Abstract
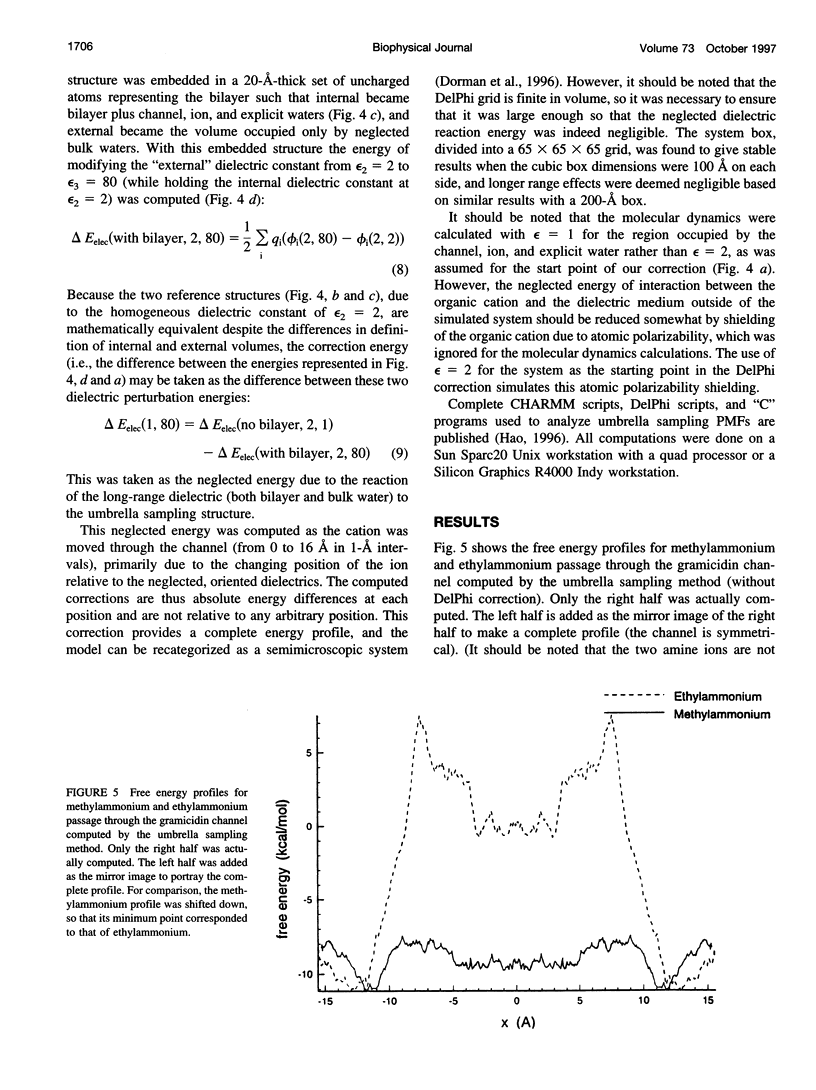
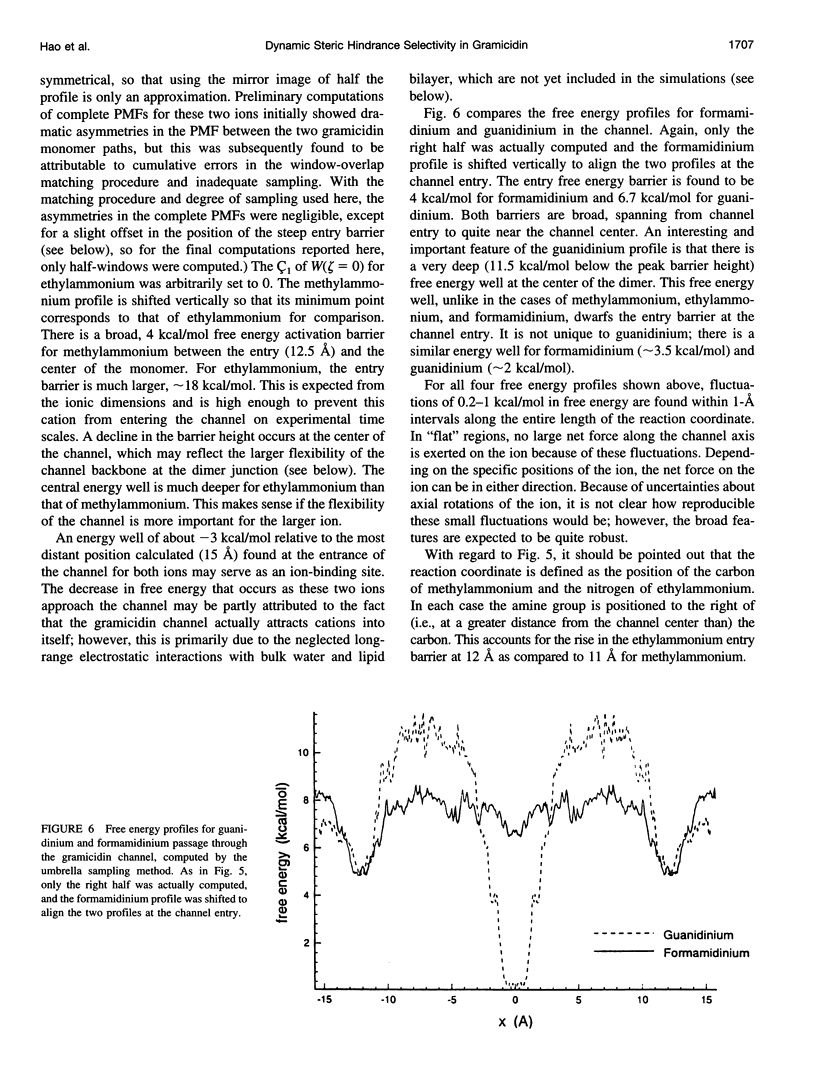
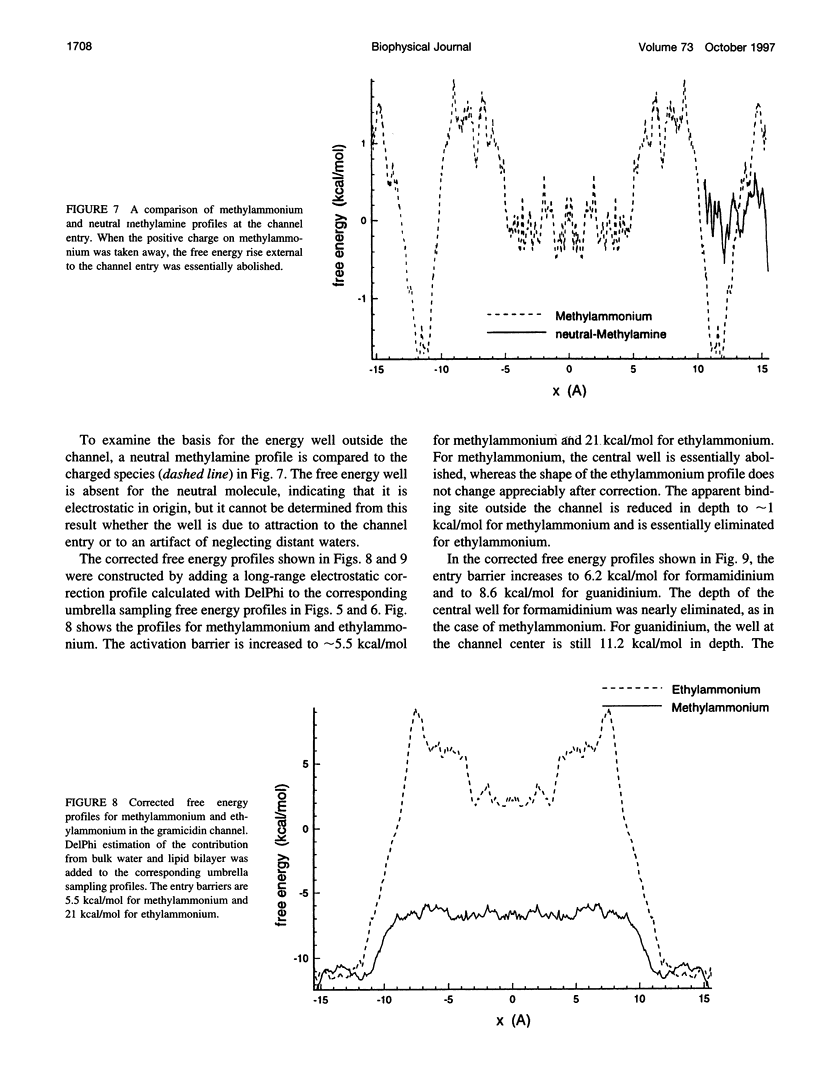
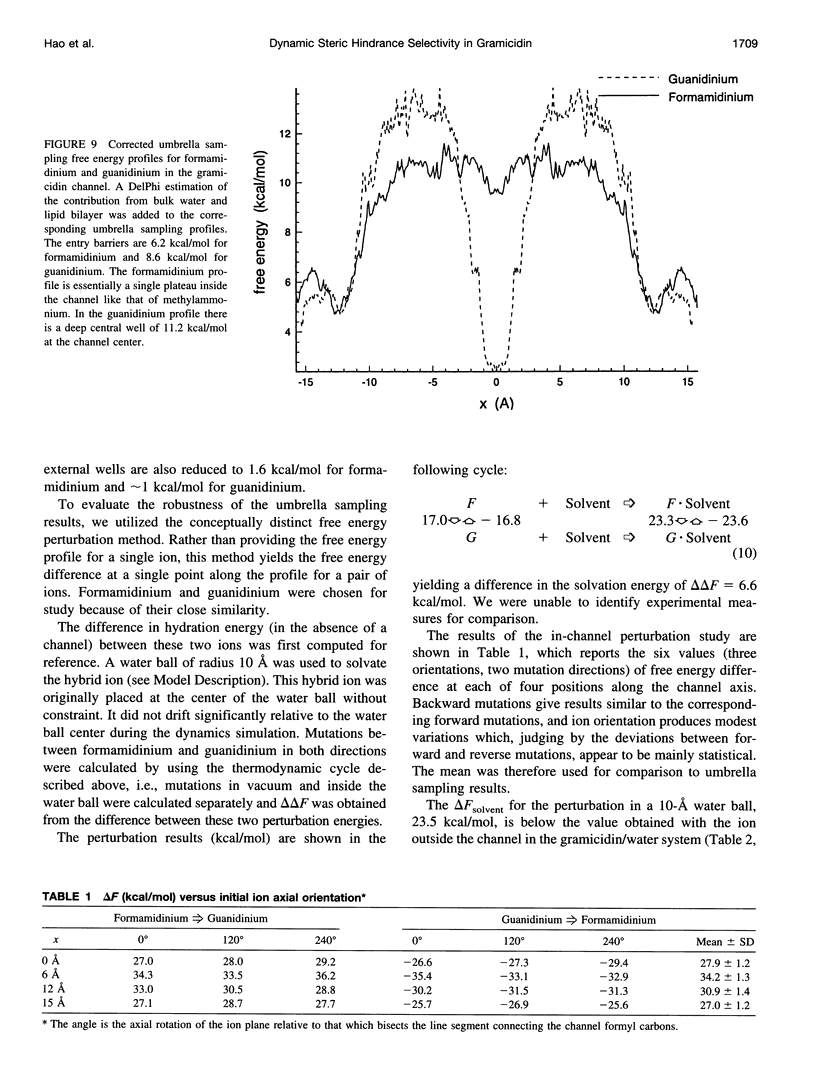
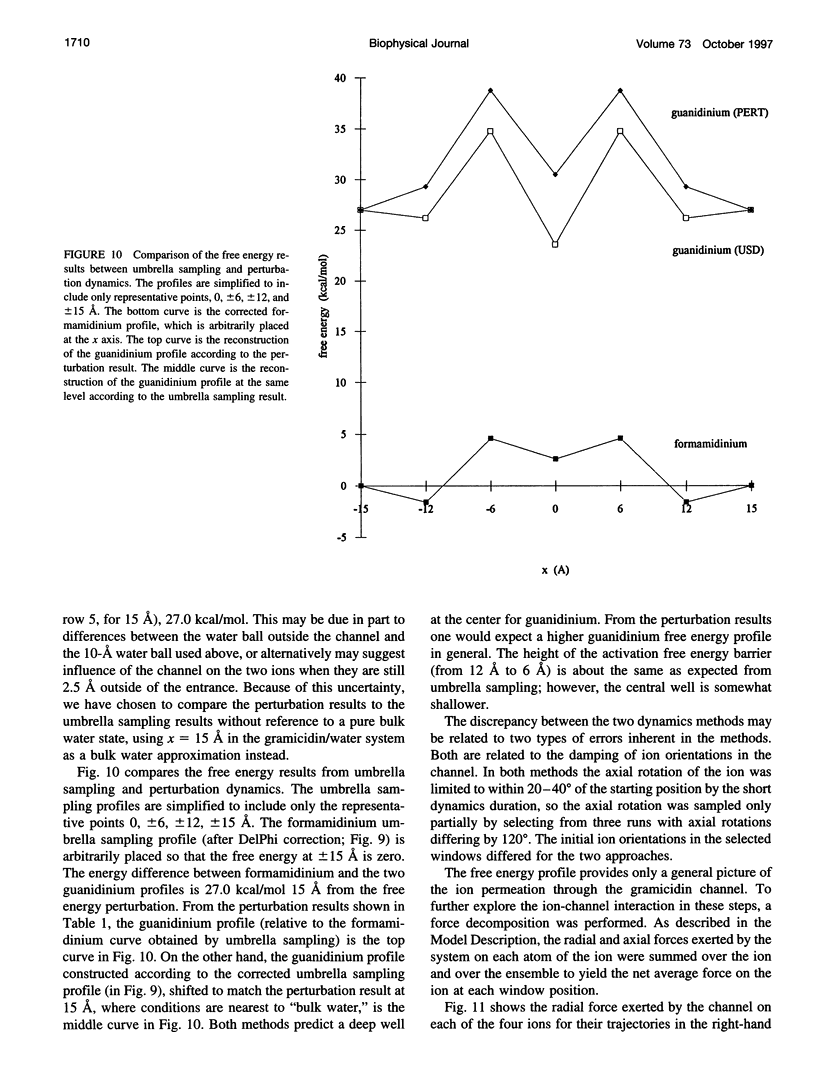
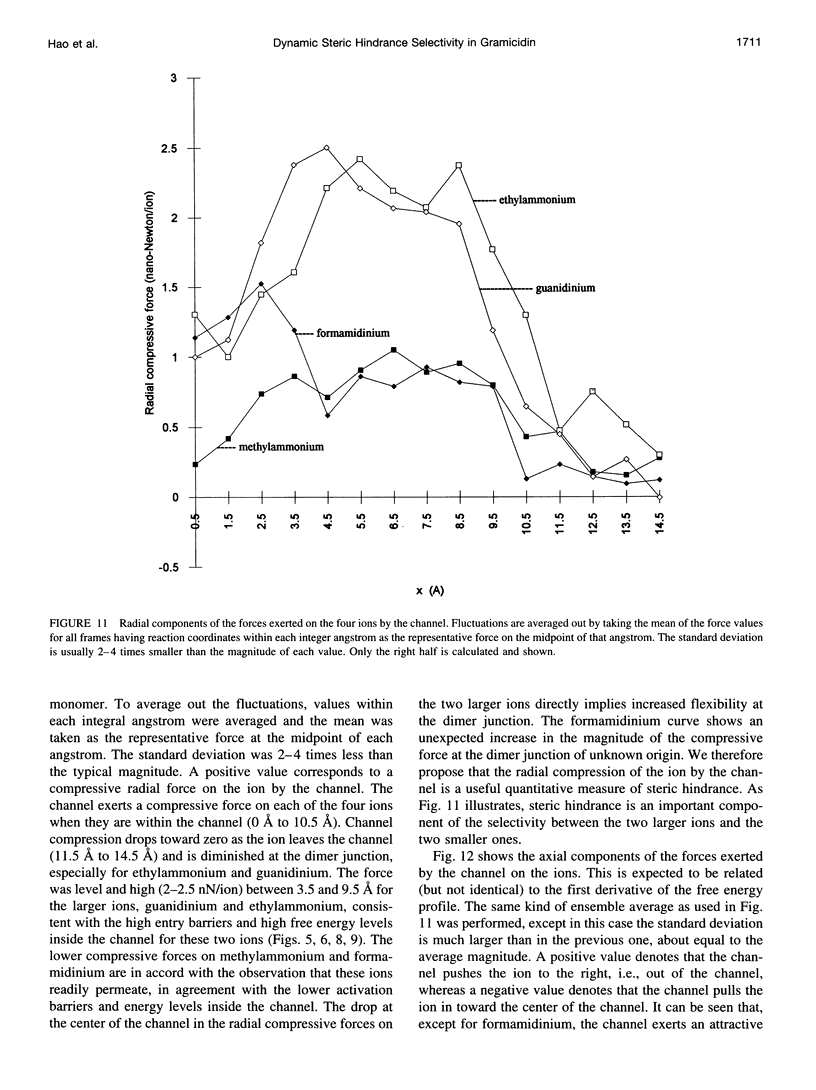
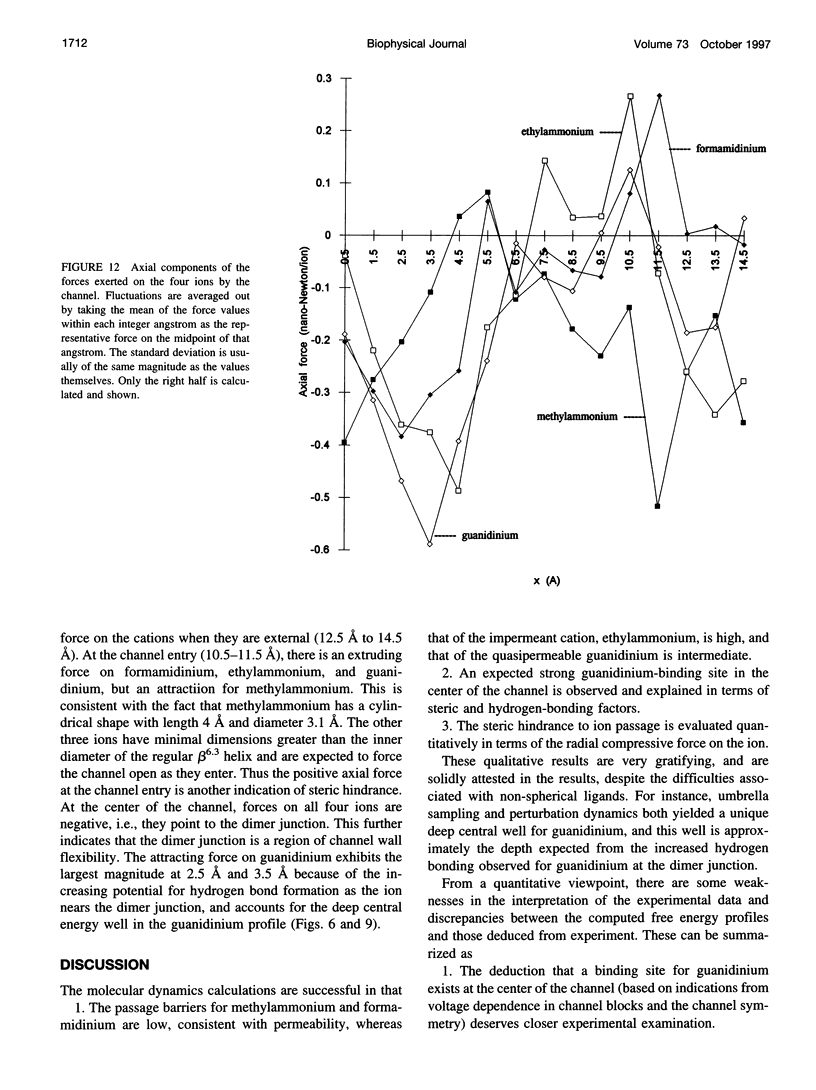
The free energy profiles for four organic cations in right-handed single-helix gramicidin A dimers were computed by using umbrella sampling molecular dynamics with CHARMM. Ion-water column translocations were facilitated by using a novel "water-tunnel" approach. The overlapping pieces of free energy profile for adjacent windows were selected from three trajectories that differed in initial ion rotation and were aligned by the method of umbrella potential differences. Neglected long-range electrostatic energies from the bulk water and the bilayer were computed with DelPhi and added to the profile. The approach was corroborated for the formamidinium-guanidinium pair by using perturbation dynamics at axial positions 0, 6, 12, and 15 A from the channel center. The barrier to ethylammonium entry was prohibitive at 21 kcal/mol, whereas for methylammonium