Abstract
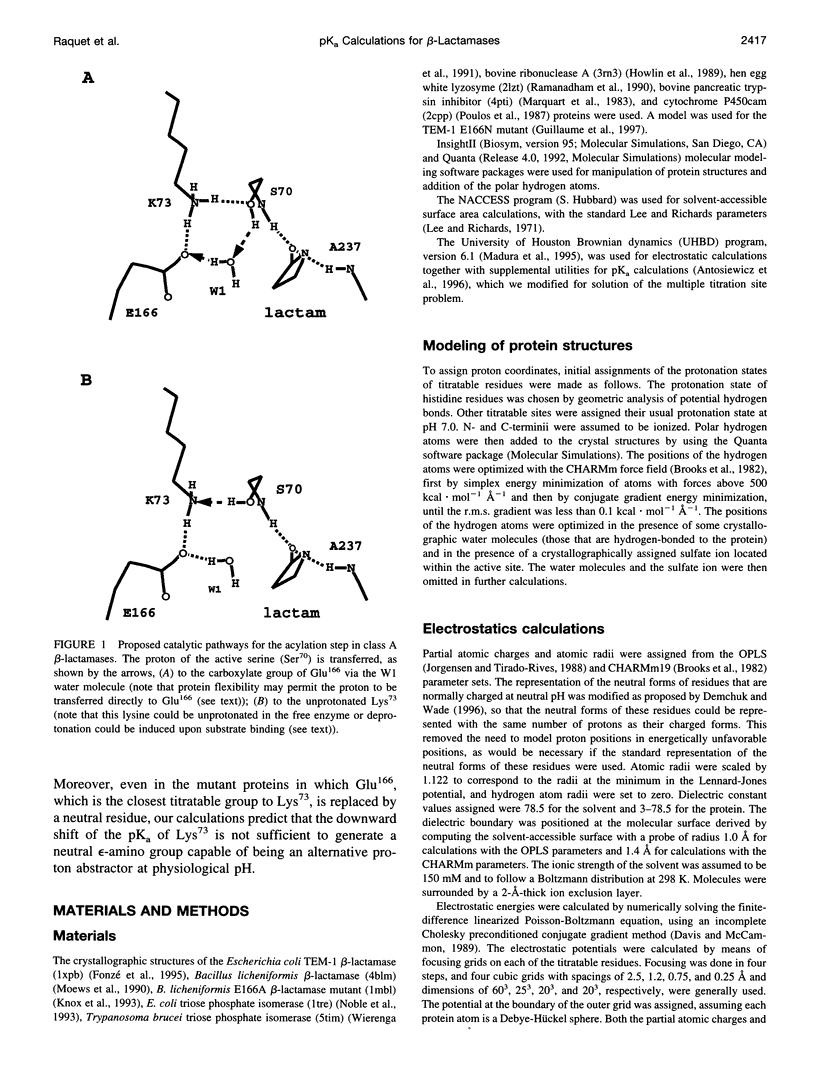
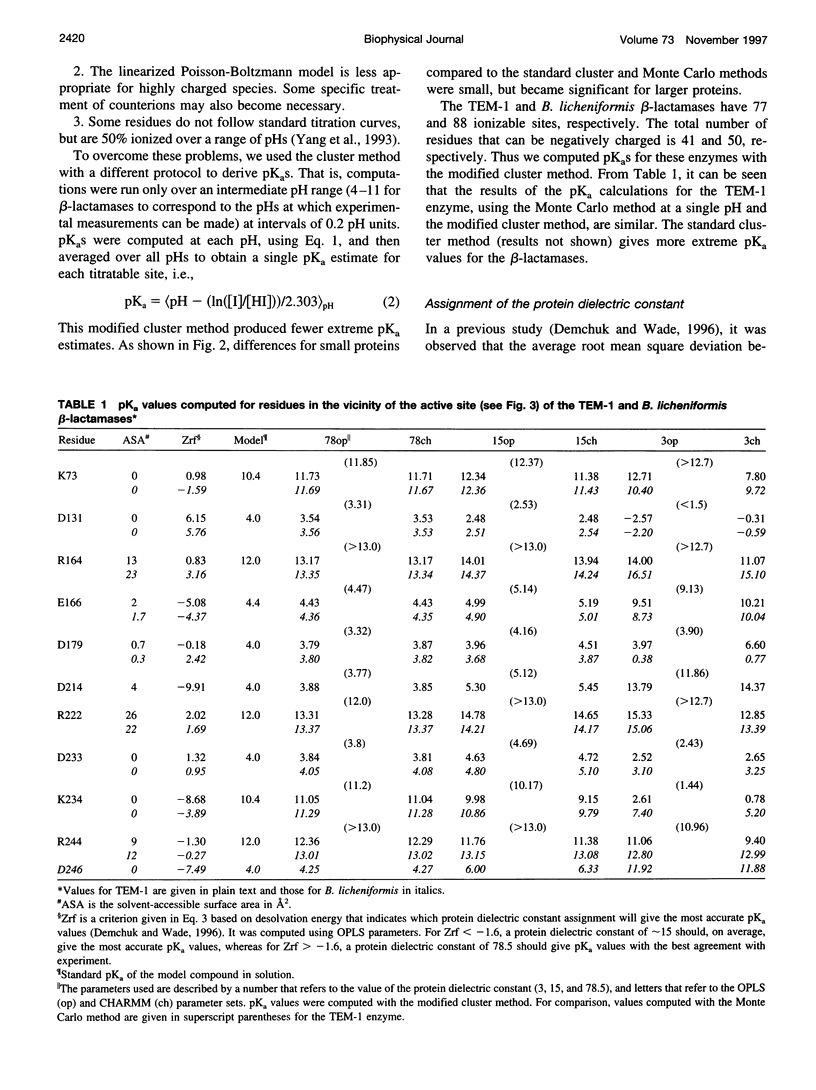
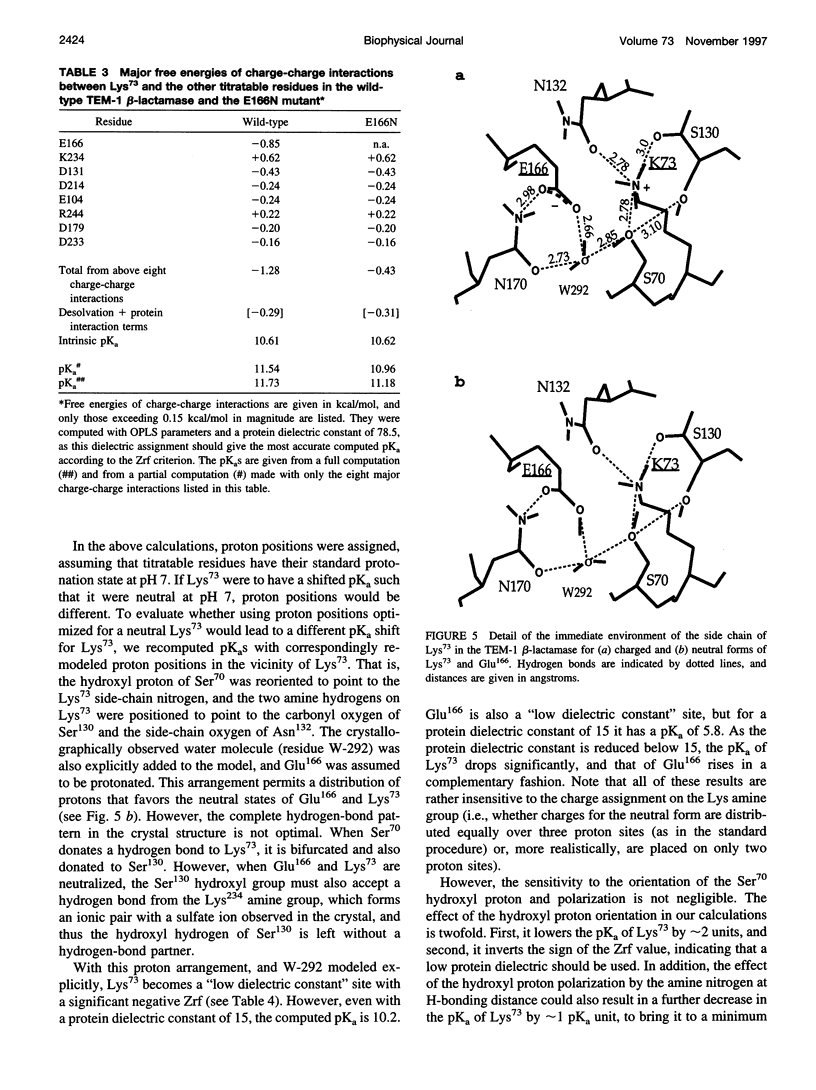
Beta-lactamases are responsible for resistance to penicillins and related beta-lactam compounds. Despite numerous studies, the identity of the general base involved in the acylation step is still unclear. It has been proposed, on the basis of a previous pKa calculation and analysis of structural data, that the unprotonated Lys73 in the active site could act as the general base. Using a continuum electrostatic model with an improved treatment of the multiple titration site problem, we calculated the pKa values of all titratable residues in the substrate-free TEM-1 and Bacillus licheniformis class A beta-lactamases. The pKa of Lys73 in both enzymes was computed to be above 10, in good agreement with recent experimental data on the TEM-1 beta-lactamase, but inconsistent with the proposal that Lys73 acts as the general base. Even when the closest titratable residue, Glu166, is mutated to a neutral residue, the predicted downward shift of the pKa of Lys73 shows that it is unlikely to act as a proton abstractor in either enzyme. These results support a mechanism in which the proton of the active Ser70 is transferred to the carboxylate group of Glu166.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Coulson A. F., Frère J. M., Ghuysen J. M., Joris B., Forsman M., Levesque R. C., Tiraby G., Waley S. G. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991 May 15;276(Pt 1):269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosiewicz J., McCammon J. A., Gilson M. K. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994 May 6;238(3):415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- Antosiewicz J., McCammon J. A., Gilson M. K. The determinants of pKas in proteins. Biochemistry. 1996 Jun 18;35(24):7819–7833. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]
