Abstract
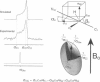
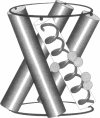
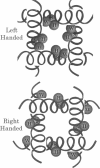
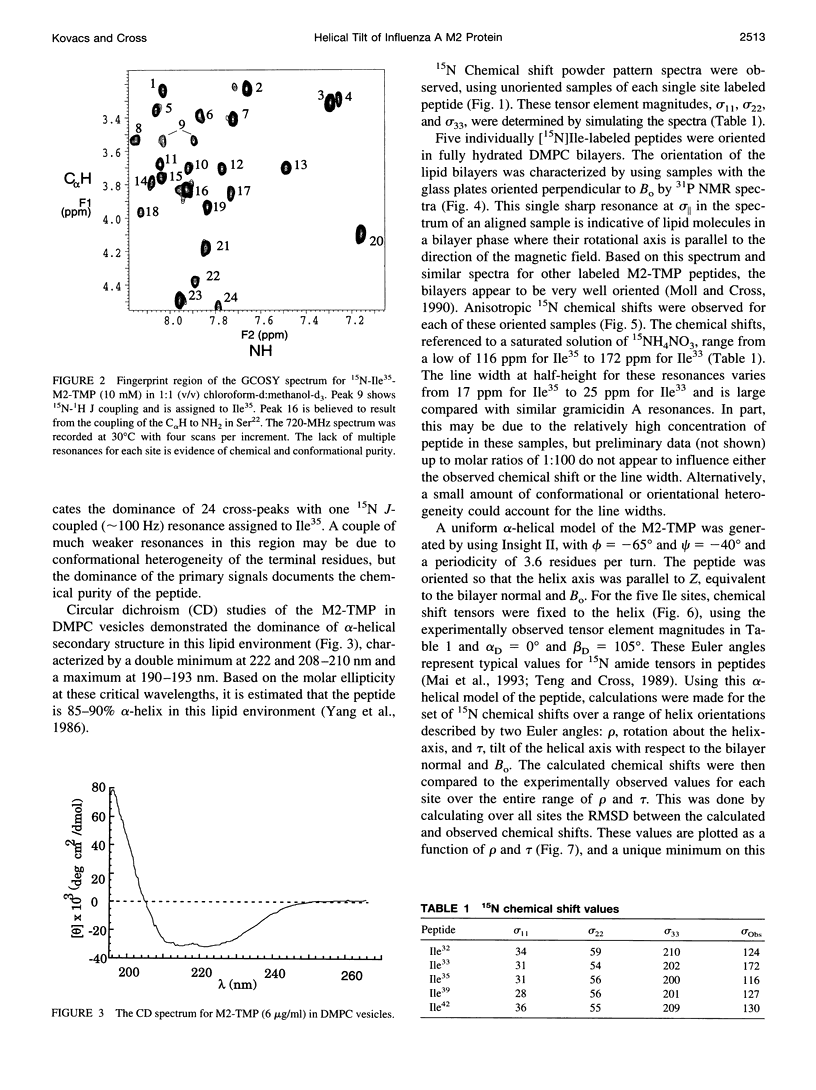
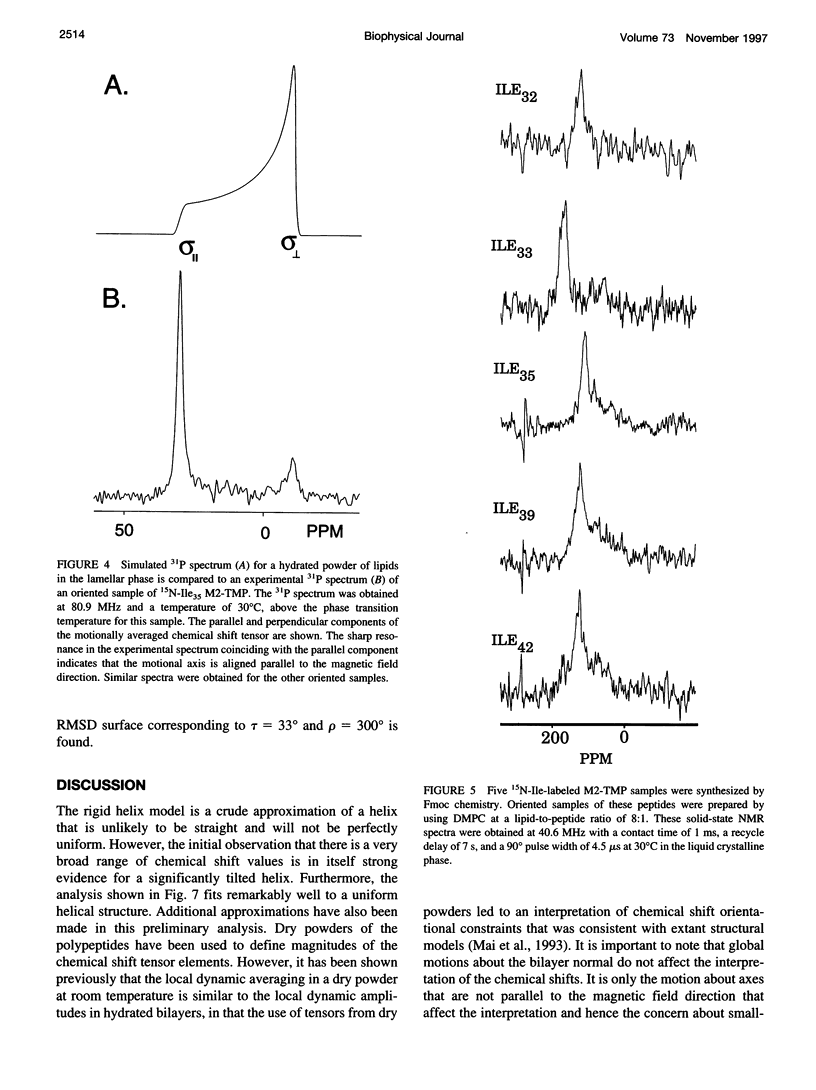
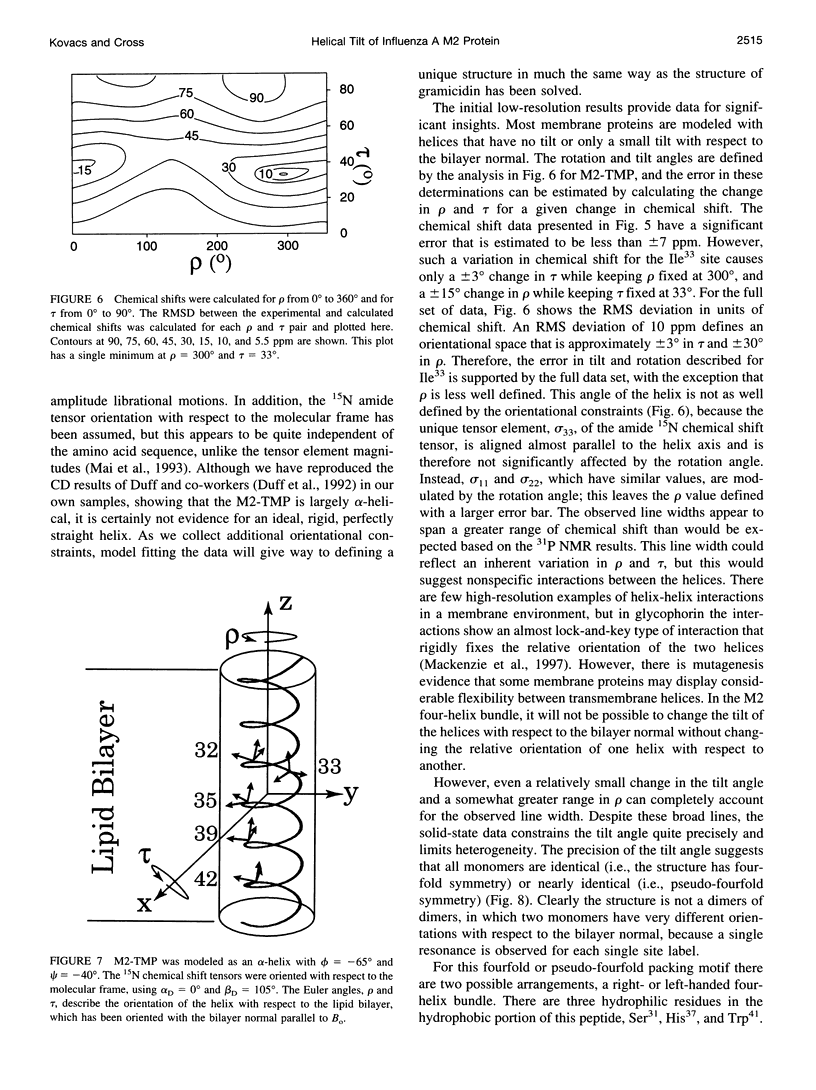
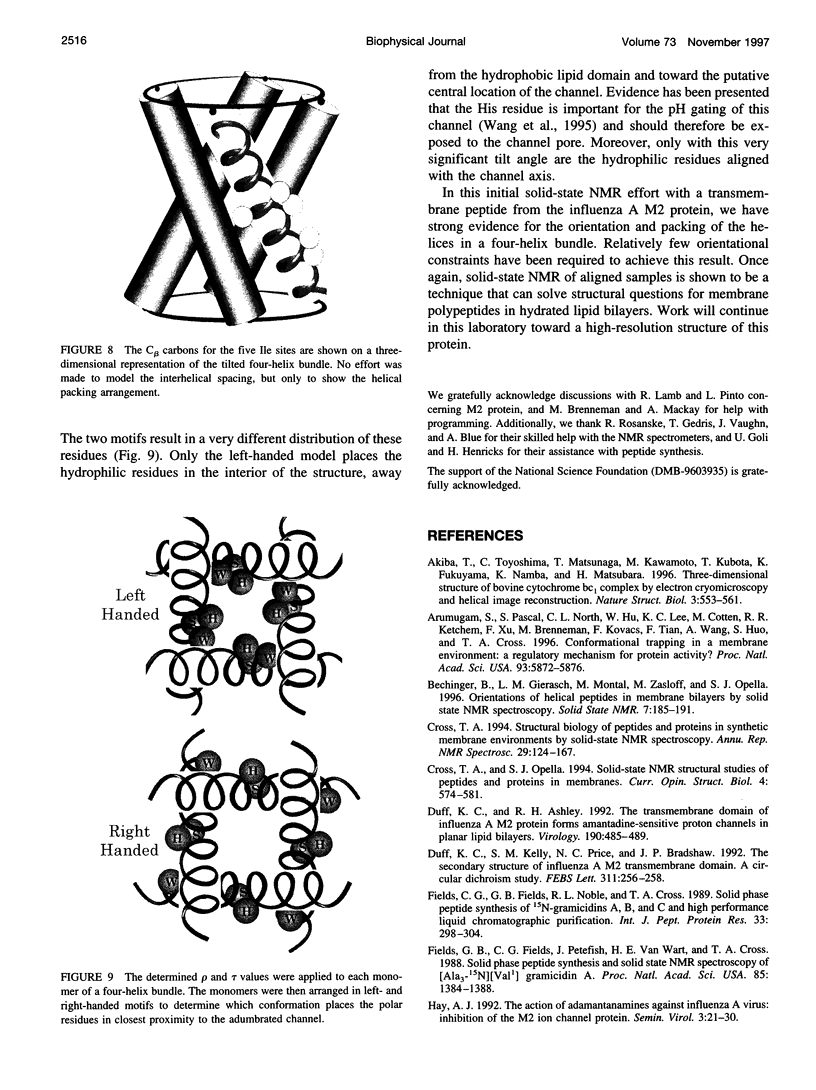
The transmembrane portion of the M2 protein from the Influenza A virus has been studied in hydrated dimyristroylphosphotidylcholine lipid bilayers with solid-state NMR. Orientational constraints were obtained from isotopically labeled peptide samples mechanically aligned between thin glass plates. 15N chemical shifts from single site labeled samples constrain the molecular frame with respect to the magnetic field. When these constraints are applied to the peptide, modeled as a uniform alpha-helix, the tilt of the helix with respect to the bilayer normal was determined to be 33 degrees +/- 3 degrees. Furthermore, the orientation about the helix axis was also determined within an error of +/- 30 degrees. These results imply that the packing of this tetrameric protein is in a left-handed four-helix bundle. Only with such a large tilt angle are the hydrophilic residues aligned to the channel axis.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiba T., Toyoshima C., Matsunaga T., Kawamoto M., Kubota T., Fukuyama K., Namba K., Matsubara H. Three-dimensional structure of bovine cytochrome bc1 complex by electron cryomicroscopy and helical image reconstruction. Nat Struct Biol. 1996 Jun;3(6):553–561. doi: 10.1038/nsb0696-553. [DOI] [PubMed] [Google Scholar]
- Arumugam S., Pascal S., North C. L., Hu W., Lee K. C., Cotten M., Ketchem R. R., Xu F., Brenneman M., Kovacs F. Conformational trapping in a membrane environment: a regulatory mechanism for protein activity? Proc Natl Acad Sci U S A. 1996 Jun 11;93(12):5872–5876. doi: 10.1073/pnas.93.12.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechinger B., Gierasch L. M., Montal M., Zasloff M., Opella S. J. Orientations of helical peptides in membrane bilayers by solid state NMR spectroscopy. Solid State Nucl Magn Reson. 1996 Dec;7(3):185–191. doi: 10.1016/0926-2040(95)01224-9. [DOI] [PubMed] [Google Scholar]