it was 5.5 kcal/mol, and the profile was quite flat through the channel, roughly consistent with conductance measurements. The profile for formamidinium was very similar to that of methylammonium. Guanidinium had a high entry barrier (deltaF = +8.6 kcal/mol) and a narrow deep central well (deltaF = -2.6 kcal/mol), qualitatively consistent with predictions from voltage-dependent potassium current block measurements. Its deep central well, contrasting with the flat profile for formamidinium, was verified with perturbation dynamics and was correlated with its high propensity to form hydrogen bonds with the channel at the dimer junction (not shared by the other three cations). Analysis of the ensemble average radial forces on the ions demonstrates that all four ions undergo compressive forces in the channel that are at maximum at the center of the monomer and relieved at the dimer junction, illustrating increased flexibility of the channel walls in the center of the channel.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aqvist J., Warshel A. Energetics of ion permeation through membrane channels. Solvation of Na+ by gramicidin A. Biophys J. 1989 Jul;56(1):171–182. doi: 10.1016/S0006-3495(89)82662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberg E., Läuger P. Temperature-dependent properties of gramicidin A channels. Biochim Biophys Acta. 1974 Oct 29;367(2):127–133. doi: 10.1016/0005-2736(74)90037-6. [DOI] [PubMed] [Google Scholar]
- Becker M. D., Koeppe R. E., 2nd, Andersen O. S. Amino acid substitutions and ion channel function. Model-dependent conclusions. Biophys J. 1992 Apr;62(1):25–27. doi: 10.1016/S0006-3495(92)81767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busath D. D. The use of physical methods in determining gramicidin channel structure and function. Annu Rev Physiol. 1993;55:473–501. doi: 10.1146/annurev.ph.55.030193.002353. [DOI] [PubMed] [Google Scholar]
- Crouzy S., Woolf T. B., Roux B. A molecular dynamics study of gating in dioxolane-linked gramicidin A channels. Biophys J. 1994 Oct;67(4):1370–1386. doi: 10.1016/S0006-3495(94)80618-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman V., Partenskii M. B., Jordan P. C. A semi-microscopic Monte Carlo study of permeation energetics in a gramicidin-like channel: the