Abstract
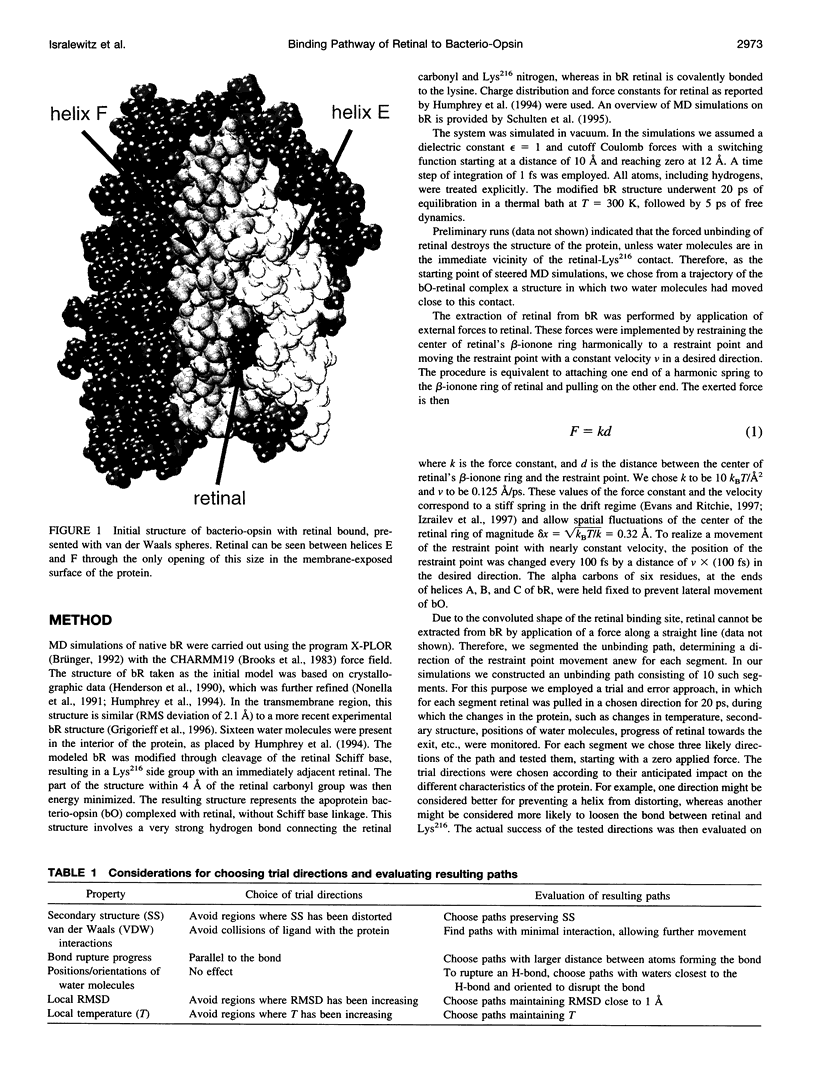
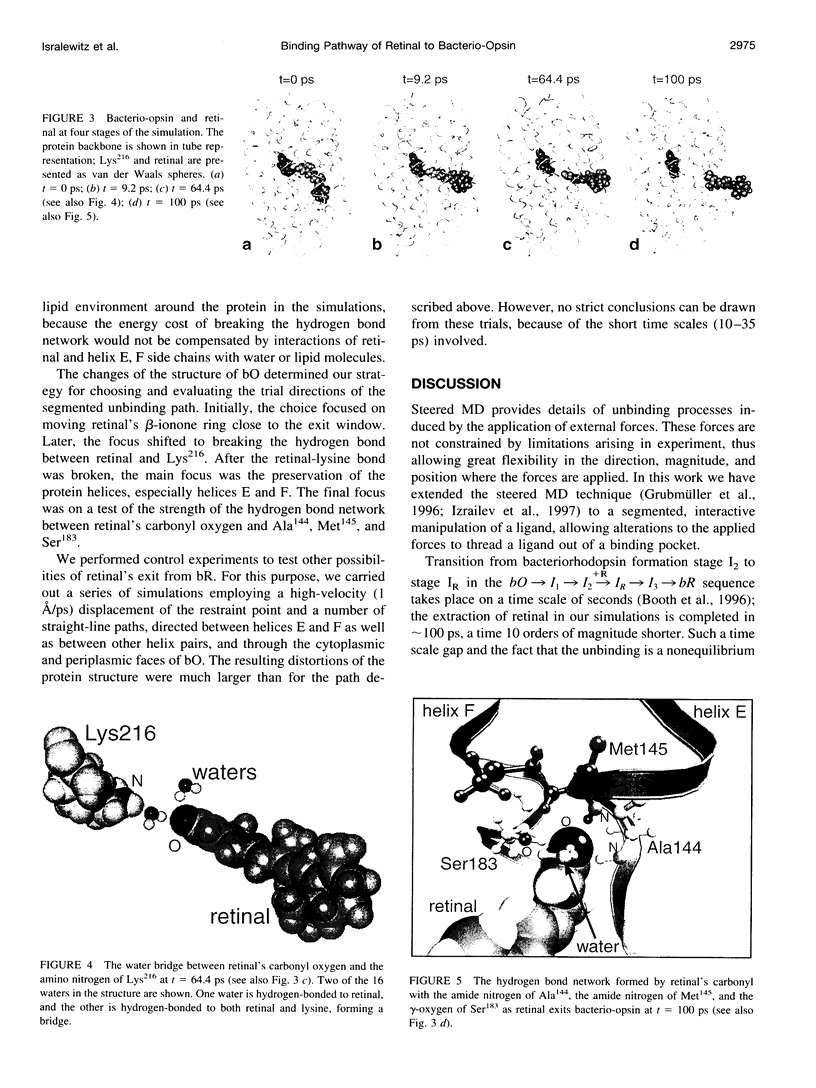
Formation of bacteriorhodopsin (bR) from apoprotein and retinal has been studied experimentally, but the actual pathway, including the point of entry, is little understood. Molecular dynamics simulations provide a surprisingly clear prediction. A window between bR helices E and F in the transmembrane part of the protein can be identified as an entry point for retinal. Steered molecular dynamics, performed by applying a series of external forces in the range of 200-1000 pN over a period of 0.2 ns to retinal, allows one to extract this chromophore from bR once the Schiff base bond to Lys216 is cleaved. Extraction proceeds until the retinal tail forms a hydrogen bond network with Ala144, Met145, and Ser183 side groups lining the exit/entry window. The manipulation induces a distortion with a fitted root mean square deviation of coordinates (ignoring retinal, water, and hydrogen atoms) of less than 1.9 A by the time the retinal carbonyl reaches the protein surface. The forces needed to extract retinal are due to friction and do not indicate significant potential barriers. The simulations therefore suggest a pathway for the binding of retinal. Water molecules are found to play a crucial role in the binding process.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbach C., Greenhalgh D. A., Khorana H. G., Hubbell W. L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balsera M., Stepaniants S., Izrailev S., Oono Y., Schulten K. Reconstructing potential energy functions from simulated force-induced unbinding processes. Biophys J. 1997 Sep;73(3):1281–1287. doi: 10.1016/S0006-3495(97)78161-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley H., Huang K. S., Radhakrishnan R., Ross A. H., Takagaki Y., Khorana H. G. Site of attachment of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2225–2229. doi: 10.1073/pnas.78.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge R. R. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim Biophys Acta. 1990 Apr 26;1016(3):293–327. doi: 10.1016/0005-2728(90)90163-x. [DOI] [PubMed] [Google Scholar]
- Booth P. J., Farooq A., Flitsch S. L. Retinal binding during folding and assembly of the membrane protein bacteriorhodopsin. Biochemistry. 1996 May 7;35(18):5902–5909. doi: 10.1021/bi960129e. [DOI] [PubMed] [Google Scholar]
- Cladera J., Torres J., Padrós E. Analysis of conformational changes in bacteriorhodopsin upon retinal removal. Biophys J. 1996 Jun;70(6):2882–2887. doi: 10.1016/S0006-3495(96)79858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch R. K. Studies of rhodopsin and bacteriorhodopsin using modified retinals. Photochem Photobiol. 1986 Dec;44(6):803–807. doi: 10.1111/j.1751-1097.1986.tb05540.x. [DOI] [PubMed] [Google Scholar]
