Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human gammaherpesvirus implicated in AIDS-related neoplasms. Previously, we demonstrated that the early lytic gene product K-bZIP is a transcriptional repressor that affects a subset of viral gene transcriptions mediated by the viral transactivator K-Rta (Y. Izumiya et al. J. Virol. 77:1441-1451, 2003). Sumoylation has emerged as an important posttranslational modification that affects the location and function of cellular and viral proteins and also plays a significant role in transcriptional repression along with Ubc9, the E2 SUMO conjugation enzyme. Here, we provide evidence that K-bZIP is sumoylated at the lysine 158 residue and associates with Ubc9 both in a cell-free system and in virus-infected BCBL-1 cells. Reporter assays showed that the expression of SUMO-specific protease 1 attenuated the transcriptional repression activity of K-bZIP. The expression of a K-bZIPK158R mutant, which was no longer sumoylated, exhibited the reduced transcriptional repression activity. This indicates that sumoylation plays an important part in the transcriptional repression activity of K-bZIP. Finally, chromatin immunoprecipitation experiments demonstrated that K-bZIP interacts with and recruits Ubc9 to specific KSHV promoters. Thus, our data indicate that K-bZIP is a SUMO adaptor, which recruits Ubc9 to specific viral target promoters, thereby exerting its transcriptional repression activity.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a member of the gammaherpesvirus family, which includes Epstein-Barr virus (EBV) and herpesvirus saimiri. KSHV infection is associated with all types of Kaposi's sarcoma (KS), including AIDS-associated KS, endemic forms of KS, and renal transplant-related KS. This virus has also been implicated in B-cell lymphoproliferative diseases such as primary effusion lymphoma and multicentric Castleman's disease (13). Like other herpesviruses, KSHV encodes both latent and lytic genes (61). Latent genes are primarily responsible for the maintenance of latency and directly involved in cell transformation. Lytic genes participate directly in viral replication (genome replication, transcription, etc.) or indirectly by providing a cellular environment conducive for viral infection (e.g., B-cell activation, immune modulation of host response, target cell recruitment) or both (7, 61, 70). Products from both latent and lytic genes of KSHV have been shown to participate in transformation and tumor progression (7, 49).
We previously reported the identification of K-bZIP/K8 (45; also see reference 24) as an early lytic cycle gene. Studies by others found that K-bZIP is among the earliest to be expressed after acute infection or reactivation of the latent genome (38, 54, 56). K-bZIP is a 237-amino-acid protein containing a basic and leucine zipper domain and is the positional and structural analogue of EBV Zta (also called ZEBRA, BZLF1, and EB1) (45). EBV Zta is a strong transcriptional factor capable of triggering EBV reactivation and lytic replication (10) and transactivates a number of viral and cellular genes (50, 78). Although transactivation is a major role assumed by EBV Zta, this gene also exhibits trans-repression activity on certain cellular promoters via either a direct or indirect mechanism (14, 51, 52). In addition to being a transcriptional factor, EBV Zta is involved in viral genome replication as an origin-binding protein (15, 63), interacts with p53, and blocks the G1/S transition of the infected cell (16). The multiple functions of EBV Zta are modulated posttranslationally by phosphorylation and sumoylation (1, 14).
The overall structure of K-bZIP is similar to EBV Zta, with the bZIP region located near the C terminus and the presumptive regulatory domain at the N terminus. The sequence homology between these two proteins, however, is limited, with 37% similarity (45). Although there are a number of biochemical properties of K-bZIP which are shared with EBV Zta, there are also important differences. Like EBV Zta, K-bZIP forms homodimers (45) and is localized in the nucleus and in PODs (for “PML [promyelocytic] oncogenic domains”) (36, 79). Attempts to show that K-bZIP directly binds DNA have not met with success, although chromatin immunoprecipitation (ChIP) experiments suggest that it is associated with the KSHV chromosome, including the regions near the replication origin and the early gene promoters (44 and this report). Unlike EBV Zta, overexpression of K-bZIP in a KSHV latent cell line is insufficient to induce lytic replication (57). K-bZIP has a relatively strong trans-repression activity (30, 31, 33, 55) and modest trans-activation activity (76) when coexpressed with other transcriptional factors. Recent evidence suggests that K-bZIP directly participates in KSHV genome replication (4), although the exact role is not clear. We and others recently showed that K-bZIP delayed G1 to S progression by at least two mechanisms: direct inhibition of cyclin-CDK2 (34) and transactivation of p21 (80).
Located upstream from K-bZIP in the KSHV genome is another early lytic gene, K-Rta/ORF50. K-Rta, the homolog of EBV BRLF1/Rta, is a potent transactivator with broad promoter specificity (6, 35, 47, 62, 68) capable of activating a large number of viral early genes, such as ORF6, K8, K12, K14, ORF57, nut-1/polyadenylated nuclear (PAN) RNA, viral interleukin-6, viral interferon regulatory factor-1, K-Rta, and thymidine kinase (9, 35, 41, 54, 62, 68, 73, 74). Overexpression of K-Rta activates a transcriptional cascade leading to lytic replication (22, 48, 54, 71). The broad promoter specificity of this viral gene comes from its ability both to bind DNA directly (69) and to associate with other DNA-binding transcriptional factors, such as RBP-Jk (42) and C/EBPα (28). Furthermore, K-Rta interacts with several transcriptional regulators, including Oct-1 (62), STAT3 (27), C/EBPα (28), CBP (25), K-RBP (75), SWI/SNF, and TRAP230 (26). Thus, K-Rta appears to be a global transcriptional factor which assembles different transcriptional complexes on different promoter sites. Perhaps related to its transcriptional potency, constitutive expression of K-Rta often leads to cell death.
We previously showed that K-bZIP and K-Rta physically associate with each other, and K-bZIP represses K-Rta transactivation in a promoter-dependent manner (33, 43) such that ORF57 and K8 promoters are affected, whereas the nut1/PAN RNA promoter is immune to this repression. We interpret this to mean that K-bZIP only targets certain K-Rta transcriptional complexes for inhibition and suggest that K-bZIP is a feedback modulator of K-Rta. This pattern of regulation may be critical to the survival of virus in the infected cell. This model is consistent with the tightly and temporally regulated activities of K-Rta and K-bZIP during viral infection (38). EBV Rta and Zta also have been shown to functionally interact with each other in a gene-specific manner (14, 59). For some genes, both viral regulators are synergistic (2, 17, 58), whereas for others, EBV Zta represses Rta activity (14, 18, 59). Interestingly, the repression activity is influenced by the phosphorylation status of Zta (14). EBV Zta trans-repression activity on RAR (retinoic acid receptor) and p53, by direct protein-protein interaction, has also been reported (66, 83). Likewise, K-bZIP was shown to repress p53 and CBP-mediated transactivation (30, 55). Taken together, the data suggest that trans-repression is an important function of the bZIP proteins of gammaherpesviruses.
In this study, we explore the repression mechanism of K-bZIP. A major finding is that K-bZIP is sumoylated and binds SUMO E2-conjugating enzyme Ubc9. Sumoylation plays an important part in K-bZIP trans-repression of K-Rta. The repression activity of K-bZIP correlates with its ability to be sumoylated and to recruit Ubc9 to the promoter site, where K-Rta resides. The data are consistent with the increasingly recognized role of SUMO in transcriptional repression (64). The potential mechanisms whereby K-bZIP represses K-Rta will be discussed.
MATERIALS AND METHODS
Plasmids.
Plasmids encoding the full-length K-bZIP (K-bZIP/wt, residues 1 to 273) and a natural spliced variant of K-bZIP, K-bZIPΔLZ, which carries a deletion of the leucine-zipper region, were cloned into pcDNA3.1 (Invitrogen). This cloning introduced a CpoI site and a Flag tag or T7 tag to the N terminus as described previously (45). The resulting plasmids were designated pFlag-K-bZIP, pT7-K-bZIP, or pT7-K-bZIPΔLZ. Full-length cDNAs of SUMO-1, SUMO-2, SUMO-3, and Ubc9 were amplified from BCBL-1 total RNA by reverse transcription-PCR (RT-PCR) with primers listed in Table 1 and cloned into the CpoI site of pcDNA-Flag- or pGEX-modified vector, which introduced a CpoI site. Our SUMO protein sequences are identical with the one previously reported (3). In order to prepare the active form of SUMO proteins, a stop codon was generated immediately after two glycines at the C-terminal region of each SUMO protein by site-directed mutagenesis (Stratagene) with primers listed in Table 2, using pGEX expression plasmids served as templates. These plasmids were designated as pGEX-SUMO-1GG, pGEX-SUMO-2GG, or pGEX-SUMO-3GG. A plasmid encoding an enzymatically inactive mutant of Ubc9 was prepared by mutating Cysteine 93 to Serine using PCR-based mutagenesis with primers listed in Table 2. pFlag-K-bZIPK158R was generated with site-directed mutagenesis (Stratagene) with primers listed in Table 2, using pFlag-K-bZIP as a template. Deletion fragments of K-bZIP were amplified by PCR using PFU-turbo (Stratagene) with primers previously described (33) and cloned into the CpoI site of pcDNA-Flag-modified vector. Plasmids containing SUMO-specific proteinase 1 (SENP1) were previously described (20), and the inactive mutant (SENP1 R630L, K631M) was generated by PCR-based mutagenesis using primers listed in Table 2. pGEX SAE1/SAE2 expression plasmid (72) was a generous gift from Ronald T. Hay (University of St. Andrews).