- Antosiewicz J., Porschke D. The nature of protein dipole moments: experimental and calculated permanent dipole of alpha-chymotrypsin. Biochemistry. 1989 Dec 26;28(26):10072–10078. doi: 10.1021/bi00452a029. [DOI] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Protonation of interacting residues in a protein by a Monte Carlo method: application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannigan J., Matagne A., Jacob F., Damblon C., Joris B., Klein D., Spratt B. G., Frère J. M. The mutation Lys234His yields a class A beta-lactamase with a novel pH-dependence. Biochem J. 1991 Sep 15;278(Pt 3):673–678. doi: 10.1042/bj2780673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damblon C., Raquet X., Lian L. Y., Lamotte-Brasseur J., Fonze E., Charlier P., Roberts G. C., Frère J. M. The catalytic mechanism of beta-lactamases: NMR titration of an active-site lysine residue of the TEM-1 enzyme. Proc Natl Acad Sci U S A. 1996 Mar 5;93(5):1747–1752. doi: 10.1073/pnas.93.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzé E., Charlier P., To'th Y., Vermeire M., Raquet X., Dubus A., Frère J. M. TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr D Biol Crystallogr. 1995 Sep 1;51(Pt 5):682–694. doi: 10.1107/S0907444994014496. [DOI] [PubMed] [Google Scholar]
- Frère J. M. Beta-lactamases and bacterial resistance to antibiotics. Mol Microbiol. 1995 May;16(3):385–395. doi: 10.1111/j.1365-2958.1995.tb02404.x. [DOI] [PubMed] [Google Scholar]
- Gilson M. K. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993 Mar;15(3):266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- Guillaume G., Vanhove M., Lamotte-Brasseur J., Ledent P., Jamin M., Joris B., Frère J. M. Site-directed mutagenesis of glutamate 166 in two beta-lactamases. Kinetic and molecular modeling studies. J Biol Chem. 1997 Feb 28;272(9):5438–5444. doi: 10.1074/jbc.272.9.5438. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Kapadia G., Blanco B., Smith T. S., Coulson A. Structural basis for the inactivation of the P54 mutant of beta-lactamase from Staphylococcus aureus PC1. Biochemistry. 1991 Oct 1;30(39):9503–9509. doi: 10.1021/bi00103a017. [DOI] [PubMed] [Google Scholar]