- Evans E., Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997 Apr;72(4):1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E., Ritchie K., Merkel R. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys J. 1995 Jun;68(6):2580–2587. doi: 10.1016/S0006-3495(95)80441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrens D. L., Altenbach C., Yang K., Hubbell W. L., Khorana H. G. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996 Nov 1;274(5288):768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- Flitsch S. L., Khorana H. G. Structural studies on transmembrane proteins. 1. Model study using bacteriorhodopsin mutants containing single cysteine residues. Biochemistry. 1989 Sep 19;28(19):7800–7805. doi: 10.1021/bi00445a041. [DOI] [PubMed] [Google Scholar]
- Florin E. L., Moy V. T., Gaub H. E. Adhesion forces between individual ligand-receptor pairs. Science. 1994 Apr 15;264(5157):415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995 Dec;23(4):566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Greenhalgh D. A., Altenbach C., Hubbell W. L., Khorana H. G. Locations of Arg-82, Asp-85, and Asp-96 in helix C of bacteriorhodopsin relative to the aqueous boundaries. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8626–8630. doi: 10.1073/pnas.88.19.8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh D. A., Farrens D. L., Subramaniam S., Khorana H. G. Hydrophobic amino acids in the retinal-binding pocket of bacteriorhodopsin. J Biol Chem. 1993 Sep 25;268(27):20305–20311. [PubMed] [Google Scholar]
- Grigorieff N., Ceska T. A., Downing K. H., Baldwin J. M., Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996 Jun 14;259(3):393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Grubmüller H., Heymann B., Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996 Feb 16;271(5251):997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- Hackett N. R., Stern L. J., Chao B. H., Kronis K. A., Khorana H. G. Structure-function studies on bacteriorhodopsin. V. Effects of amino acid substitutions in the putative helix F. J Biol Chem. 1987 Jul 5;262(19):9277–9284. [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Ho M. T., Massey J. B., Pownall H. J., Anderson R. E., Hollyfield J. G. Mechanism of vitamin A movement between rod outer segments, interphotoreceptor retinoid-binding protein, and liposomes. J Biol Chem. 1989 Jan 15;264(2):928–935. [PubMed] [Google Scholar]
- Huang K. S., Bayley H., Liao M. J., London E., Khorana H. G. Refolding of an integral membrane protein. Denaturation, renaturation, and reconstitution of intact bacteriorhodopsin and two proteolytic fragments. J Biol Chem. 1981 Apr 25;256(8):3802–3809. [PubMed] [Google Scholar]
- Humphrey W., Logunov I., Schulten K., Sheves M. Molecular dynamics study of bacteriorhodopsin and artificial pigments. Biochemistry. 1994 Mar 29;33(12):3668–3678. doi: 10.1021/bi00178a025. [DOI] [PubMed] [Google Scholar]
- Izrailev S., Stepaniants S., Balsera M., Oono Y., Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys J. 1997 Apr;72(4):1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana H. G. Bacteriorhodopsin, a membrane protein that uses light to translocate protons. J Biol Chem. 1988 Jun 5;263(16):7439–7442. [PubMed] [Google Scholar]