- Duff K. C., Ashley R. H. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992 Sep;190(1):485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- Duff K. C., Kelly S. M., Price N. C., Bradshaw J. P. The secondary structure of influenza A M2 transmembrane domain. A circular dichroism study. FEBS Lett. 1992 Oct 26;311(3):256–258. doi: 10.1016/0014-5793(92)81114-2. [DOI] [PubMed] [Google Scholar]
- Fields C. G., Fields G. B., Noble R. L., Cross T. A. Solid phase peptide synthesis of 15N-gramicidins A, B, and C and high performance liquid chromatographic purification. Int J Pept Protein Res. 1989 Apr;33(4):298–303. doi: 10.1111/j.1399-3011.1989.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Fields G. B., Fields C. G., Petefish J., Van Wart H. E., Cross T. A. Solid-phase peptide synthesis and solid-state NMR spectroscopy of [Ala3-15N][Val1]gramicidin A. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1384–1388. doi: 10.1073/pnas.85.5.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger L. J., Lamb R. A. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991 Jul;183(1):32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- Holsinger L. J., Shaughnessy M. A., Micko A., Pinto L. H., Lamb R. A. Analysis of the posttranslational modifications of the influenza virus M2 protein. J Virol. 1995 Feb;69(2):1219–1225. doi: 10.1128/jvi.69.2.1219-1225.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S., Ostermeier C., Ludwig B., Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995 Aug 24;376(6542):660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Ketchem R. R., Lee K. C., Huo S., Cross T. A. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. J Biomol NMR. 1996 Jul;8(1):1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W., Wang D. N., Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994 Feb 17;367(6464):614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Zebedee S. L., Richardson C. D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985 Mar;40(3):627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- MacKenzie K. R., Prestegard J. H., Engelman D. M. A transmembrane helix dimer: structure and implications. Science. 1997 Apr 4;276(5309):131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- Mai W., Hu W., Wang C., Cross T. A. Orientational constraints as three-dimensional structural constraints from chemical shift anisotropy: the polypeptide backbone of gramicidin A in a lipid bilayer. Protein Sci. 1993 Apr;2(4):532–542. doi: 10.1002/pro.5560020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell P. A., Shon K., Kim Y., Opella S. J. fd coat protein structure in membrane environments. J Mol Biol. 1993 Oct 5;233(3):447–463. doi: 10.1006/jmbi.1993.1523. [DOI] [PubMed] [Google Scholar]
- Moll F., 3rd, Cross T. A. Optimizing and characterizing alignment of oriented lipid bilayers containing gramicidin D. Biophys J. 1990 Feb;57(2):351–362. doi: 10.1016/S0006-3495(90)82536-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortishire-Smith R. J., Pitzenberger S. M., Burke C. J., Middaugh C. R., Garsky V. M., Johnson R. G. Solution structure of the cytoplasmic domain of phopholamban: phosphorylation leads to a local perturbation in secondary structure. Biochemistry. 1995 Jun 13;34(23):7603–7613. doi: 10.1021/bi00023a006. [DOI] [PubMed] [Google Scholar]
- North C. L., Barranger-Mathys M., Cafiso D. S. Membrane orientation of the N-terminal segment of alamethicin determined by solid-state 15N NMR. Biophys J. 1995 Dec;69(6):2392–2397. doi: 10.1016/S0006-3495(95)80108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks G. D., Lamb R. A. Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell. 1991 Feb 22;64(4):777–787. doi: 10.1016/0092-8674(91)90507-u. [DOI] [PubMed] [Google Scholar]