TABLE 1.
Primers used for cloning
| Primer | Sequence (5′-3′)a |
|---|---|
| SUMO-1 F | aacggtccg ATG TCT GAC CAG GAG GCA AAA |
| SUMO-1 R | aacggaccg CTA AAC TGT TGA ATG ACC CCC |
| SUMO-2 F | aacggtccg ATG GCC GAC GAA AAG CCC AAG |
| SUMO-2 R | aacggaccg TCA GTA GAC ACC TCC CGT CTG |
| SUMO-3 F | aacggtccg ATG TCC GAG GAG AAG CCC AAG |
| SUMO-3 R | aacggaccg CTA GAA ACT GTG CCC TGC CAG |
| Ubc9 F | aacggtccg ATG TCG GGG ATC GCC CTC AGC |
| Ubc9 R | aacggaccg TTA TGA GGG CGC AAA CTT CTT G |
In all sequences, the italic lowercase nucleotides represent restriction enzyme sites used for cloning the PCR products.
TABLE 2.
Primers used for mutagenesis
| Primer | Sequence (5′-3′)a |
|---|---|
| SUMO-1GG-F | ACG GGA GGT TAA TCA ACA GTT TAC GGT CCG TGA |
| SUMO-1GG-R | AAC TGT TGA TTA ACC TCC CGT TTG TTC CTG ATA |
| SUMO-2GG-F | CG GGA GGT TAA TAC TGA CGG TCC GTG AAT TCA TC |
| SUMO-2GG-R | CCG TCA GTA TTA ACC TCC CGT CTG CTG TTG GAA C |
| SUMO-3GG-F | ACG GGA GGT TAG CCG GAG AGC AGC CTG GCA GG |
| SUMO-3GG-R | GCT CTC CGG CTA ACC TCC CGT CTG CTG CTG GAA C |
| SENP1 RK/LM | CCA TAC TTC CTGATG CGG ATG GTC TGG GAG |
| SENP1 RK/LM | CTC CCA GAC CAT CCG CATCAG GAA GTA TGG |
| K-bZIP158K/R-F | TCT GTA GTT AGG GCC GAA GTA TGT GAT CAG TCA |
| K-bZIP 158K/R-R | TTC GGC CCT AAC TAC AGA CGC AGG CAC G |
| Ubc9 C93S-F | GGG ACA GTG TCC CTG TCC ATC TTA GAG GAG GA |
| Ubc9 C93S-R | GGA CAG GGA CAC TGT CCC CGA AGG GTA CAC AT |
A mutation was introduced in the underlined sequence.
Cell culture.
Human embryonic kidney epithelial 293 cells and 293T cells were grown in monolayer cultures in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in the presence of 5% CO2. The TREx-K-Rta BCBL-1 cell line had been generated by Nakamura et al. (54) and was cultured in RPMI 1640 supplemented with 20% FBS, 100 μg/ml of blasticidin (Invitrogen), and 100 μg/ml of hygromycin (Invitrogen).
Immunoprecipitation and immunoblot analyses.
TREx-K-Rta BCBL-1 cells were rinsed in ice-cold phosphate-buffered saline (PBS), and 1 × 107 cells were lysed in EBC lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 200 μM Na2VO4, 1 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with a protease inhibitor cocktail (Roche). After centrifugation (15,000 × g for 10 min at 4°C), 20 μl of protein A and protein G Sepharose beads (Upstate) were added to the supernatants and preincubated overnight at 4°C. Five-hundred micrograms of each of the cleared supernatants was reacted with 3 μg of anti-Ubc9 (Santa Cruz) or anti-hemagglutinin (HA) tag (Babco) for 3 h at 4°C with gentle rotation. The immune complex was then captured by the addition 20 μl of a protein A and protein G Sepharose bead mixture and was rocked for an additional 2 h at 4°C. Beads were washed four times with EBC buffer and boiled for 5 min in 20 μl of 2× sodium dodecyl sulfate (SDS) sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromphenol blue). Protein samples from total cell lysates (50 μg/lane) or immunoprecipitates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to a polyvinylidene fluoride membrane (Millipore) using a semidry transfer apparatus (Amersham Pharmacia). 293T cells were cotransfected with 2 μg of pT7-K-bZIP or pT7-K-bZIPΔLZ and 3 μg of pFlag-Ubc9 or pFlag-empty expression plasmids using FuGENE 6 (Roche) according to the supplier's recommendations. The cells were harvested 48 h after transfection and lysed in EBC buffer. Five-hundred micrograms of cell lysates was immunoprecipitated with the addition of 25 μl of anti-Flag antibody-conjugated agarose (Sigma). Beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× SDS sample buffer. Protein samples from total cell lysates (50 μg/lane) or immunoprecipitates were subjected to SDS-12% PAGE and then transferred as described above. After blocking for 1 h at room temperature with 5% skim milk in TBST (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 0.05% Tween 20), the membranes were incubated with primary antibodies for 2 h at room temperature. The membrane was washed with TBST three times for 10 min each at room temperature and incubated with horseradish peroxidase-conjugated antibodies for 1 h at room temperature. Membranes were washed three times with TBST and visualized with enhanced chemiluminescence reagent (Amersham-Pharmacia). Final dilutions of the primary antibodies for immunoblotting were 1 μg/ml of anti-K-bZIP immunoglobulin G (IgG), 1 μg/ml of anti-K-Rta, 1 μg/ml of anti-actin (Santa Cruz), 1:3,000 anti-T7 tag antibody (Novagen), 1 μg/ml of anti-SUMO-1 (Zymed Laboratories), and 1 μg/ml of anti-SUMO-3 (Zymed Laboratories) containing 5% skim milk. The anti-K-bZIP and anti-K-Rta rabbit sera were raised against purified glutathione S-transferase (GST)-K-bZIP full-length or GST-K-Rta (residues 400 to 604) protein, respectively. Purification of rabbit IgG was performed with a standard procedure. Protein amount was examined by bicinchoninic acid (BCA) protein assay, and purity of IgG (>90%) was confirmed by SDS-PAGE. For SUMO-modified proteins, the cells were washed twice with PBS and lysed with lysis buffer (50 mM Tris-Hcl [pH 6.8], 2% SDS, 10% glycerol, 20 μM N-ethylmaleimide [Sigma]) supplemented with a protease inhibitor cocktail (Roche). The cells were briefly sonicated and centrifuged, and the supernatant was used for immunoblot analysis. For immunoprecipitation of SUMO-modified K-bZIP, after lysing the cells as described above the lysate was incubated at 95°C for 10 min and cleared by centrifugation at 15,000 × g for 10 min at room temperature. The lysate was then diluted 1:10 in dilution buffer (50 mM HEPES, pH 7.0, 250 mM NaCl, 0.1% NP-40, and protease inhibitors) and immunoprecipitated with 4 μg of anti-K-bZIP rabbit IgG or preinoculated rabbit IgG.
Immunofluorescence assay.
Forty-eight hours after doxycycline treatment, TREx-K-Rta BCBL-1 cells were fixed with 3.7% formaldehyde in PBS for 5 min at room temperature and subsequently treated with 1.0% Triton X-100 followed by 1.0% NP-40 in PBS for 10 min each at room temperature. After washing twice with 0.2% Tween 20 in PBS, cells were smeared on a coverslip (Fisher). After treatment with blocking solution containing 2% bovine serum albumin (BSA; [Fisher]) in PBS, cells on the coverslip were incubated with anti-K-bZIP rabbit IgG (1:2,000) in PBS-2% BSA for 1 h at room temperature. After washing four times with PBS, Alexa Fluor 555-conjugated goat anti-rabbit IgG F(ab′)2 (1:5,000) (Molecular Probes) in blocking solution was applied and allowed to react for 1 h at room temperature.