origin of cation selectivity. Biophys J. 1996 Jan;70(1):121–134. doi: 10.1016/S0006-3495(96)79554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin J. T., Providence L. L., Koeppe R. E., 2nd, Andersen O. S. Formation of non-beta 6.3-helical gramicidin channels between sequence-substituted gramicidin analogues. Biophys J. 1992 Apr;62(1):145–159. doi: 10.1016/S0006-3495(92)81801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman G., Horn R. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J Membr Biol. 1983;76(3):197–225. doi: 10.1007/BF01870364. [DOI] [PubMed] [Google Scholar]
- Etchebest C., Pullman A. Energy profile of Cs+ in gramicidin A in the presence of water. Problem of the ion selectivity of the channel. J Biomol Struct Dyn. 1988 Apr;5(5):1111–1125. doi: 10.1080/07391102.1988.10506452. [DOI] [PubMed] [Google Scholar]
- Etchebest C., Ranganathan S., Pullman A. The gramicidin A channel: comparison of the energy profiles of Na+, K+ and Cs+. Influence of the flexibility of the ethanolamine end chain on the profiles. FEBS Lett. 1984 Aug 6;173(2):301–306. doi: 10.1016/0014-5793(84)80795-4. [DOI] [PubMed] [Google Scholar]
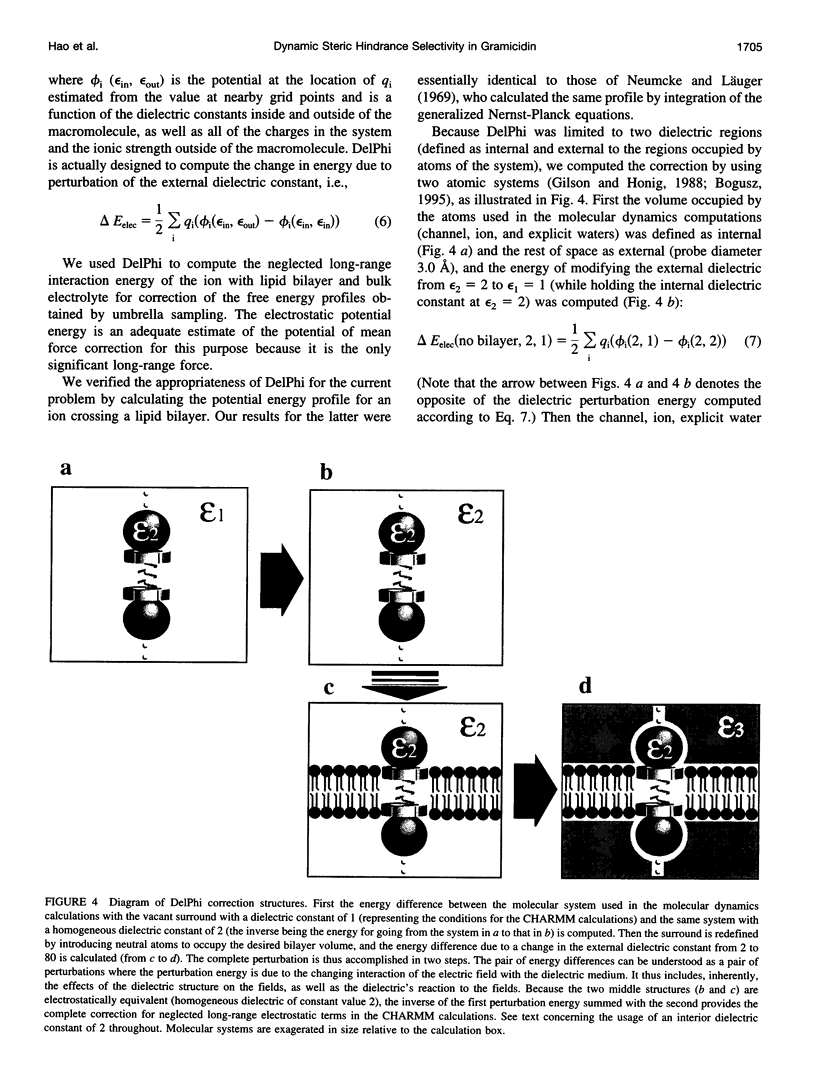
- Gilson M. K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Hemsley G., Busath D. Small iminium ions block gramicidin channels in lipid bilayers. Biophys J. 1991 Apr;59(4):901–907. doi: 10.1016/S0006-3495(91)82303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P. C. Ion-water and ion-polypeptide correlations in a gramicidin-like channel. A molecular dynamics study. Biophys J. 1990 Nov;58(5):1133–1156. doi: 10.1016/S0006-3495(90)82456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchem R. R., Lee K. C., Huo S., Cross T. A. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. J Biomol NMR. 1996 Jul;8(1):1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- Kim K. S., Vercauteren D. P., Welti M., Chin S., Clementi E. Interaction of K+ ion with the solvated gramicidin A transmembrane channel. Biophys J. 1985 Mar;47(3):327–335. doi: 10.1016/S0006-3495(85)83923-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper I., Hagstrom R., Fine R., Sharp K., Honig B. Focusing of electric fields in the active site of Cu-Zn superoxide dismutase: effects of ionic strength and amino-acid modification. Proteins. 