- Pinto L. H., Holsinger L. J., Lamb R. A. Influenza virus M2 protein has ion channel activity. Cell. 1992 May 1;69(3):517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Prosser R. S., Daleman S. I., Davis J. H. The structure of an integral membrane peptide: a deuterium NMR study of gramicidin. Biophys J. 1994 May;66(5):1415–1428. doi: 10.1016/S0006-3495(94)80932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi T., Tu Q., Pinto L. H., Lamb R. A. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc Natl Acad Sci U S A. 1997 May 13;94(10):5000–5005. doi: 10.1073/pnas.94.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R., Separovic F., Milne T. J., Whittaker A., Bennett F. M., Cornell B. A., Makriyannis A. Structure and orientation of the pore-forming peptide, melittin, in lipid bilayers. J Mol Biol. 1994 Aug 19;241(3):456–466. doi: 10.1006/jmbi.1994.1520. [DOI] [PubMed] [Google Scholar]
- Sugrue R. J., Hay A. J. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991 Feb;180(2):617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosteson M. T., Pinto L. H., Holsinger L. J., Lamb R. A. Reconstitution of the influenza virus M2 ion channel in lipid bilayers. J Membr Biol. 1994 Oct;142(1):117–126. doi: 10.1007/BF00233389. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996 May 24;272(5265):1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Wang C., Lamb R. A., Pinto L. H. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995 Oct;69(4):1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Takeuchi K., Pinto L. H., Lamb R. A. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993 Sep;67(9):5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. T., Wu C. S., Martinez H. M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]