293 cells were grown on coverslips in 6-well plates. Expression vector, pcDNA Flag-K-bZIP, or pcDNA Flag-K-bZIPK158R was transfected into 293 cells. Forty-eight hours after transfection, cells were fixed with 3.7% formaldehyde-PBS for 5 min at room temperature and then rinsed with PBS three times and subsequently treated with 1.0% Triton X-100 followed by 1.0% NP-40 in PBS for 10 min, each at room temperature. After washing twice with 0.2% Tween 20 in PBS, coverslips were treated with blocking solution. The cells were incubated with 0.5 μg/ml of anti-K-bZIP rabbit serum (1:2,000) and anti-PML mouse monoclonal antibody (1 μg/ml; PG-M3 [SantaCruz]) in blocking solution for 1 h at room temperature. After washing four times with PBS, Alexa Fluor 555-conjugated goat anti-rabbit IgG F(ab′)2 (1:5,000) (Molecular Probes) and Alexa Fluor 488-conjugated goat anti-mouse IgG F(ab′)2 (1:5,000) (Molecular Probes) in blocking solution were applied and allowed to react for 1 h at room temperature. After washing twice with TBST and once with PBS, coverslips were air dried and mounted on glass slides (Fisher). Imaging was viewed with an Olympus BX61. Images were captured with a CCD camera operated with Slide Book Software (Intelligent Imaging Innovations, Inc.).
Preparation and purification of GST fusion proteins.
GST fusion proteins were expressed in Escherichia coli strain BL21 transformed with pGEX-Ubc9, pGEX-SUMO-1GG, pGEX-SUMO-2GG, pGEX-SUMO-3GG, pGEX SAE1/SAE2, or pGEX4T-2. The GST fusion proteins were purified using glutathione-Sepharose beads (Amersham-Pharmacia) by standard procedures. After induction, bacterial cells (500 ml) were cultured in Luria broth for each construct. Protein expression was induced for 3 h with 1 mM (final concentration) isopropylthio-β-d-galactoside. Bacterial cells were washed once in PBS and then lysed with BugBuster (Novagen) supplemented with a protease inhibitor cocktail (Roche). After clearing by centrifugation at 8,000 × g for 10 min at 4°C, glutathione-Sepharose beads (500 μl of a 1:1 slurry in PBS) were added to the lysates for affinity purification. After incubation for 1 h at 4°C with rotation, the beads were washed four times in PBS containing 1% Triton X-100 and 1% sarcosyl. The GST fusion proteins were cleaved by biotinylated thrombin (Novagen) while bound on glutathione-Sepharose beads. Biotinylated thrombin was captured by streptavidin-conjugated agarose beads (Novagen). Purified proteins were dialyzed against HEPES buffer (20 mM HEPES [pH 7.4], 1 mM dithiothreitol [DTT]), and protein amounts were measured by BCA protein assay (Pierce). Purity of the proteins was confirmed by standard SDS-PAGE. For GST pull-down assay, the proteins immobilized on the glutathione-agarose beads were measured by Coomassie blue staining, using BSA as a protein standard.
In vitro SUMO conjugation assays.
SUMO conjugation assays were performed in 40-μl volumes. The substrates were in vitro translated (IVT) by using the TNT quick-coupled reticulocyte lysate system (Promega). Two microliters of labeled translation product was incubated for 2 h at 37°C in an ATP-regenerating buffer (50 mM Tris [pH 7.5], 5 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate [USB, Cleveland Ohio], 3.5 U/ml of creatine kinase [USB], 0.6 U/ml of inorganic pyrophosphatase [USB]) containing 100 ng/ml of E1 (SAE1/SAE2), 50 μg/ml of E2 (Ubc9), and 200 μg/ml of SUMO-GG. After termination of the reaction with SDS sample buffer containing β-mercaptoethanol, reaction products were fractionated by SDS-PAGE. The gel was dried and analyzed by PhosphorImager (Bio-Rad).
Pulse-chase analysis.
293 cells were seeded in a 6-cm2 dish and transfected with 4 μg of pFlag-K-bZIPwt or pFlag-K-bZIPK158R. Transfected cells were metabolically labeled for 4 h with 50 μCi of [35S]methionine and [35S]cysteine (Amersham) after 24 h posttransfection in DMEM supplemented with 10% PBS-dialyzed FBS. After labeling, medium was removed and cells were washed three times with 1 ml of DMEM containing unlabeled methionine and cysteine. Cells were harvested in 1 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1% Triton X-100, 1 mM NaF, 1 mM Na2VO4, 1 mM PMSF) supplemented with a protease inhibitor cocktail (Roche) and immunoprecipitated with 3 μg of an anti-K-bZIP IgG. Immunoprecipitates were subjected to SDS-PAGE and transferred to a membrane as described above. Transferred membrane was quantified by PhosphorImager, and signal intensity was measured by using QuantityOne software (Bio-Rad).
In vitro interaction assay.
GST-protein beads, containing approximately 2.0 μg of proteins, were resuspended in binding buffer (20 mM HEPES [pH 7.9], 150 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM DTT, 0.1% NP-40, 10% glycerol; supplemented before use with 1 mg/ml BSA, 0.5 mM PMSF, and 1× protease inhibitor cocktail), and incubated for 30 min at 4°C with 10 μl of IVT proteins, which were labeled with [35S]methionine using the TNT coupled transcription and translation system (Promega). The beads were washed four times with the binding buffer and resuspended and boiled in 2× SDS sample buffer. After proteins were separated by SDS-PAGE, radiolabeled polypeptides retained on the beads were visualized by autoradiography.
Chromatin-immunoprecipitation assay.
After 48 h with or without induction of viral reactivation with doxycyclin (1 μg/ml), 107 of TREx-K-Rta BCBL-1 cells were fixed with 1% formaldehyde at room temperature for 10 min and washed with ice-cold PBS. Cells were washed in Buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Cell pellets were collected by centrifugation and washed in Buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Two-hundred-microliter cell pellets were resuspended in 1 ml of lysis buffer (0.5% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail [Roche]) and sonicated four times for 30 s with 0.5-s pulses (Fisher 550 Sonic Dismembrator). Cell debris was removed by centrifugation, and the chromatin solutions were diluted five times in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris, pH 8.1, 1× protease inhibitor cocktail). A sample of total chromatin was collected to serve as a total input DNA control. Chromatin fragments were immunoprecipitated with anti-K-bZIP rabbit serum (1:100), preinoculated rabbit serum (1:100), or anti-Ubc9 antibody (1:100 [Santa Cruz]) overnight at 4°C with gentle rotation. Immunocomplexes were recovered and eluted as described before (8). After reverse cross-linking at 65°C overnight, the DNA fragments were purified with a QIAquick PCR Purification kit (QIAGEN), after adjusting pH with 3 M sodium acetate (pH 7.0), and eluted with 100 μl of 1× Tris-EDTA buffer, pH 8.0.
Southern blotting was performed using KSHV cosmid clones GB11, GA29, Not39, Not33, and GA2. The cosmid clones GB11, GA29, and GA2 are generous gifts from Ren Sun, University of California at Los Angeles. Two NotI fragments (nucleotides 43172 to 76999 and 77000 to 116203; GenBank accession number U75698) were cloned into the SuperCosI vector (Stratagene) after digestion of the KSHV genomic DNA with NotI (New England Biolabs). Each cosmid clone was digested with three different restriction enzymes overnight at 37°C. DNA fragments were separated on a 0.8% agarose gel. The gel was depurinated by incubation in depurination buffer (0.25 M HCl), followed by denaturation in the buffer (1.5 M NaOH, 0.5 M NaCl) for 20 min each. After denaturation, the restriction fragments were transferred to a nylon membrane (Biodyne; Pall Gelman Laboratory) by standard procedures. The DNA was immobilized on the membrane by drying at room temperature for 1 h and UV cross-linking. Immunoprecipitated DNA fragments were radiolabeled with [α32-P]dATP using a Strip-EZ DNA kit (Ambion), and hybridization was performed in ULTRAhyb buffer (Ambion) as recommended by the supplier. Equal amounts of probe (1 × 106 cpm/ml) were used for each hybridization.
In order to confirm the results of Southern blotting, low-cycle-number PCR amplification was performed with recombinant Taq polymerase (Invitrogen). DNA input was either of total-input DNA (control), preimmune-, or K-bZIP-immunoprecipitated DNA fractions. All primer sequences are provided in Table 3.
TABLE 3.
Primers used for ChIP assay
| Primer | Sequence (5′-3′) |
|---|---|
| K1-ORF6 pro.-F | TTGGACTACTAACCGCTGTCGCC |
| K1-ORF6 pro.-R | TGGTCACCGCCCCTAAAATAG |
| ORF9 pro.-F | ATCGGAAAAACGGTGGTGAAC |
| ORF9 pro.-R | CCTTCTGCCTCTCCTCTTGTTGTAG |
| Origin region-F | CCAAAGAAGCAGAGAAAACCGAG |
| Origin region-R | TATGTAGGGAAGAGGTGGGGAACG |
| ORF57 pro.-F | CCTCCTCTGAGTTTGACGAATCG |
| ORF57 pro.-R | CGTGTTGTTCTGCCGTATTGTAGG |
| K-bZIP pro.-F | ATTACCACCCCCCAGGATGTG |
| K-bZIP pro.-R | CCTTGCGAACACTTCAGTCTCG |
| ORF36 pro.-F | CGCCATTCGCTACTTCTCGG |
| ORF36 pro.-R | CGTCCTGAAACTGCTTCTCAAACAC |
| PAN RNA pro.-F | GGGTTTGACCACGGTTACTGATAGG |
| PAN RNA pro.-R | CCATTTTTGGAAGCCACGCC |
| ORF57 coding region-F | GCTTTCGTGGAGGAACAAATGAC |
| ORF57 coding region-R | CGTTTAGTAGCCCCATCACATCC |
Dual luciferase reporter assays.
