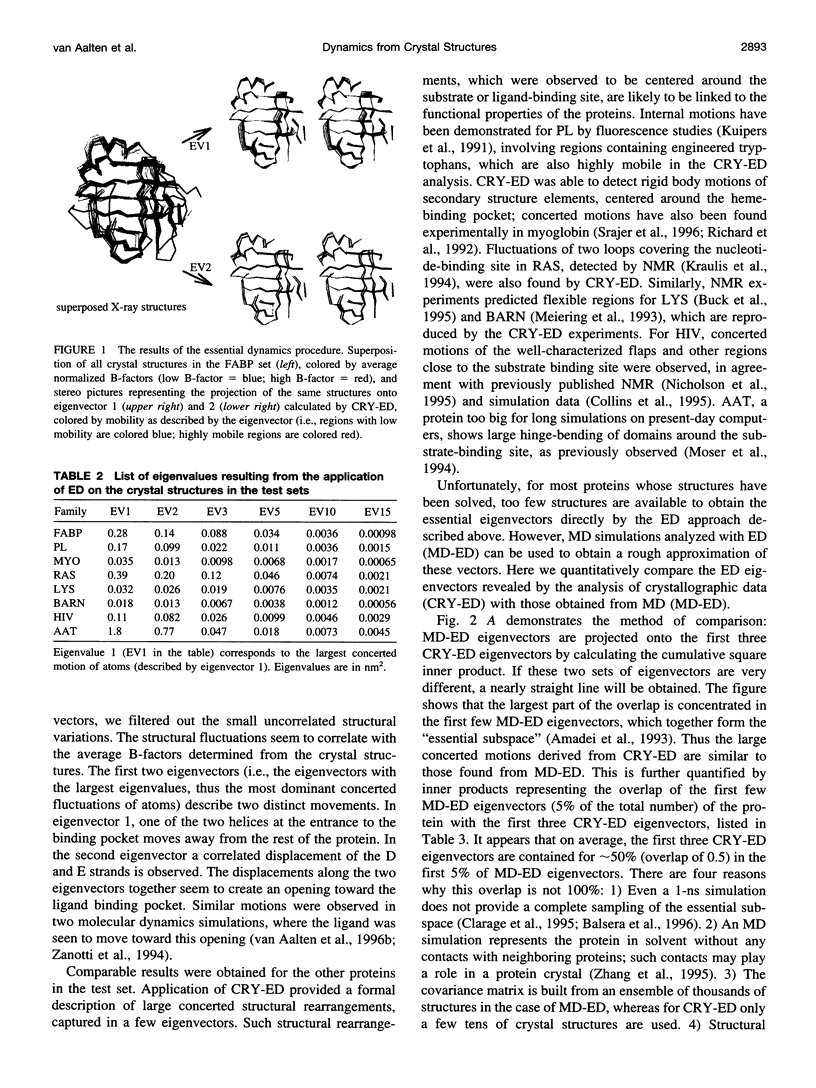
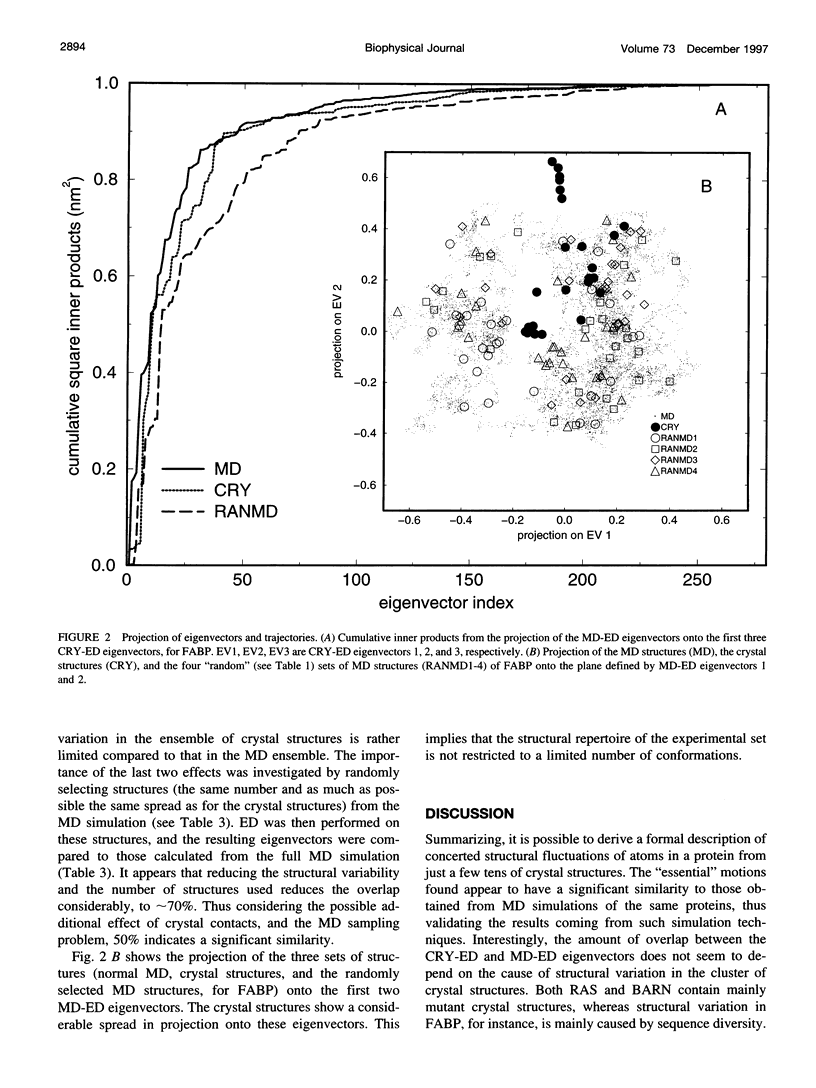
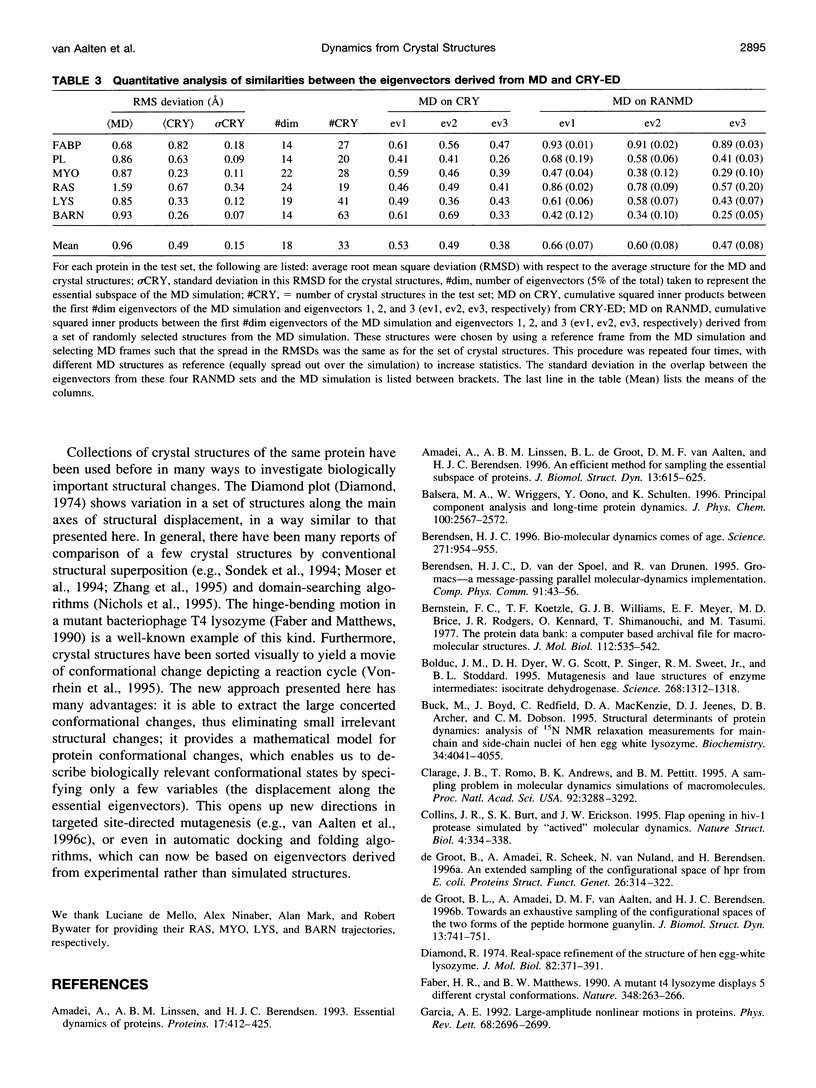
Abstract
A method is presented to mathematically extract concerted structural transitions in proteins from collections of crystal structures. The "essential dynamics" procedure is used to filter out small-amplitude fluctuations from such a set of structures; the remaining large conformational changes describe motions such as those important for the uptake/release of substrate/ligand and in catalytic reactions. The method is applied to sets of x-ray structures for a number of proteins, and the results are compared with the results from essential dynamics as applied to molecular dynamics simulations of those proteins. A significant degree of similarity is found, thereby providing a direct experimental basis for the application of such simulations to the description of large concerted motions in proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amadei A., Linssen A. B., Berendsen H. J. Essential dynamics of proteins. Proteins. 1993 Dec;17(4):412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- Amadei A., Linssen A. B., de Groot B. L., van Aalten D. M., Berendsen H. J. An efficient method for sampling the essential subspace of proteins. J Biomol Struct Dyn. 1996 Feb;13(4):615–625. doi: 10.1080/07391102.1996.10508874. [DOI] [PubMed] [Google Scholar]
- Berendsen H. J. Bio-molecular dynamics comes of age. Science. 1996 Feb 16;271(5251):954–955. doi: 10.1126/science.271.5251.954. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bolduc J. M., Dyer D. H., Scott W. G., Singer P., Sweet R. M., Koshland D. E., Jr, Stoddard B. L. Mutagenesis and Laue structures of enzyme intermediates: isocitrate dehydrogenase. Science. 1995 Jun 2;268(5215):1312–1318. doi: 10.1126/science.7761851. [DOI] [PubMed] [Google Scholar]
- Buck M., Boyd J., Redfield C., MacKenzie D. A., Jeenes D. J., Archer D. B., Dobson C. M. Structural determinants of protein dynamics: analysis of 15N NMR relaxation measurements for main-chain and side-chain nuclei of hen egg white lysozyme. Biochemistry. 1995 Mar 28;34(12):4041–4055. doi: 10.1021/bi00012a023. [DOI] [PubMed] [Google Scholar]