- Howlin B., Moss D. S., Harris G. W. Segmented anisotropic refinement of bovine ribonuclease A by the application of the rigid-body TLS model. Acta Crystallogr A. 1989 Dec 1;45(Pt 12):851–861. doi: 10.1107/s0108767389009177. [DOI] [PubMed] [Google Scholar]
- Ishiguro M., Imajo S. Modeling study on a hydrolytic mechanism of class A beta-lactamases. J Med Chem. 1996 May 24;39(11):2207–2218. doi: 10.1021/jm9506027. [DOI] [PubMed] [Google Scholar]
- Jacob F., Joris B., Lepage S., Dusart J., Frère J. M. Role of the conserved amino acids of the 'SDN' loop (Ser130, Asp131 and Asn132) in a class A beta-lactamase studied by site-directed mutagenesis. Biochem J. 1990 Oct 15;271(2):399–406. doi: 10.1042/bj2710399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joris B., Ghuysen J. M., Dive G., Renard A., Dideberg O., Charlier P., Frère J. M., Kelly J. A., Boyington J. C., Moews P. C. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988 Mar 1;250(2):313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J. R., Moews P. C., Escobar W. A., Fink A. L. A catalytically-impaired class A beta-lactamase: 2 A crystal structure and kinetics of the Bacillus licheniformis E166A mutant. Protein Eng. 1993 Jan;6(1):11–18. doi: 10.1093/protein/6.1.11. [DOI] [PubMed] [Google Scholar]
- Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. Mechanism of acyl transfer by the class A serine beta-lactamase of Streptomyces albus G. Biochem J. 1991 Oct 1;279(Pt 1):213–221. doi: 10.1042/bj2790213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Matagne A., Frère J. M. Contribution of mutant analysis to the understanding of enzyme catalysis: the case of class A beta-lactamases. Biochim Biophys Acta. 1995 Jan 19;1246(2):109–127. doi: 10.1016/0167-4838(94)00177-i. [DOI] [PubMed] [Google Scholar]
- Moews P. C., Knox J. R., Dideberg O., Charlier P., Frère J. M. Beta-lactamase of Bacillus licheniformis 749/C at 2 A resolution. Proteins. 1990;7(2):156–171. doi: 10.1002/prot.340070205. [DOI] [PubMed] [Google Scholar]
- Noble M. E., Zeelen J. P., Wierenga R. K., Mainfroid V., Goraj K., Gohimont A. C., Martial J. A. Structure of triosephosphate isomerase from Escherichia coli determined at 2.6 A resolution. Acta Crystallogr D Biol Crystallogr. 1993 Jul 1;49(Pt 4):403–417. doi: 10.1107/S0907444993002628. [DOI] [PubMed] [Google Scholar]