Reporter plasmids were constructed by inserting promoter regions (74) upstream of the firefly luciferase coding region (Luc) in the pGL3-Basic vector (Promega). 293 cells were seeded in 12-well plates at 1 × 105/well in 1.5 ml of DMEM supplemented with 10% FBS and incubated at 37°C with 5% CO2. For each well, an equal amount of plasmid DNA, including the reporter and the control/expression plasmid, were transfected using the Fugene6 reagent following the manufacturer's protocol (Roche). All wells were cotransfected with a control reporter, pRL-cytomegalovirus (CMV) Renilla luciferase plasmid (Promega), which served as an internal control to normalize for variation in transfection efficiency. Cell lysates were prepared 48 h after transfection with 1× Passive Lysis Buffer (Promega). Dual luciferase assay was performed according to the manufacturer's protocol using a Lumat LB 9501 Luminometer (Wallac Inc.). At least three independent experiments were performed in each setting.
RESULTS
K-bZIP is sumoylated in vitro.
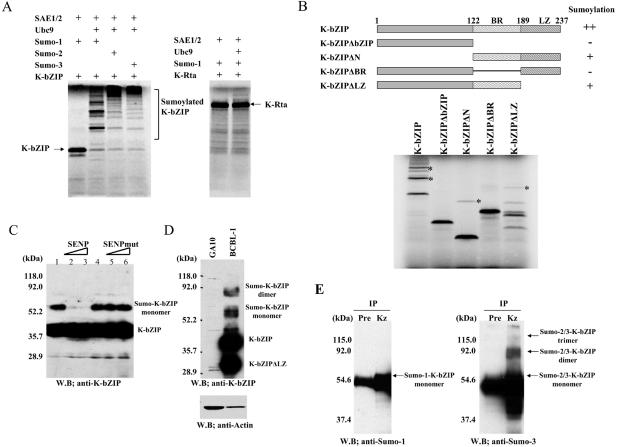
The observation that K-bZIP possesses transcriptional repression activity and is localized in PML bodies or PODs (a nuclear structure replete with sumoylated molecules) prompted us to investigate the possibility that K-bZIP is sumoylated. First, the potential of K-bZIP to be sumoylated was tested in a cell-free system using in vitro-translated (IVT)-K-bZIP and purified components of SUMO activating enzymes (SAE1/SAE2) and SUMO-conjugating enzyme (Ubc9) (21). This reaction was supplemented with Sumo1-GG, Sumo2-GG, or Sumo3-GG, the activated forms of SUMO proteins, and ATP was provided. As shown in Fig. 1A, K -bZIP was efficiently modified by either of the three SUMO peptides to the extent that the majority of K-bZIP were in the conjugated form. By contrast, K-Rta, another transcriptional factor encoded by KSHV, exhibited very little, if any, sumoylation (Fig. 1A). To identify the domain(s) responsible for sumoylation, the sumoylation potential of a series of K-bZIP mutants was tested. The K-bZIP mutant with the basic region deleted (ΔBasic) and the mutant without bZIP domain (ΔbZIP) were not sumoylated (Fig. 1B). This finding suggests that the major sumoylation site(s) resides in the basic domain. Interestingly, K-bZIPΔLZ, with the leucine zipper domain deleted, or K-bZIPΔN, the N-terminal deletion mutant, was sumoylated at much lower efficiency than full-length K-bZIP; this finding suggests that the native, or dimer, conformation of K-bZIP may be important for the sumoylation (Fig. 1B).
FIG. 1.
(A) SUMO conjugation in vitro. K-bZIP but not K-Rta is efficiently modified by SUMO. In vitro-translated (IVT)-K-bZIP was incubated with purified recombinant proteins E1 (SAE1/SAE2), E2 (Ubc9), and each activated form of SUMO protein (SUMO-1GG, SUMO-2GG, or SUMO-3GG). The reaction was carried out in the presence of the ATP regenerating system. Reactions without Ubc9 served as negative controls. (B) Mapping of SUMO-modified region. Deletion mutants of IVT-K-bZIP were incubated in the SUMO reaction. Basic region (amino acids 122 to 189) of K-bZIP is important for the sumoylation (*). (C) Sumoylation of K-bZIP in vivo. K-bZIP transfected into 293T cells displays a high-molecular-weight band when probed with anti-K-bZIP antibody, and which disappeared upon cotransfection with increasing amounts of SUMO-specific protease (SENP) but not an SENP mutant (SENPmut) lacking protease activity. (D) Sumoylated K-bZIP was detected in TREx-K-Rta BCBL-1 activated by K-Rta expression. Protein extract (100 μg/lane) was loaded. K-bZIP was detected with anti-K-bZIP antibody. KSHV negative B-cell line (GA10) was the negative control. An alternatively spliced K-bZIP (K-bZIPΔLZ) and sumoylated form of K-bZIP was also observed. Actin protein served as an internal control for the amount of protein on membrane and was detected by anti-actin goat serum. (E) K-bZIP is modified by both SUMO-1 and SUMO-2/3. BCBL-1 cells (1 mg) induced viral reactivation by K-Rta expression and lysed in SDS and were immunoprecipitated (IP) with 4 μg of preinoculated rabbit IgG (Pre) or anti-K-bZIP rabbit IgG (Kz). Immunoprecipitates were separated on a SDS-9% PAGE and immunoblotted (W.B.) with indicated antibodies. Anti-SUMO-3 polyclonal antibody detects both SUMO-2 and SUMO-3 proteins because of significant identity.
K-bZIP is sumoylated in vivo.
Sumoylation of K-bZIP was studied in vivo using 293T cells by transfection of K-bZIP expression vector (Fig. 1C), which utilized the endogenous SUMO ligation system. High-molecular-weight K-bZIP, with an increased size expected for monosumoylation, was readily detected (lanes 1 and 4). To demonstrate that this modification was due to sumoylation, SENP1, a SUMO-specific proteinase 1, was cotransfected with K-bZIP at a dose of 0.5 (lane 2) or 1.0 μg (lane 3). The putative sumoylated bands gradually disappeared (Fig. 1C, lanes 2 and 3). In contrast, cotransfection with a plasmid carrying inactive SENP1 (SENPmut) (lanes 5 and 6) did not affect the intensity of the high-molecular-weight bands (Fig. 1C). These data suggest that the modification of K-bZIP in transfected cells was largely, if not completely, due to sumoylation.
Sumoylation of endogenous K-bZIP was examined during the reactivation of the latent KSHV genome. In order to archive higher specificity, we employed the cell line, TREx-K-Rta BCBL-1, developed by Nakamura et al. (54), where the K-Rta gene is under the control of a tetracycline (Tet)-inducible promoter. Induction of K-Rta with the Tet analog, doxycycline (Dox), activated K-bZIP expression, and the level of K-bZIP reached a peak at about 48 h after induction. At that time, cells were harvested, and K-bZIP was probed with specific antibody (Fig. 1D). Bands corresponding to full-length and the type II splice variant of K-bZIP were identified, together with the mono- and disumoylated forms of K-bZIP. A significant fraction of K-bZIP in BCBL-1 was sumoylated, reaching a level higher than reported for most cellular factors without cotransfection of exogenous SUMO (46, 53).
We also investigated the type(s) of SUMO peptide conjugated to the K-bZIP in naturally infected cells. The BCBL-1 cell lysates, after Dox induction, were immunoprecipitated with anti-K-bZIP antibody, followed by blotting with either anti-SUMO-1 monoclonal antibody or anti-SUMO-3 rabbit polyclonal antibody (which recognizes both SUMO-2 and SUMO-3 moieties, due to their high degree of homology). Immunoprecipitation with preimmune rabbit IgG served as a negative control. Figure 1E showed that both SUMO-1 and SUMO-2/3 were covalently attached to K-bZIP during reactivation. Because of the dual specificity of the antibody, we were unable to determine whether the modification is due to SUMO-2, SUMO-3, or both. Our data are also consistent with the structures of the different SUMO species, in that SUMO-1 usually conjugates with the substrate in the monomer form, whereas SUMO-2 and -3 are themselves sumoylated and thus can form dimers and trimers (72).
Taken together, these results indicate that K-bZIP is sumoylated in vitro, in vivo, and during viral reactivation. Our data also showed that K-bZIP is able to conjugate with both SUMO-1 and SUMO-2/3.
Mapping the sumoylation site of K-bZIP.