- Clarage J. B., Romo T., Andrews B. K., Pettitt B. M., Phillips G. N., Jr A sampling problem in molecular dynamics simulations of macromolecules. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3288–3292. doi: 10.1073/pnas.92.8.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. R., Burt S. K., Erickson J. W. Flap opening in HIV-1 protease simulated by 'activated' molecular dynamics. Nat Struct Biol. 1995 Apr;2(4):334–338. doi: 10.1038/nsb0495-334. [DOI] [PubMed] [Google Scholar]
- Diamond R. Real-space refinement of the structure of hen egg-white lysozyme. J Mol Biol. 1974 Jan 25;82(3):371–391. doi: 10.1016/0022-2836(74)90598-1. [DOI] [PubMed] [Google Scholar]
- Faber H. R., Matthews B. W. A mutant T4 lysozyme displays five different crystal conformations. Nature. 1990 Nov 15;348(6298):263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- García AE. Large-amplitude nonlinear motions in proteins. Phys Rev Lett. 1992 Apr 27;68(17):2696–2699. doi: 10.1103/PhysRevLett.68.2696. [DOI] [PubMed] [Google Scholar]
- Genick U. K., Borgstahl G. E., Ng K., Ren Z., Pradervand C., Burke P. M., Srajer V., Teng T. Y., Schildkamp W., McRee D. E. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science. 1997 Mar 7;275(5305):1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- Jäger J., Moser M., Sauder U., Jansonius J. N. Crystal structures of Escherichia coli aspartate aminotransferase in two conformations. Comparison of an unliganded open and two liganded closed forms. J Mol Biol. 1994 Jun 3;239(2):285–305. doi: 10.1006/jmbi.1994.1368. [DOI] [PubMed] [Google Scholar]
- Kraulis P. J., Domaille P. J., Campbell-Burk S. L., Van Aken T., Laue E. D. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994 Mar 29;33(12):3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- Kuipers O. P., Vincent M., Brochon J. C., Verheij H. M., de Haas G. H., Gallay J. Insight into the conformational dynamics of specific regions of porcine pancreatic phospholipase A2 from a time-resolved fluorescence study of a genetically inserted single tryptophan residue. Biochemistry. 1991 Sep 10;30(36):8771–8785. doi: 10.1021/bi00100a008. [DOI] [PubMed] [Google Scholar]
- Meiering E. M., Bycroft M., Lubienski M. J., Fersht A. R. Structure and dynamics of barnase complexed with 3'-GMP studied by NMR spectroscopy. Biochemistry. 1993 Oct 19;32(41):10975–10987. doi: 10.1021/bi00092a006. [DOI] [PubMed] [Google Scholar]
