Abstract
We propose a partially flexible, double-helical model for describing the conformational and dynamic properties of DNA. In this model, each nucleotide is represented by one element (bead), and the known geometrical features of the double helix are incorporated in the equilibrium conformation. Each bead is connected to a few neighbor beads in both strands by means of stiff springs that maintain the connectivity but still allow for some extent of flexibility and internal motion. We have used Brownian dynamics simulation to sample the conformational space and monitor the overall and internal dynamics of short DNA pieces, with up to 20 basepairs. From Brownian trajectories, we calculate the dimensions of the helix and estimate its persistence length. We obtain translational diffusion coefficient and various rotational relaxation times, including both overall rotation and internal motion. Although we have not carried out a detailed parameterization of the model, the calculated properties agree rather well with experimental data available for those oligomers.
Full text
PDF

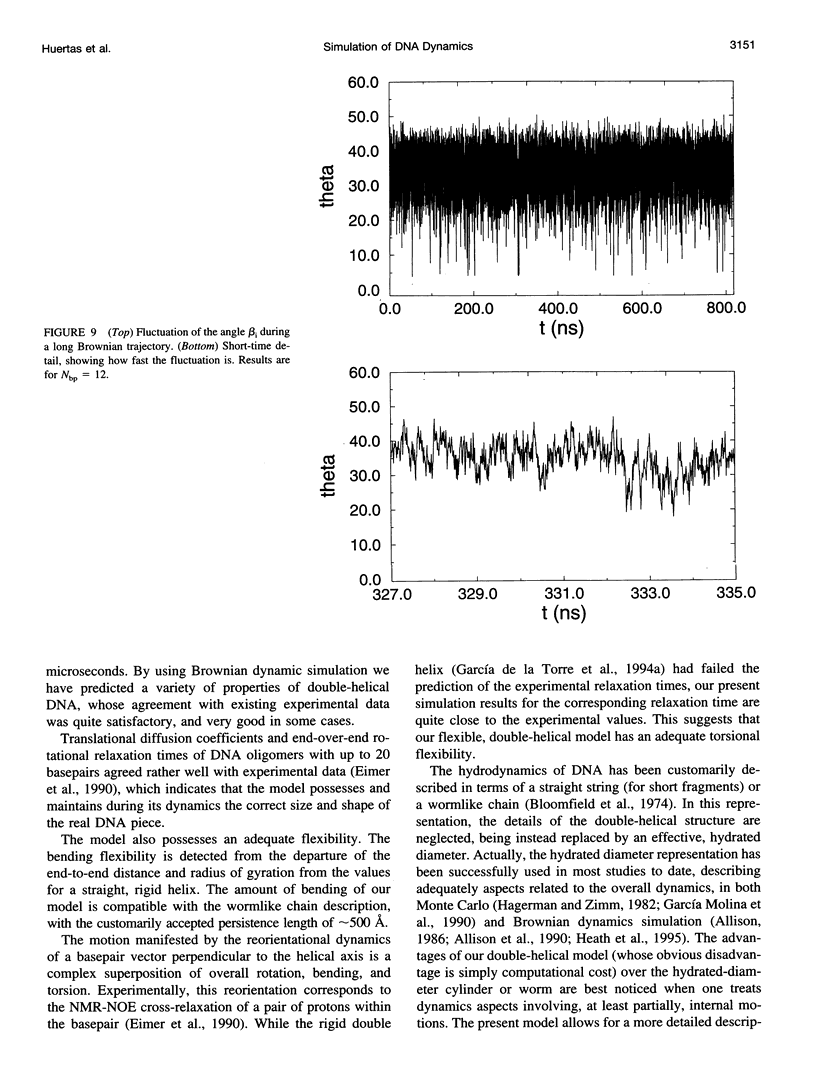
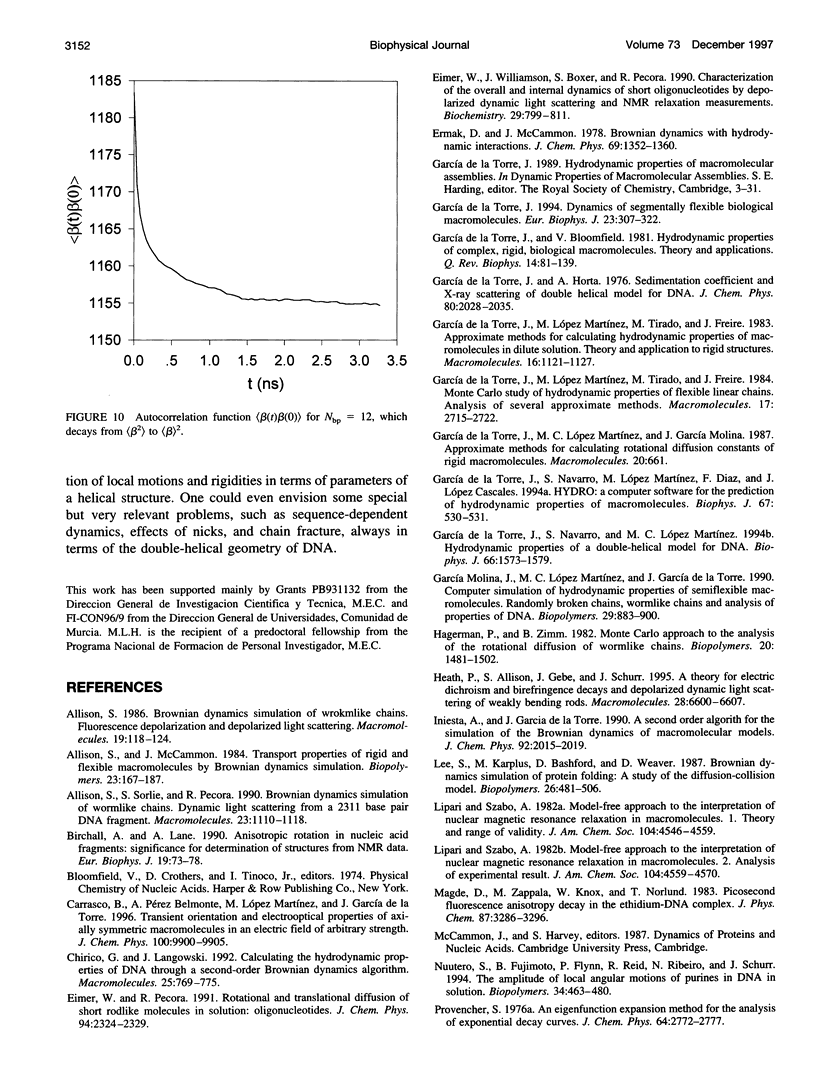

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchall A. J., Lane A. N. Anisotropic rotation in nucleic acid fragments: significance for determination of structures from NMR data. Eur Biophys J. 1990;19(2):73–78. doi: 10.1007/BF00185089. [DOI] [PubMed] [Google Scholar]
- Eimer W., Williamson J. R., Boxer S. G., Pecora R. Characterization of the overall and internal dynamics of short oligonucleotides by depolarized dynamic light scattering and NMR relaxation measurements. Biochemistry. 1990 Jan 23;29(3):799–811. doi: 10.1021/bi00455a030. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J. G., Bloomfield V. A. Hydrodynamic properties of complex, rigid, biological macromolecules: theory and applications. Q Rev Biophys. 1981 Feb;14(1):81–139. doi: 10.1017/s0033583500002080. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J., Navarro S., Lopez Martinez M. C., Diaz F. G., Lopez Cascales J. J. HYDRO: a computer program for the prediction of hydrodynamic properties of macromolecules. Biophys J. 1994 Aug;67(2):530–531. doi: 10.1016/S0006-3495(94)80512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia de la Torre J., Navarro S., Lopez Martinez M. C. Hydrodynamic properties of a double-helical model for DNA. Biophys J. 1994 May;66(5):1573–1579. doi: 10.1016/S0006-3495(94)80949-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Molina J. J., López Martínez M. C., García de la Torre J. Computer simulation of hydrodynamic properties of semiflexible macromolecules: randomly broken chains, wormlike chains, and analysis of properties of DNA. Biopolymers. 1990 May-Jun;29(6-7):883–900. doi: 10.1002/bip.360290603. [DOI] [PubMed] [Google Scholar]
- Keller J. B., Rubinow S. I. Swimming of flagellated microorganisms. Biophys J. 1976 Feb;16(2 Pt 1):151–170. doi: 10.1016/s0006-3495(76)85672-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Karplus M., Bashford D., Weaver D. Brownian dynamics simulation of protein folding: a study of the diffusion-collision model. Biopolymers. 1987 Apr;26(4):481–506. doi: 10.1002/bip.360260404. [DOI] [PubMed] [Google Scholar]
- Nuutero S., Fujimoto B. S., Flynn P. F., Reid B. R., Ribeiro N. S., Schurr J. M. The amplitude of local angular motion of purines in DNA in solution. Biopolymers. 1994 Apr;34(4):463–480. doi: 10.1002/bip.360340404. [DOI] [PubMed] [Google Scholar]
- Schneller W., Weaver D. L. Simulation of alpha-helix-coil transitions in simplified polyvaline: equilibrium properties and Brownian dynamics. Biopolymers. 1993 Oct;33(10):1519–1535. doi: 10.1002/bip.360331004. [DOI] [PubMed] [Google Scholar]
- Schurr J. M., Fujimoto B. S. The amplitude of local angular motions of intercalated dyes and bases in DNA. Biopolymers. 1988 Oct;27(10):1543–1569. doi: 10.1002/bip.360271003. [DOI] [PubMed] [Google Scholar]
- Smith S. B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996 Feb 9;271(5250):795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Wade R. C., Davis M. E., Luty B. A., Madura J. D., McCammon J. A. Gating of the active site of triose phosphate isomerase: Brownian dynamics simulations of flexible peptide loops in the enzyme. Biophys J. 1993 Jan;64(1):9–15. doi: 10.1016/S0006-3495(93)81335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre J. G. Hydrodynamics of segmentally flexible macromolecules. Eur Biophys J. 1994;23(5):307–322. doi: 10.1007/BF00188655. [DOI] [PubMed] [Google Scholar]