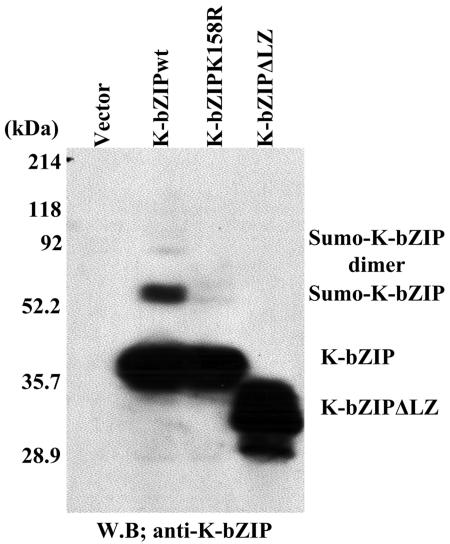
Inspection of sequences within the basic domain reveals that amino acids 157 to 160 (VKAE) of K-bZIP matches the SUMO consensus (φKXE, with φ being a hydrophobic residue). The putative conjugation site is lysine-158, and this motif is located in the basic region of the molecule, consistent with the sumoylation mapping data described above. Using PCR-based site-directed mutagenesis, the lysine-158 of K-bZIP was mutated to arginine, and the K158R mutant was transfected into 293T cells. The K158R mutant protein synthesized in transfected 293T cells was not sumoylated (Fig. 2), indicating that lysine 158 was the predominant, if not the sole, in vivo target site of sumoylation. Interestingly, a natural splice variant of K-bZIP, K-bZIPΔLZ lacking the leucine-zipper domain (45), was not sumoylated in vivo, despite the presence of the VKAE motif, suggesting that the consensus motif is not the sole structural determinant of the SUMO reaction.
FIG. 2.
Identification of sumoylation site. K-bZIP wild type, K-bZIP K158R, or K-bZIPΔLZ was transfected into 293T cells. Protein extract (50 μg/lane) lysed in SDS was loaded. K-bZIP was detected with anti-K-bZIP antibody. W.B., Western blotting.
Subcellular distribution of the K-bZIP sumoylation mutant.
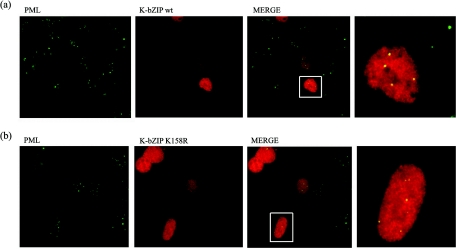
One of the major functions of sumoylation is to change the subcellular distribution. Sumoylation is important for nuclear and PML translocation of certain proteins (12, 32, 84). Previous studies showed that K-bZIP is colocalized with PML in POD, a structure enriched with sumoylated molecules (36, 79). In this study, the ability of the K-bZIP mutant to colocalize with PML was examined. Wild-type K-bZIP and K158R mutant were individually transfected into 293 cells, and expressed proteins were visualized by Alexa Fluor 555-conjugated antibodies against anti-K-bZIP rabbit IgG. As shown in Fig. 3, the overall subcellular distribution of K-bZIP and K158R were similar. Furthermore, K158R mutant and PML colocalized in POD, like the wild type, indicating that sumoylation was not essential for K-bZIP translocation into PML; this finding is similar to the analyses reported for EBV Zta (1) and CMV IE2 (3).
FIG. 3.
Localization of K-bZIP wt (a) and K-bZIPK158R (b) with PML in 293 cells. Immunofluorescence analysis was performed by using anti-K-bZIP rabbit serum and anti-PML mouse monoclonal antibody. K-bZIP (red) and PML (green) were detected with Alexa Fluor 555-conjugated anti-rabbit IgG and Alexa Fluor 488-conjugated anti-mouse IgG. Fluorescence images were collected separately and overlaid by a computer. The rightmost panel shows enlarged images of indicated cells. These panels are representative of 10 different fields. (magnification, ×600).
K-bZIP protein stability is unaffected by sumoylation.
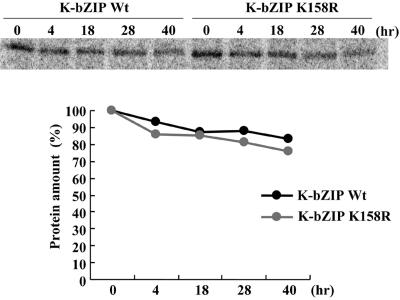
SUMO modification has been implicated in the modulation of protein stability (64). To explore whether SUMO modification affects K-bZIP stability, we conducted a pulse-chase experiment. 293 cells transfected with K-bZIP wild type or the K-bZIPK158R were labeled with [35S]methionine and [35S]cysteine for a 4-h pulse from 24 to 28 h posttransfection, and cells were chased for 4, 18, 28, or 40 h. No appreciable difference in the synthesis and reduction in the amount of labeled protein during the time course was observed (Fig. 4). This result suggested that sumoylation does not affect the overall stability of K-bZIP.
FIG. 4.
Determination of the stability of the K-bZIP wild type and the K-bZIPK158R mutant. (A) K-bZIP wild type and K-bZIPK158R were transfected into 293 cells and cells were labeled with [35S]methionine and [35S]cysteine for a 4-h pulse from 24 to 28 h posttransfection. Cells were chased with medium containing nonlabeled amino acids for the indicated time periods. K-bZIP proteins were immunoprecipitated with anti-K-bZIP antibody and subjected to SDS-PAGE followed by autoradiography. (B) Quantification of the labeled protein. Protein amount was quantified by PhosphorImager analysis of the dried SDS-PAGE gel with Quantity One (Bio-Rad). Black indicates the K-bZIP wild type, while gray refers to K-bZIPK158R.
Sumoylation and K-bZIP mediated transcriptional repression.
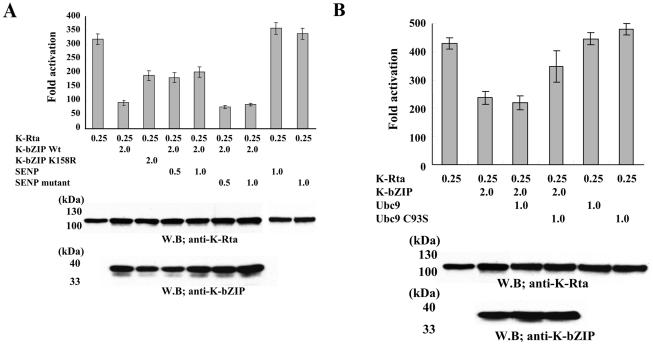
Another major function of sumoylation is transcriptional repression. Increasing evidence suggests that transcriptional factors, when modified by sumoylation, often become repressors (29, 60). Our previous work showed that K-bZIP is a transcriptional repressor of K-Rta by physical interaction with K-Rta (33, 43). Accordingly, repression activity of K-bZIP was examined by assessing K-Rta-mediated activation of the ORF57 promoter (33). When cotransfected with K-bZIP, the transactivation of the ORF57 promoter by K-Rta was drastically reduced to about 25%, a result consistent with our previous work (33). If the K158R mutant was used, transactivation activity of K-Rta remained high (60% of the full K-Rta activity). This suggests that sumoylation of K158 is important for K-bZIP's repression activity. To ensure that sumoylation, but not other modifications of lysine-158, were responsible, SENP1, a SUMO-specific protease, was transfected at two different doses (0.5 and 1 μg) with K-Rta and wild-type K-bZIP. The addition of SENP1 also relieved the repression activity of K-bZIP, while SENP1 alone had little effect on K-Rta. As an additional control, a SENP mutant with impaired protease activity was used; K-bZIP repression activity was restored (Fig. 5A). That these variations in activity are not due to changes in the amount of proteins was shown by immunoblots of the reaction mixtures with K-bZIP and K-Rta antibodies. These data, taken together, indicate that sumoylation of K-bZIP is largely, but not exclusively, responsible for the repression activity on K-Rta.
FIG. 5.
(A) Repression via sumoylation. K-Rta activation of the ORF57 promoter (leftmost bar) was repressed by K-bZIP wild type but had less repression by K-bZIP K158R. Cotransfection with the SENP1 wild type but not an inactive mutant diminished the repression. 293 cells were cotransfected with K-Rta, ORF57 promoter, and the indicated plasmids. K-Rta and K-bZIP amounts were examined by immunoblotting with anti-K-Rta or anti-K-bZIP antibody. (B) Ubc9 dominant-negative relived K-bZIP repression. 293 cells were cotransfected with indicated plasmids. Protein amounts of K-Rta and K-bZIP were examined by immunoblotting with indicated antibodies.
Requirement of Ubc9 activity in K-bZIP-mediated repression.
