Abstract
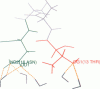
We have studied the winter flounder antifreeze protein (AFP) and two of its mutants using molecular dynamics simulation techniques. The simulations were performed under four conditions: in the gas phase, solvated by water, adsorbed on the ice (2021) crystal plane in the gas phase and in aqueous solution. This study provided details of the ice-binding pattern of the winter flounder AFP. Simulation results indicated that the Asp, Asn, and Thr residues in the AFP are important in ice binding and that Asn and Thr as a group bind cooperatively to the ice surface. These ice-binding residues can be collected into four distinct ice-binding regions: Asp-1/Thr-2/Asp-5, Thr-13/Asn-16, Thr-24/Asn-27, and Thr-35/Arg-37. These four regions are 11 residues apart and the repeat distance between them matches the ice lattice constant along the (1102) direction. This match is crucial to ensure that all four groups can interact with the ice surface simultaneously, thereby, enhancing ice binding. These Asx (x = p or n)/Thr regions each form 5-6 hydrogen bonds with the ice surface: Asn forms about three hydrogen bonds with ice molecules located in the step region while Thr forms one to two hydrogen bonds with the ice molecules in the ridge of the (2021) crystal plane. Both the distance between Thr and Asn and the ordering of the two residues are crucial for effective ice binding. The proper sequence is necessary to generate a binding surface that is compatible with the ice surface topology, thus providing a perfect "host/guest" interaction that simultaneously satisfies both hydrogen bonding and van der Waals interactions. The results also show the relation among binding energy, the number of hydrogen bonds, and the activity. The activity is correlated to the binding energy, and in the case of the mutants we have studied the number of hydrogen bonds. The greater the number of the hydrogen bonds the greater the antifreeze activity. The roles van der Waals interactions and the hydrophobic effect play in ice binding are also highlighted. For the latter it is demonstrated that the surface of ice has a clathratelike structure which favors the partitioning of hydrophobic groups to the surface of ice. It is suggested that mutations that involve the deletion of hydrophobic residues (e.g., the Leu residues) will provide insight into the role the hydrophobic effect plays in partitioning these peptides to the surface of ice.
Full text
PDF






















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arav A., Rubinsky B., Fletcher G., Seren E. Cryogenic protection of oocytes with antifreeze proteins. Mol Reprod Dev. 1993 Dec;36(4):488–493. doi: 10.1002/mrd.1080360413. [DOI] [PubMed] [Google Scholar]
- Bash P. A., Singh U. C., Langridge R., Kollman P. A. Free energy calculations by computer simulation. Science. 1987 May 1;236(4801):564–568. doi: 10.1126/science.3576184. [DOI] [PubMed] [Google Scholar]
- Burcham T. S., Osuga D. T., Chino H., Feeney R. E. Analysis of antifreeze glycoproteins in fish serum. Anal Biochem. 1984 May 15;139(1):197–204. doi: 10.1016/0003-2697(84)90405-6. [DOI] [PubMed] [Google Scholar]
- Chakrabartty A., Ananthanarayanan V. S., Hew C. L. Structure-function relationships in a winter flounder antifreeze polypeptide. I. Stabilization of an alpha-helical antifreeze polypeptide by charged-group and hydrophobic interactions. J Biol Chem. 1989 Jul 5;264(19):11307–11312. [PubMed] [Google Scholar]
- Chakrabartty A., Yang D. S., Hew C. L. Structure-function relationship in a winter flounder antifreeze polypeptide. II. Alteration of the component growth rates of ice by synthetic antifreeze polypeptides. J Biol Chem. 1989 Jul 5;264(19):11313–11316. [PubMed] [Google Scholar]
- Chou K. C. Energy-optimized structure of antifreeze protein and its binding mechanism. J Mol Biol. 1992 Jan 20;223(2):509–517. doi: 10.1016/0022-2836(92)90666-8. [DOI] [PubMed] [Google Scholar]
- Davies P. L., Hew C. L. Biochemistry of fish antifreeze proteins. FASEB J. 1990 May;4(8):2460–2468. doi: 10.1096/fasebj.4.8.2185972. [DOI] [PubMed] [Google Scholar]
- DeVries A. L., Komatsu S. K., Feeney R. E. Chemical and physical properties of freezing point-depressing glycoproteins from Antarctic fishes. J Biol Chem. 1970 Jun 10;245(11):2901–2908. [PubMed] [Google Scholar]