- Mello L. V., van Aalten D. M., Findlay J. B. Comparison of ras-p21 bound to GDP and GTP: differences in protein and ligand dynamics. Protein Eng. 1997 Apr;10(4):381–387. doi: 10.1093/protein/10.4.381. [DOI] [PubMed] [Google Scholar]
- Moffat K. Time-resolved macromolecular crystallography. Annu Rev Biophys Biophys Chem. 1989;18:309–332. doi: 10.1146/annurev.bb.18.060189.001521. [DOI] [PubMed] [Google Scholar]
- Nichols W. L., Rose G. D., Ten Eyck L. F., Zimm B. H. Rigid domains in proteins: an algorithmic approach to their identification. Proteins. 1995 Sep;23(1):38–48. doi: 10.1002/prot.340230106. [DOI] [PubMed] [Google Scholar]
- Nicholson L. K., Yamazaki T., Torchia D. A., Grzesiek S., Bax A., Stahl S. J., Kaufman J. D., Wingfield P. T., Lam P. Y., Jadhav P. K. Flexibility and function in HIV-1 protease. Nat Struct Biol. 1995 Apr;2(4):274–280. doi: 10.1038/nsb0495-274. [DOI] [PubMed] [Google Scholar]
- Phillips G. N., Jr Comparison of the dynamics of myoglobin in different crystal forms. Biophys J. 1990 Feb;57(2):381–383. doi: 10.1016/S0006-3495(90)82540-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard L., Genberg L., Deak J., Chiu H. L., Miller R. J. Picosecond phase grating spectroscopy of hemoglobin and myoglobin: energetics and dynamics of global protein motion. Biochemistry. 1992 Nov 10;31(44):10703–10715. doi: 10.1021/bi00159a010. [DOI] [PubMed] [Google Scholar]
- Romo T. D., Clarage J. B., Sorensen D. C., Phillips G. N., Jr Automatic identification of discrete substates in proteins: singular value decomposition analysis of time-averaged crystallographic refinements. Proteins. 1995 Aug;22(4):311–321. doi: 10.1002/prot.340220403. [DOI] [PubMed] [Google Scholar]
- Smith L. J., Mark A. E., Dobson C. M., van Gunsteren W. F. Comparison of MD simulations and NMR experiments for hen lysozyme. Analysis of local fluctuations, cooperative motions, and global changes. Biochemistry. 1995 Aug 29;34(34):10918–10931. doi: 10.1021/bi00034a026. [DOI] [PubMed] [Google Scholar]
- Sondek J., Lambright D. G., Noel J. P., Hamm H. E., Sigler P. B. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature. 1994 Nov 17;372(6503):276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- Srajer V., Teng T., Ursby T., Pradervand C., Ren Z., Adachi S., Schildkamp W., Bourgeois D., Wulff M., Moffat K. Photolysis of the carbon monoxide complex of myoglobin: nanosecond time-resolved crystallography. Science. 