To demonstrate the role of Ubc9 in K-bZIP-mediated repression, we took advantage of a catalytically inactive mutant Ubc9C93S, shown previously to behave in a dominant-negative manner against endogenous Ubc9 (19). If Ubc9 is involved in K-bZIP-mediated repression of K-Rta, coexpression of Ubc9C93S should reduce the repression effect. When cotransfected with K-Rta and K-bZIP, Ubc9C93S, but not Ubc9 wild type, largely restored the ability of K-Rta to transactivate the ORF57 promoter, indicating that Ubc9 activity is required to achieve the full repression activity of K-bZIP (Fig. 5B). It is noteworthy that expression of Ubc9 (or UbcC93C), in the absence of K-bZIP, has no effect on K-Rta's transactivation activity. This reinforces the notion that Ubc9 is recruited to the K-Rta complex by K-bZIP.
Genome-wide scanning for K-bZIP binding sites on the KSHV chromatin.
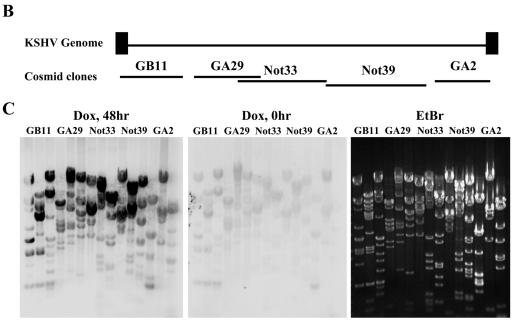
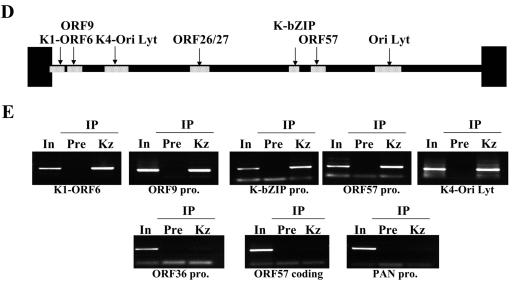
Previously, we showed that K-bZIP suppresses the transcriptional activation of a subset of K-Rta target genes. For instance, the transcription of ORF57 and K-bZIP, but not nut-1/PAN RNA, was suppressed by overexpressed K-bZIP (33). If K-bZIP association with K-Rta was important for this process, K-bZIP should be recruited to ORF57 and K-bZIP promoters but not the nut-1/PAN RNA promoter. Taking advantage of a chromatin immunoprecipitation-based genome-wide scanning approach we developed (40), the major binding sites of K-bZIP on the KSHV chromatin were analyzed during reactivation of the lytic replication cycle. The K-Rta inducible cell line, TREx-K-Rta BCBL-1, was used (54). As shown previously, upon K-Rta induction by Dox, a complete cycle of KSHV replication was triggered, following a well-ordered KSHV gene expression program, including the immediate induction of K-bZIP expression. A significant advantage of this system over the 12-O-tetradecanoyl phorbol-13-acetate induction protocol is that almost every cell is induced (based on immunostaining of K-bZIP protein; Fig. 6A). This approach greatly increased the sensitivity of ChIP analysis, which uses radiolabeled DNA fragments, associated with K-bZIP chromatin, to probe a Southern blot of restriction enzyme-digested Cosmid clones that span almost the entire KSHV genome (Fig. 6B). Based on relative hybridization intensities of restriction fragments (compare to those of the ethidium bromide-stained gel), the major K-bZIP binding sites can be mapped. K-bZIP antibody (Kz) was used to generate K-bZIP ChIP. Preinoculation rabbit serum (Pre) served as a negative control. Without Dox induction, little K-bZIP was present and only basal hybridization was detected, attesting to the specificity of the antibodies. Forty-eight hours after Dox induction, bands with strong hybridization intensity, above the general background, were identified (Fig. 6C). This result suggests low-affinity binding of K-bZIP at numerous sites along the KSHV genome, in addition to its high-affinity binding sites. The high-intensity hybridization regions were mapped to six sites: K1-ORF6 promoter, ORF9 promoter, K4 promoter/KSHV-Ori lytic DNA replication, ORF26-ORF27 region, K-bZIP promoter, and ORF57 promoter. As predicted, ORF57 and K-bZIP promoters, but not the nut-1/PAN RNA promoter, were among the hybridized regions. In addition, the two origins of lytic-phase DNA replication, shown by others to be K-bZIP binding sites (4, 44), were also identified (Fig. 6D). These results were confirmed by analysis of PCR products generated with specific primer sets corresponding to these regions (Fig. 6E). Primer sets for the ORF57 coding regions and other promoters such as nut-1/PAN RNA promoter were also included as negative controls. The PCR results are in complete agreement with the Southern blot results, thus substantiating the mapping data and indicating that K-bZIP exerts its repression activity by direct association with the transcription complex.
FIG.6.
Identification of the K-bZIP binding sites on the KSHV chromosome by the ChIP method. (A) Reactivation by K-Rta induction. K-bZIP expression was analyzed by immunofluorescence assay using anti-K-bZIP antibody (red). DNA was stained with TOPRO-3 (Molecular Probes; blue). (B) Schematic diagram of the KHSV genome. The five cosmids (GB11, GA29, Not33, Not39, and GA2) spanning the KSHV genome are indicated. (C) Scanning of K-bZIP binding sites. Cosmids were digested with the restriction enzymes and separated on agarose gel (0.8%). The gel was stained with ethidium bromide (EtBr; right panel) and Southern blotted with radiolabeled probes derived from ChIP before (Dox, 0 h) and after 48 h of KSHV reactivation (Dox 48 h). The DNA associated with K-bZIP chromatin was radiolabeled as described in Materials and Methods. (D) Schematic diagram of K-bZIP binding sites. Putative major K-bZIP binding sites are summarized in the diagram. (E) PCR verification of K-bZIP binding sites. PCR primers were designed for the K-bZIP associated regions. Other KSHV promoters and coding region served as controls (ORF36 promoter, PAN promoter, ORF57 coding region). For each primer set, a PCR with the total input DNA (In) before immunoprecipitation was carried out. ChIP fragments of preimmune serum (Pre) or anti-K-bZIP (Kz) after 48 h of KSHV reactivation were subjected to PCR analyses to confirm Southern blotting results.
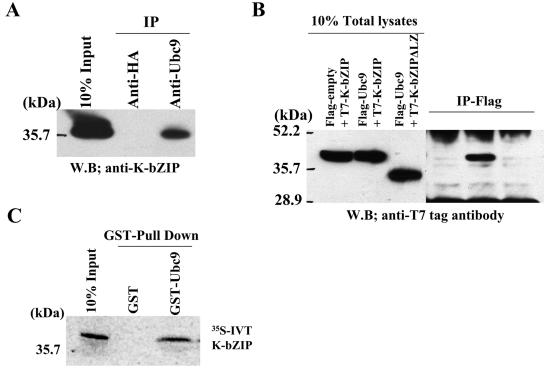
K-bZIP association with Ubc9 in vitro and in vivo.
Mechanistically, there are two ways to explain how sumoylation of K-bZIP results in the repression of the K-Rta transcriptional complex. The first is that the SUMO moieties directly hinder assembly of the productive transcription machinery, as proposed by Holmstrom et al. (29). The second is that the sumoylation site of K-bZIP recruits E2 conjugating enzyme Ubc9 to the K-Rta complex. Recently, Ubc9 was identified as a strong transcriptional corepressor (37). These two mechanisms are not mutually exclusive, and most likely K-bZIP utilizes both mechanisms for transcriptional repression. In the first case, K-bZIP is a corepressor itself, whereas in the second, K-bZIP serves as an adaptor, more akin to the role of an E3 SUMO ligase. To test the second mechanism, the association of endogenous K-bZIP with Ubc9 was examined in BCBL-1 cells. TREx-K-Rta-BCBL-1 was treated with Dox to induce K-bZIP expression, and cell extracts were isolated at 48 h. Ubc9 was immunoprecipitated with specific antibody, followed by immunoblotting with antibody against K-bZIP. Additional unrelated antibodies were used as controls. As shown in Fig. 7A, K -bZIP coprecipitated with Ubc9 in vivo. As an independent test of the association between K-bZIP and Ubc9 and to rule out the possibility that the observed association in BCBL-1 cells was due to nonspecificity of the polyclonal antibody, we constructed a T7-tagged K-bZIP and a Flag-tagged Ubc9 expression vector. We also included a T7-tagged construct of the natural, spliced variant of K-bZIP (K-bZIPΔLZ), which as shown above is not sumoylated and lacks repression ability (43). These constructs were cotransfected into 293T cells. At 48 h after transfection, the cells were harvested and lysed. Immunoprecipitation was carried out with Flag-antibody-conjugated agarose. As shown in Fig. 7B, Flag-Ubc9, but not Flag-empty alone (as encoded by Flag-empty, which bears only the Flag epitope), pulled down T7-K-bZIP. Furthermore, Flag-Ubc9 did not coprecipitate with T7-K-bZIPΔLZ, demonstrating the specificity of its association. The data also suggest that the leucine-zipper domain is required for this association, which may explain why K-bZIPΔLZ is not sumoylated in vivo (Fig. 2).
FIG. 7.