- Khorana H. G. Two light-transducing membrane proteins: bacteriorhodopsin and the mammalian rhodopsin. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1166–1171. doi: 10.1073/pnas.90.4.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. P., Khorana H. G. Mechanism of light-dependent proton translocation by bacteriorhodopsin. J Bacteriol. 1993 Mar;175(6):1555–1560. doi: 10.1128/jb.175.6.1555-1560.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Proton transfer and energy coupling in the bacteriorhodopsin photocycle. J Bioenerg Biomembr. 1992 Apr;24(2):169–179. doi: 10.1007/BF00762675. [DOI] [PubMed] [Google Scholar]
- London E., Khorana H. G. Denaturation and renaturation of bacteriorhodopsin in detergents and lipid-detergent mixtures. J Biol Chem. 1982 Jun 25;257(12):7003–7011. [PubMed] [Google Scholar]
- Marti T., Otto H., Mogi T., Rösselet S. J., Heyn M. P., Khorana H. G. Bacteriorhodopsin mutants containing single substitutions of serine or threonine residues are all active in proton translocation. J Biol Chem. 1991 Apr 15;266(11):6919–6927. [PubMed] [Google Scholar]
- Marti T., Rösselet S. J., Otto H., Heyn M. P., Khorana H. G. The retinylidene Schiff base counterion in bacteriorhodopsin. J Biol Chem. 1991 Oct 5;266(28):18674–18683. [PubMed] [Google Scholar]
- McCaslin D. R., Tanford C. Effects of detergent micelles on the recombination reaction of opsin and 11-cis-retinal. Biochemistry. 1981 Sep 1;20(18):5207–5212. doi: 10.1021/bi00521a017. [DOI] [PubMed] [Google Scholar]
- Miercke L. J., Betlach M. C., Mitra A. K., Shand R. F., Fong S. K., Stroud R. M. Wild-type and mutant bacteriorhodopsins D85N, D96N, and R82Q: purification to homogeneity, pH dependence of pumping, and electron diffraction. Biochemistry. 1991 Mar 26;30(12):3088–3098. doi: 10.1021/bi00226a016. [DOI] [PubMed] [Google Scholar]
- Mogi T., Marti T., Khorana H. G. Structure-function studies on bacteriorhodopsin. IX. Substitutions of tryptophan residues affect protein-retinal interactions in bacteriorhodopsin. J Biol Chem. 1989 Aug 25;264(24):14197–14201. [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Chao B. H., Khorana H. G. Structure-function studies on bacteriorhodopsin. VIII. Substitutions of the membrane-embedded prolines 50, 91, and 186: the effects are determined by the substituting amino acids. J Biol Chem. 1989 Aug 25;264(24):14192–14196. [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Hackett N. R., Khorana H. G. Bacteriorhodopsin mutants containing single tyrosine to phenylalanine substitutions are all active in proton translocation. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5595–5599. doi: 10.1073/pnas.84.16.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Marti T., Chao B. H., Khorana H. G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Meentzen M., Schuhmann L. Reversible dissociation of the purple complex in bacteriorhodopsin and identification of 13-cis and all-trans-retinal as its chromophores. Eur J Biochem. 1973 Dec 17;40(2):453–463. doi: 10.1111/j.1432-1033.1973.tb03214.x. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Schuhmann L., Gruber H. Light-dependent reaction of bacteriorhodopsin with hydroxylamine in cell suspensions of Halobacterium halobium: demonstration of an apo-membrane. FEBS Lett. 1974 Aug 30;44(3):257–261. doi: 10.1016/0014-5793(74)81152-x. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Schuhmann L. Reconstitution of bacteriorhodopsin. FEBS Lett. 1974 Aug 30;44(3):262–265. doi: 10.1016/0014-5793(74)81153-1. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Tittor J., Bamberg E. A unifying concept for ion translocation by retinal proteins. J Bioenerg Biomembr. 1992 Apr;24(2):181–191. doi: 10.1007/BF00762676. [DOI] [PubMed] [Google Scholar]
- Popot J. L., Engelman D. M. Membrane protein folding and oligomerization: the two-stage model. Biochemistry. 1990 May 1;29(17):4031–4037. doi: 10.1021/bi00469a001. [DOI] [PubMed] [Google Scholar]
- Renthal R., McMillan K., Guerra L., Garcia M. N., Rangel R., Jen C. M. Long-range effects on the retinal chromophore of bacteriorhodopsin caused by surface carboxyl group modification. Biochemistry. 1995 Jun 20;34(24):7869–7878. doi: 10.1021/bi00024a011. [DOI] [PubMed] [Google Scholar]
- Rousso I., Brodsky I., Lewis A., Sheves M. The role of water in retinal complexation to bacterio-opsin. J Biol Chem. 1995 Jun 9;270(23):13860–13868. doi: 10.1074/jbc.270.23.13860. [DOI] [PubMed] [Google Scholar]
- Stern L. J., Khorana H. G. Structure-function studies on bacteriorhodopsin. X. Individual substitutions of arginine residues by glutamine affect chromophore formation, photocycle, and proton translocation. J Biol Chem. 1989 Aug 25;264(24):14202–14208. [PubMed] [Google Scholar]
- Subramaniam S., Greenhalgh D. A., Rath P., Rothschild K. J., Khorana H. G. Replacement of leucine-93 by alanine or threonine slows down the decay of the N and O intermediates in the photocycle of bacteriorhodopsin: implications for proton uptake and 13-cis-retinal----all-trans-retinal reisomerization. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6873–6877. doi: 10.1073/pnas.88.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K., Block S. M. Biological applications of optical forces. Annu Rev Biophys Biomol Struct. 1994;23:247–285. doi: 10.1146/annurev.bb.23.060194.001335. [DOI] [PubMed] [Google Scholar]
- Szuts E. Z., Harosi F. I. Solubility of retinoids in water. Arch Biochem Biophys. 1991 Jun;287(2):297–304. doi: 10.1016/0003-9861(91)90482-x. [DOI] [PubMed] [Google Scholar]
- Towner P., Gaertner W., Walckhoff B., Oesterhelt D., Hopf H. Regeneration of rhodopsin and bacteriorhodopsin. The role of retinal analogues as inhibitors. Eur J Biochem. 1981 Jul;117(2):353–359. doi: 10.1111/j.1432-1033.1981.tb06345.x. [DOI] [PubMed] [Google Scholar]
- Unger V. M., Hargrave P. A., Baldwin J. M., Schertler G. F. Arrangement of rhodopsin transmembrane alpha-helices. Nature. 1997 Sep 11;389(6647):203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- Zingoni J., Or Y. S., Crouch R. K., Chang C. H., Govindjee R., Ebrey T. G. Effect of variation of retinal polyene side-chain length on formation and function of bacteriorhodopsin analogue pigments. Biochemistry. 1986 Apr 22;25(8):2022–2027. doi: 10.1021/bi00356a028. [DOI] [PubMed] [Google Scholar]