- Poulos T. L., Finzel B. C., Howard A. J. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987 Jun 5;195(3):687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- Ramanadham M., Sieker L. C., Jensen L. H. Refinement of triclinic lysozyme: II. The method of stereochemically restrained least squares. Acta Crystallogr B. 1990 Feb 1;46(Pt 1):63–69. doi: 10.1107/s0108768189009195. [DOI] [PubMed] [Google Scholar]
- Raquet X., Vanhove M., Lamotte-Brasseur J., Goussard S., Courvalin P., Frère J. M. Stability of TEM beta-lactamase mutants hydrolyzing third generation cephalosporins. Proteins. 1995 Sep;23(1):63–72. doi: 10.1002/prot.340230108. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 A resolution. Nature. 1992 Oct 22;359(6397):700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- Swarén P., Maveyraud L., Guillet V., Masson J. M., Mourey L., Samama J. P. Electrostatic analysis of TEM1 beta-lactamase: effect of substrate binding, steep potential gradients and consequences of site-directed mutations. Structure. 1995 Jun 15;3(6):603–613. doi: 10.1016/s0969-2126(01)00194-0. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Noble M. E., Vriend G., Nauche S., Hol W. G. Refined 1.83 A structure of trypanosomal triosephosphate isomerase crystallized in the presence of 2.4 M-ammonium sulphate. A comparison with the structure of the trypanosomal triosephosphate isomerase-glycerol-3-phosphate complex. J Mol Biol. 1991 Aug 20;220(4):995–1015. doi: 10.1016/0022-2836(91)90368-g. [DOI] [PubMed] [Google Scholar]
- Yang A. S., Gunner M. R., Sampogna R., Sharp K., Honig B. On the calculation of pKas in proteins. Proteins. 1993 Mar;15(3):252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- You T. J., Bashford D. Conformation and hydrogen ion titration of proteins: a continuum electrostatic model with conformational flexibility. Biophys J. 1995 Nov;69(5):1721–1733. doi: 10.1016/S0006-3495(95)80042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]