K-bZIP association with Ubc9. K-bZIP expressed in BCBL-1 was coprecipitated with Ubc9 by Ubc9 antibodies. Anti-HA antibody was used as a negative control. (B) Association between K-bZIP and Ubc9 in cotransfected 293T cells. 293T cells were cotransfected with the indicated plasmids. Cell lysates were precipitated with Flag antibody-conjugated agarose, and coimmunoprecipitation of K-bZIP was detected by using anti-T7 tag antibody. The expression of T7-tagged K-bZIP in one-tenth of total cell lysates used in coimmunoprecipitation is shown in the same blot as a control. (C) K-bZIP associated with Ubc9 in vitro. GST-Ubc9 but not GST precipitates in vitro-translated (IVT)-K-bZIP. The inputs are one-tenth of the lysates used for the binding study.
Finally, to show that the association between K-bZIP and Ubc9 is direct, purified GST-Ubc9 was incubated with in vitro-translated K-bZIP (IVT-K-bZIP). As shown in Fig. 7C, GST-Ubc9 but not GST alone efficiently pulled down IVT-K-bZIP. Thus, by three different approaches, we have demonstrated here that K-bZIP associates with Ubc9.
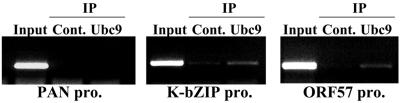
Ubc9 recruitment to the ORF57 and K-bZIP promoters by K-bZIP.
The studies described above have shown that Ubc9 was associated with K-bZIP. Recruitment of Ubc9 to the ORF57 and K-bZIP promoters was also analyzed using anti-Ubc9 antibody. Using the ChIP protocol described earlier, Ubc9 was recruited to the ORF57 promoter and to the K-bZIP promoter but not to the nut-1/PAN RNA promoter (Fig. 8).
FIG. 8.
Recruitment of Ubc9 to K-bZIP binding sites. ChIP assay was performed by using anti-Ubc9 antibody or rabbit normal serum (control serum). PCR showed recruitment of Ubc9 to K-bZIP-associated chromatin.
DISCUSSION
A growing list of transcriptional factors has been shown to be sumoylated, including glucocorticoid receptor, Sp3, Smad4, p53, and c-jun (29, 46, 53, 60). Although the effects of sumoylation on transcriptional factors are multiple, this posttranslational modification usually causes a reduction of the transcriptional activity of a transactivator or an increase of the repression activity of a trans-repressor. Previously, we demonstrated that K-bZIP is a strong transcriptional repressor for K-Rta (33). In this study, we present data suggesting that a major mechanism of K-bZIP mediated repression is via sumoylation. These include that K-bZIP is sumoylated in vitro and in vivo and that the in vivo sumoylation site maps to the lysine at 158. Importantly, the sumoylation of K-bZIP was detected not only in transient transfected 293T cells but also in naturally infected BCBL-1 cells during reactivation. Both types of SUMO peptides (SUMO-1 and SUMO-2/3) were found to be conjugated to K-bZIP endogenously in naturally infected BCBL-1 cells. The fraction of sumoylated K-bZIP in BCBL-1 cells was close to 10%, a figure significantly higher than most other transcriptional factors (46, 53). K-bZIP thus joins the growing list of sumoylated herpesvirus immediate early proteins, which include human cytomegalovirus IE1, IE2, (3, 81), EBV Zta (1) and Rta (5), and human herpesvirus-6 IE1 (23).
We showed that mutation of lysine-158 to arginine virtually eliminated SUMO modification of K-bZIP in vivo, with a concomitant loss of much of the repression activity. Because lysine is a potential site for other posttranslational modifications, including acetylation and ubiquitination, it was necessary to demonstrate that the removal of sumoylation, not other types of modification, was responsible for the loss of K-bZIP repression activity. In fact, we found that wild-type K-bZIP does not appear to be acetylated in living cells (unpublished data). Furthermore, the expression of SENP1 effectively removed the sumoylation from K-bZIP and significantly compromised the ability of K-bZIP to repress K-Rta transactivation, whereas the expression of enzymatically inactive SENP1 did not do so under the same conditions. These data, taken together, strongly suggest that sumoylation plays a primary role in K-bZIP-mediated repression.
How does sumoylation contribute to the repression activity of K-bZIP? Sumoylation regulates the translocation of target molecules to different subcellular compartments, thereby affecting their functions. K-bZIP is localized primarily in nucleoplasm but also in PODs, a nuclear structure rich in sumoylated molecules such as PML, SP100, p53, and Daxx (12, 32, 84). Indeed, the sumoylation of PML has been shown to be crucial for its structural formation (84). When the subcellular locations of transiently transfected K-bZIP wild type and K158R mutant were compared, no difference was detected. This result, however, needs to be interpreted with caution, because sumoylated K-bZIP only represents 10% of the population, so its location may be masked by the unmodified form, which appeared as a diffusive pattern in the nucleus. Nevertheless, the result tends to suggest that sumoylation was not required for K-bZIP localization to POD, an observation reminiscent of p53 and EBV Zta (1, 39). In a recent report, Adamson and Kenney (1) showed that overexpression of EBV Zta wild type, but not a sumoylation mutant, disrupts POD and suggests that Zta may compete with PML for limiting quantities of SUMO peptides, preventing the formation of PODs. Our work does not directly address this issue. At least at the overexpression level studied here, K-bZIP did not disrupt the POD structure.
Alternatively, sumoylation has been shown to affect protein stability (11, 77), which is a potential contribution to the repression activity of K-bZIP. We examined the possibility by comparing the half-life of K-bZIP to that of the sumoylation mutant K158R. No significant difference was observed, indicating that protein stability is not a major contributing factor to the repression activity of K-bZIP.
A third possible mechanism, whereby SUMO mediates repression, is by virtue of its ability to recruit Ubc9, the E2 SUMO-conjugating enzyme. Ubc9 binds avidly to SUMO moieties and thus can be recruited by sumoylated K-bZIP to the K-Rta transcriptional complex. We showed that Ubc9 binds K-bZIP and is recruited to the viral promoter sites, where K-bZIP and K-Rta assemble. The ability of Ubc9 to catalyze sumoylation of the proximal transcriptional factors or histone 4 is one mechanism of repression function (29, 46, 53, 60, 65). In this regard, K-bZIP functions like a SUMO ligase or SUMO adaptor to bring Ubc9 to its potential substrate. In addition, a recent report showed that Ubc9 also exhibited conjugation-independent repression function (37). Our Ubc9 dominant-negative mutant experiment (Fig. 5B) suggested that the Ubc9 conjugation activity is crucial in mediating K-bZIP repression. We therefore favor the former hypothesis.
Aside from recruitment of Ubc9, the SUMO peptide itself may exhibit repression activity by the recruitment of transcriptional repressors (67). As a preliminary test of this notion, we attached SUMO directly to the N terminus of K-Rta, which induces sharp reduction of K-Rta transcriptional activity (data not shown). By contrast, introducing a GST peptide of similar size as SUMO showed little effect. This finding suggests that the SUMO moiety itself is a repressor. A recent report indicated that EBV Rta is naturally sumoylated (5). We were not able to demonstrate sumoylation of KSHV Rta (Fig. 1A), suggesting that the regulation of K-Rta by sumoylation is via K-bZIP. Recently, a SUMO-binding motif (V/I-X-V/I-V/I) has been identified, and this motif is present in HDAC2 and -6 (67). Recruitment of HDAC2 and -6 to a transcriptional factor would decrease its trans-activation potential and increase repression activity. This is indeed the case for Elk-1 and p300 (19, 82). These transcriptional factors become repressors upon sumoylation, and the sumoylated forms of Elk-1 and p300 recruit, respectively, HDAC2 (82) and HDAC6 (19). It is thus conceivable that sumoylated K-bZIP serves as a platform to recruit a corepressor(s) to K-Rta, resulting in the repression of K-Rta activity. This hypothesis and the one discussed in the last paragraph (recruitment of Ubc9) are not mutually exclusive and differ only in details, as both rely on the sumoylation of K-bZIP to bring in corepressor, be it HDAC or Ubc9.
K-Rta is a central regulator of KSHV lytic replication and is a potent activator for a large number of KSHV promoters (9, 41, 47, 68, 73, 74). K-Rta acts on these viral promoters by either direct binding to the enhancer sequences or via the association with other transcriptional factors, such as RBJκ, Oct-1, and SP-1 (9, 42, 62). Presumably, depending on the sequence motif and promoter context, different transcriptional complexes are assembled. We previously showed that K-bZIP only targets a subset of the K-Rta complexes for repression. In addition, K-bZIP represses K-Rta-mediated activation of both the K-bZIP and the ORF57 promoter but not the nut-1/PAN RNA promoter (33, 43). Our ChIP data agree with such selectivity: K-bZIP was recruited to the K-bZIP and ORF57 promoters but not the nut-1/PAN RNA promoter. Perhaps, noncoincidentally, the K-Rta-responsive elements for these two promoters are very similar (74). The cosmid-based ChIP approach allows us to quickly scan the entire viral genome for high-affinity binding sites. The specificity of this approach was confirmed by the PCRs of hybridized regions. This approach was useful for mapping of high-affinity binding sites but may have missed weak binding sites due to the hybridization background. With this study, we have now used this approach to map the chromatin binding sites of two herpesvirus bZIP proteins: the Meq of Marek's disease virus (40) and K-bZIP of KSHV. This approach can be readily extended to other herpesviruses to study the transcriptional and replication factor assembly sites, as well as the sites of histone modifications, during lytic and latent phases.