1986 Sep;1(1):47–59. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- Koeppe R. E., 2nd, Providence L. L., Greathouse D. V., Heitz F., Trudelle Y., Purdie N., Andersen O. S. On the helix sense of gramicidin A single channels. Proteins. 1992 Jan;12(1):49–62. doi: 10.1002/prot.340120107. [DOI] [PubMed] [Google Scholar]
- Levitt D. G. Comparison of Nernst-Planck and reaction rate models for multiply occupied channels. Biophys J. 1982 Mar;37(3):575–587. [PMC free article] [PubMed] [Google Scholar]
- Levitt D. G. General continuum theory for multiion channel. I. Theory. Biophys J. 1991 Feb;59(2):271–277. doi: 10.1016/S0006-3495(91)82220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läuger P. Ion transport through pores: a rate-theory analysis. Biochim Biophys Acta. 1973 Jul 6;311(3):423–441. doi: 10.1016/0005-2736(73)90323-4. [DOI] [PubMed] [Google Scholar]
- Myers V. B., Haydon D. A. Ion transfer across lipid membranes in the presence of gramicidin A. II. The ion selectivity. Biochim Biophys Acta. 1972 Aug 9;274(2):313–322. doi: 10.1016/0005-2736(72)90179-4. [DOI] [PubMed] [Google Scholar]
