR-16: a potential biomarker for predicting melanoma prognosis. J Investig Dermatol 136(5):985–993 [DOI] [PubMed] [Google Scholar]
- Hanash SM, Baik CS, Kallioniemi O (2011) Emerging molecular biomarkers–blood-based strategies to detect and monitor cancer. Nat Rev Clin Oncol 8(3):142–150 [DOI] [PubMed] [Google Scholar]
- Johansen JS, Calatayud D, Albieri V, Schultz NA, Dehlendorff C, Werner J, Jensen BV, Pfeiffer P, Bojesen SE, Giese N, Nielsen KR, Nielsen SE, Yilmaz M, Hollander NH, Andersen KK (2016) The potential diagnostic value of serum microRNA signature in patients with pancreatic cancer. Int J Cancer 139(10):2312–2324 [DOI] [PubMed] [Google Scholar]
- Ke Y, Zhao W, Xiong J, Cao R (2013) Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS lett 587(18):3153–3157 [DOI] [PubMed] [Google Scholar]
- Li S, Zhang H, Wang X, Qu Y, Duan J, Liu R, Deng T, Ning T, Zhang L, Bai M, Zhou L, Wang X, Ge S, Ying G, Ba Y (2016) Direct targeting of HGF by miR-16 regulates proliferation and migration in gastric cancer. Tumour Biol [DOI] [PubMed]
- Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X (2012) Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer 131(3):683–691 [DOI] [PubMed] [Google Scholar]
- Liu X, Luo HN, Tian WD, Lu J, Li G, Wang L, Zhang B, Liang BJ, Peng XH, Lin SX, Peng Y, Li XP (2013) Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol Ther 14(12):1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclellan SA, Lawson J, Baik J, Guillaud M, Poh CF, Garnis C (2012) Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med 1(2):268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Muller SC, Ellinger J (2011) Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology 77(5):1265 e9–e16 [DOI] [PubMed] [Google Scholar]
- Meng X, Joosse SA, Muller V, Trillsch F, Milde-Langosch K, Mahner S, Geffken M, Pantel K, Schwarzenbach H (2015) Diagnostic and prognostic potential of serum miR-7, miR-16, miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer patients. Br J Cancer 113(9):1358–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Liang X, Zhang M, Zhang Y, Mei S, Liu J, Liu J, Su X, Cao S, Zhong X, Li Y, Sun J, Liu Q, Jiang X, Che Y, Yang R (2013) Multiple tumor-associated microRNAs modulate the survival and longevity of dendritic cells by targeting YWHAZ and Bcl2 signaling pathways. J Immunol 190(5):2437–2446 [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105(30):10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M (2011) Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol 45(4):355–360 [DOI] [PubMed] [Google Scholar]
- Ren C, Chen H, Han C, Fu D, Wang D, Shen M (2016) High expression of miR-16 and miR-451 predicating better prognosis in patients with gastric cancer. J Cancer Res Clin Oncol 142(12):2489–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, Ma X, Han S, Zhang Z (2012) Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci 57(4):897–904 [DOI] [PubMed] [Google Scholar]
- Svoronos AA, Engelman DM, Slack FJ (2016) OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res 76(13):3666–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA 65(2):87–108 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Wu Z, Sun R, Jin H, Ma J, Liu L, Ling R, Yi J, Wang L, Bian J, Chen J, Li N, Yuan S, Yun J (2014) Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med Oncol 31(12):298 [DOI] [PubMed] [Google Scholar]
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, Group Q- (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Int Med 155(8):529–536 [DOI] [PubMed] [Google Scholar]
- Wittmann J, Jack HM (2010) Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta 1806(2):200–207 [DOI] [PubMed] [Google Scholar]
- Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D (2008) miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer 123(2):372–379 [DOI] [PubMed] [Google Scholar]
- Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, Kou T, Kusaka T, Sasaki Y, Yagi M, Higurashi T, Yukawa N, Amanuma Y, Kikuchi O, Muto M, Ueno Y, Nakajima A, Chiba T, Boland CR, Goel A (2015) Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res 21(18):4234–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma Y, Chen Y, Pan F, Wang K, Ni J, Jin W, He X, Su H, Cui D (2015) Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics 5(7):733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Li X, Hu G, Gong AY, Drescher KM, Chen XM (2012) miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PloS one 7(1):e30772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, Wu Y, Qi L, Fan Y, Chen Y, Ding Y, Xu J, Qian J, Huang Z, Wang T, Zhu D, Shu Y, Liu P (2015) Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep 5:11251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Ren C, Han J, Ding Y, Du J, Dai N, Dai J, Ma H, Hu Z, Shen H, Xu Y, Jin G (2014) A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer 110(9):2291–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.