- Feeney R. E., Burcham T. S., Yeh Y. Antifreeze glycoproteins from polar fish blood. Annu Rev Biophys Biophys Chem. 1986;15:59–78. doi: 10.1146/annurev.bb.15.060186.000423. [DOI] [PubMed] [Google Scholar]
- Hansen T. N., Carpenter J. F. Calorimetric determination of inhibition of ice crystal growth by antifreeze protein in hydroxyethyl starch solutions. Biophys J. 1993 Jun;64(6):1843–1850. doi: 10.1016/S0006-3495(93)81555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hew C. L., Wang N. C., Yan S., Cai H., Sclater A., Fletcher G. L. Biosynthesis of antifreeze polypeptides in the winter flounder. Characterization and seasonal occurrence of precursor polypeptides. Eur J Biochem. 1986 Oct 15;160(2):267–272. doi: 10.1111/j.1432-1033.1986.tb09966.x. [DOI] [PubMed] [Google Scholar]
- Jia Z., DeLuca C. I., Chao H., Davies P. L. Structural basis for the binding of a globular antifreeze protein to ice. Nature. 1996 Nov 21;384(6606):285–288. doi: 10.1038/384285a0. [DOI] [PubMed] [Google Scholar]
- Jorgensen H., Mori M., Matsui H., Kanaoka M., Yanagi H., Yabusaki Y., Kikuzono Y. Molecular dynamics simulation of winter flounder antifreeze protein variants in solution: correlation between side chain spacing and ice lattice. Protein Eng. 1993 Jan;6(1):19–27. doi: 10.1093/protein/6.1.19. [DOI] [PubMed] [Google Scholar]
- Kenward K. D., Altschuler M., Hildebrand D., Davies P. L. Accumulation of type I fish antifreeze protein in transgenic tobacco is cold-specific. Plant Mol Biol. 1993 Oct;23(2):377–385. doi: 10.1007/BF00029012. [DOI] [PubMed] [Google Scholar]
- Knight C. A., Cheng C. C., DeVries A. L. Adsorption of alpha-helical antifreeze peptides on specific ice crystal surface planes. Biophys J. 1991 Feb;59(2):409–418. doi: 10.1016/S0006-3495(91)82234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C. A., DeVries A. L., Oolman L. D. Fish antifreeze protein and the freezing and recrystallization of ice. Nature. 1984 Mar 15;308(5956):295–296. doi: 10.1038/308295a0. [DOI] [PubMed] [Google Scholar]
- McDonald S. M., Brady J. W., Clancy P. Molecular dynamics simulations of a winter flounder "antifreeze" polypeptide in aqueous solution. Biopolymers. 1993 Oct;33(10):1481–1503. doi: 10.1002/bip.360331002. [DOI] [PubMed] [Google Scholar]
- Myers J. K., Pace C. N. Hydrogen bonding stabilizes globular proteins. Biophys J. 1996 Oct;71(4):2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain R. H. Protein structure. Helices of antifreeze. Nature. 1988 May 19;333(6170):207–208. doi: 10.1038/333207a0. [DOI] [PubMed] [Google Scholar]
- Raymond J. A., DeVries A. L. Adsorption inhibition as a mechanism of freezing resistance in polar fishes. Proc Natl Acad Sci U S A. 1977 Jun;74(6):2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F., Yang D. S. Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature. 1995 Jun 1;375(6530):427–431. doi: 10.1038/375427a0. [DOI] [PubMed] [Google Scholar]
- Sönnichsen F. D., DeLuca C. I., Davies P. L., Sykes B. D. Refined solution structure of type III antifreeze protein: hydrophobic groups may be involved in the energetics of the protein-ice interaction. Structure. 1996 Nov 15;4(11):1325–1337. doi: 10.1016/s0969-2126(96)00140-2. [DOI] [PubMed] [Google Scholar]
- Teeter M. M. Water structure of a hydrophobic protein at atomic resolution: Pentagon rings of water molecules in crystals of crambin. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6014–6018. doi: 10.1073/pnas.81.19.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D., Laursen R. A. A D-antifreeze polypeptide displays the same activity as its natural L-enantiomer. FEBS Lett. 1993 Feb 8;317(1-2):31–34. doi: 10.1016/0014-5793(93)81485-i. [DOI] [PubMed] [Google Scholar]
- Wen D., Laursen R. A. A model for binding of an antifreeze polypeptide to ice. Biophys J. 1992 Dec;63(6):1659–1662. doi: 10.1016/S0006-3495(92)81750-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen D., Laursen R. A. Structure-function relationships in an antifreeze polypeptide. The role of neutral, polar amino acids. J Biol Chem. 1992 Jul 15;267(20):14102–14108. [PubMed] [Google Scholar]
- Yang D. S., Sax M., Chakrabartty A., Hew C. L. Crystal structure of an antifreeze polypeptide and its mechanistic implications. Nature. 1988 May 19;333(6170):232–237. doi: 10.1038/333232a0. [DOI] [PubMed] [Google Scholar]
- Yeh Yin, Feeney Robert E. Antifreeze Proteins: Structures and Mechanisms of Function. Chem Rev. 1996 Mar 28;96(2):601–618. doi: 10.1021/cr950260c. [DOI] [PubMed] [Google Scholar]