- Neumcke B., Läuger P. Nonlinear electrical effects in lipid bilayer membranes. II. Integration of the generalized Nernst-Planck equations. Biophys J. 1969 Sep;9(9):1160–1170. doi: 10.1016/S0006-3495(69)86443-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrup S. H., Pear M. R., Lee C. Y., McCammon J. A., Karplus M. Dynamical theory of activated processes in globular proteins. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4035–4039. doi: 10.1073/pnas.79.13.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Karplus M. Ion transport in a model gramicidin channel. Structure and thermodynamics. Biophys J. 1991 May;59(5):961–981. doi: 10.1016/S0006-3495(91)82311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Karplus M. Molecular dynamics simulations of the gramicidin channel. Annu Rev Biophys Biomol Struct. 1994;23:731–761. doi: 10.1146/annurev.bb.23.060194.003503. [DOI] [PubMed] [Google Scholar]
- Seoh S. A., Busath D. D. Formamidinium-induced dimer stabilization and flicker block behavior in homo- and heterodimer channels formed by gramicidin A and N-acetyl gramicidin A. Biophys J. 1993 Nov;65(5):1817–1827. doi: 10.1016/S0006-3495(93)81239-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoh S. A., Busath D. Gramicidin tryptophans mediate formamidinium-induced channel stabilization. Biophys J. 1995 Jun;68(6):2271–2279. doi: 10.1016/S0006-3495(95)80409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
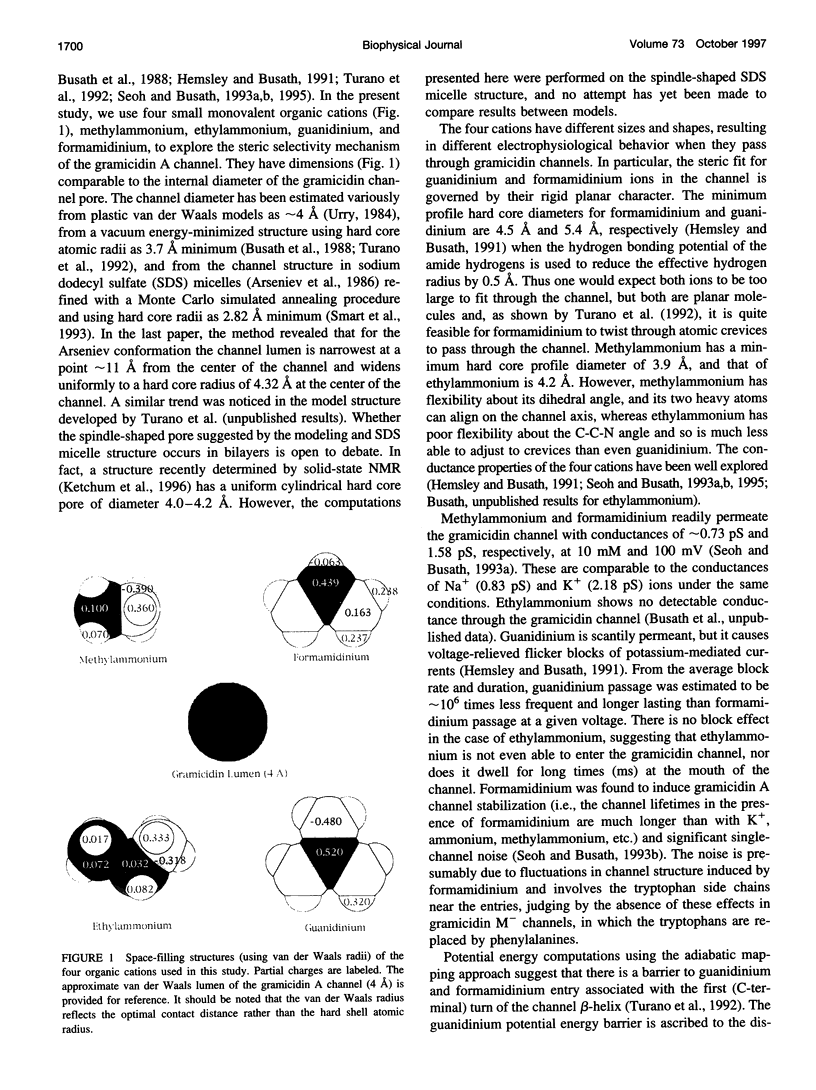
- Seoh S. A., Busath D. The permeation properties of small organic cations in gramicidin A channels. Biophys J. 1993 Apr;64(4):1017–1028. doi: 10.1016/S0006-3495(93)81467-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O. S., Goodfellow J. M., Wallace B. A. The pore dimensions of gramicidin A. Biophys J. 1993 Dec;65(6):2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano B., Pear M., Busath D. Gramicidin channel selectivity. Molecular mechanics calculations for formamidinium, guanidinium, and acetamidinium. Biophys J. 1992 Jul;63(1):152–161. doi: 10.1016/S0006-3495(92)81574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban B. W., Hladky S. B. Ion transport in the simplest single file pore. Biochim Biophys Acta. 1979 Jul 5;554(2):410–429. doi: 10.1016/0005-2736(79)90381-x. [DOI] [PubMed] [Google Scholar]