1996 Dec 6;274(5293):1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994 Nov 11;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C., Schlauderer G. J., Schulz G. E. Movie of the structural changes during a catalytic cycle of nucleoside monophosphate kinases. Structure. 1995 May 15;3(5):483–490. doi: 10.1016/s0969-2126(01)00181-2. [DOI] [PubMed] [Google Scholar]
- Zanotti G., Feltre L., Spadon P. A possible route for the release of fatty acid from fatty acid-binding protein. Biochem J. 1994 Jul 15;301(Pt 2):459–463. doi: 10.1042/bj3010459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. J., Wozniak J. A., Matthews B. W. Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme. J Mol Biol. 1995 Jul 21;250(4):527–552. doi: 10.1006/jmbi.1995.0396. [DOI] [PubMed] [Google Scholar]
- de Groot B. L., Amadei A., Scheek R. M., van Nuland N. A., Berendsen H. J. An extended sampling of the configurational space of HPr from E. coli. Proteins. 1996 Nov;26(3):314–322. doi: 10.1002/(SICI)1097-0134(199611)26:3<314::AID-PROT7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- de Groot B. L., Amadei A., van Aalten D. M., Berendsen H. J. Toward an exhaustive sampling of the configurational spaces of the two forms of the peptide hormone guanylin. J Biomol Struct Dyn. 1996 Apr;13(5):741–751. doi: 10.1080/07391102.1996.10508888. [DOI] [PubMed] [Google Scholar]
- van Aalten D. M., Amadei A., Bywater R., Findlay J. B., Berendsen H. J., Sander C., Stouten P. F. A comparison of structural and dynamic properties of different simulation methods applied to SH3. Biophys J. 1996 Feb;70(2):684–692. doi: 10.1016/S0006-3495(96)79608-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten D. M., Amadei A., Linssen A. B., Eijsink V. G., Vriend G., Berendsen H. J. The essential dynamics of thermolysin: confirmation of the hinge-bending motion and comparison of simulations in vacuum and water. Proteins. 1995 May;22(1):45–54. doi: 10.1002/prot.340220107. [DOI] [PubMed] [Google Scholar]
- van Aalten D. M., Findlay J. B., Amadei A., Berendsen H. J. Essential dynamics of the cellular retinol-binding protein--evidence for ligand-induced conformational changes. Protein Eng. 1995 Nov;8(11):1129–1135. doi: 10.1093/protein/8.11.1129. [DOI] [PubMed] [Google Scholar]