In addition to K-bZIP and ORF57 promoters, K-bZIP was also recruited to the lytic replication origin, in agreement with the results of Lin et al. (44) and AuCoin et al. (4). Curiously, K-bZIP also binds strongly to the general region covered by K1 and ORF27 promoters, raising the issue of whether these are also target genes of K-bZIP. Our preliminary data showed that these promoters indeed are modulated by K-bZIP (unpublished data).
In summary, we showed that K-bZIP was a sumoylated protein and sumoylation was important for its transcriptional repression activity. K-bZIP also binds Ubc9 E2, SUMO-conjugating enzyme, and as such may serve as a viral “SUMO ligase” to bring Ubc9 to modify the chromatin structure surrounding the transcriptional complex where K-bZIP resides. We also mapped the high-affinity binding sites of K-bZIP in the KSHV genome; these sites include the lytic replication origin and several viral promoters modulated by K-bZIP. The information presented here provides a framework to understand the complex role of K-bZIP and its posttranslational modification by sumoylation in KSHV replication.
Acknowledgments
We thank Ron Hay for providing the pGEXSAE1/SAE2 expression plasmid and Ren Sun for KSHV cosmid clones.
Y.I. is supported by the California Universitywide AIDS research program (UARP [F03-D-206]). This work was supported by grants from UARP and the National Institutes of Health (CA111185 to H.J.K.).
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, A. L., and S. C. Kenney. 1998. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology 251:187-197. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J.-H., Y. Xu, W. J. Jang, M. J. Matunis, and G. S. Hayward. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 75:3859-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2003. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 5.Chang, L. K., Y. H. Lee, T. S. Cheng, Y. R. Hong, P. J. Lu, J. J. Wang, W. H. Wang, C. W. Kuo, S. S. Li, and S. T. Liu. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803-38812. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 12.Doucas, V., M. Tini, D. A. Egan, and R. M. Evans. 1999. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA 96:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dukers, N. H., and G. Rezza. 2003. Human herpesvirus 8 epidemiology: what we do and do not know. AIDS 17:1717-1730. [DOI] [PubMed] [Google Scholar]
- 14.El-Guindy, A. S., and G. Miller. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 78:7634-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flemington, E. K. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J. Virol. 75:4475-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis, A., T. Ragoczy, L. Gradoville, L. Heston, A. El-Guindy, Y. Endo, and G. Miller. 1999. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of early lytic cycle genes and synergy with the Epstein-Barr virus R transactivator. J. Virol. 73:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giot, J. F., I. Mikaelian, M. Buisson, E. Manet, I. Joab, J. C. Nicolas, and A. Sergeant. 1991. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 19:1251-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 20.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 21.Gong, L., T. Kamitani, K. Fujise, L. S. Caskey, and E. T. Yeh. 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272:28198-28201. [DOI] [PubMed] [Google Scholar]
- 22.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gravel, A., V. Dion, N. Cloutier, J. Gosselin, and L. Flamand. 2004. Characterization of human herpesvirus 6 variant B immediate-early 1 protein modifications by small ubiquitin-related modifiers. J. Gen. Virol. 85:1319-1328. [DOI] [PubMed] [Google Scholar]
- 24.Gruffat, H., S. Portes-Sentis, A. Sergeant, and E. Manet. 1999. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J. Gen. Virol. 80:557-561. [DOI] [PubMed] [Google Scholar]
- 25.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 28.Hayward, G. S. 2003. Initiation of angiogenic Kaposi's sarcoma lesions. Cancer Cell. 3:1-3. [DOI] [PubMed] [Google Scholar]
- 29.Holmstrom, S., M. E. Van Antwerp, and J. A. Iniguez-Lluhi. 2003. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. USA 100:15758-15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang, S., Y. Gwack, H. Byun, C. Lim, and J. Choe. 2001. The Kaposi's sarcoma-associated herpesvirus K8 protein interacts with CREB-binding protein (CBP) and represses CBP-mediated transcription. J. Virol. 75:9509-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang, S., D. Lee, Y. Gwack, H. Min, and J. Choe. 2003. Kaposi's sarcoma-associated herpesvirus K8 protein interacts with hSNF5. J. Gen. Virol. 84:665-676. [DOI] [PubMed] [Google Scholar]
- 32.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izumiya, Y., S.-F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H.-J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izumiya, Y., S.-F. Lin, T. J. Ellison, A. M. Levy, G. L. Mayeur, C. Izumiya, and H.-J. Kung. 2003. Cell cycle regulation by Kaposi's sarcoma-associated herpesvirus K-bZIP: direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J. Virol. 77:9652-9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katano, H., K. Ogawa-Goto, H. Hasegawa, T. Kurata, and T. Sata. 2000. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology 286:446-455. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi, S., H. Shibata, I. Kurihara, K. Yokota, N. Suda, I. Saito, and T. Saruta. 2004. Ubc9 interacts with chicken ovalbumin upstream promoter-transcription factor I and represses receptor-dependent transcription. J. Mol. Endocrinol. 32:69-86. [DOI] [PubMed] [Google Scholar]
- 38.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwek, S. S., J. Derry, A. L. Tyner, Z. Shen, and A. V. Gudkov. 2001. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 20:2587-2599. [DOI] [PubMed] [Google Scholar]
- 40.Levy, A. M., Y. Izumiya, P. Brunovskis, L. Xia, M. S. Parcells, S. M. Reddy, L. Lee, H. W. Chen, and H.-J. Kung. 2003. Characterization of the chromosomal binding sites and dimerization partners of the viral oncoprotein Meq in Marek's disease virus-transformed T cells. J. Virol. 77:12841-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang, Y., and D. Ganem. 2004. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J. Virol. 78:6818-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao, W., Y. Tang, S.-F. Lin, H.-J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, S.-F., D. R. Robinson, G. Miller, and H.-J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long, J., G. Wang, D. He, and F. Liu. 2004. Repression of Smad4 transcriptional activity by SUMO modification. Biochem. J. 379:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin, D. F., B. D. Kuppermann, R. A. Wolitz, A. G. Palestine, H. Li, C. A. Robinson, and the Roche Ganciclovir Study Group. 1999. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. N. Engl. J. Med. 340:1063-1070. [DOI] [PubMed] [Google Scholar]
- 50.Mauser, A., E. Holley-Guthrie, A. Zanation, W. Yarborough, W. Kaufmann, A. Klingelhutz, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J. Virol. 76:12543-12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morrison, T. E., and S. C. Kenney. 2004. BZLF1, an Epstein-Barr virus immediate-early protein, induces p65 nuclear translocation while inhibiting p65 transcriptional function. Virology 328:219-232. [DOI] [PubMed] [Google Scholar]
- 52.Morrison, T. E., A. Mauser, A. Klingelhutz, and S. C. Kenney. 2004. Epstein-Barr virus immediate-early protein BZLF1 inhibits tumor necrosis factor alpha-induced signaling and apoptosis by downregulating tumor necrosis factor receptor 1. J. Virol. 78:544-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller, S., M. Berger, F. Lehembre, J. S. Seeler, Y. Haupt, and A. Dejean. 2000. c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem. 275:13321-13329. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura, H., M. Lu, Y. Gwack, J. Souvlis, S. L. Zeichner, and J. U. Jung. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polson, A. G., L. Huang, D. M. Lukac, J. D. Blethrow, D. O. Morgan, A. L. Burlingame, and D. Ganem. 2001. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J. Virol. 75:3175-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragoczy, T., and G. Miller. 1999. Role of the epstein-barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73:9858-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 61.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 65.Shiio, Y., and R. N. Eisenman. 2004. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100:13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sista, N. D., C. Barry, K. Sampson, and J. Pagano. 1995. Physical and functional interaction of the Epstein-Barr virus BZLF1 transactivator with the retinoic acid receptors RAR alpha and RXR alpha. Nucleic Acids Res. 23:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song, J., L. K. Durrin, T. A. Wilkinson, T. G. Krontiris, and Y. Chen. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA 101:14373-14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song, M. J., H. J. Brown, T.-T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by Rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun, R., S.-F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun, R., S.-F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tatham, M. H., E. Jaffray, Q. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 73.Ueda, K., K. Ishikawa, K. Nishimura, S. Sakakibara, E. Do, and K. Yamanishi. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) replication and transcription factor activates the K9 (vIRF) gene through two distinct cis elements by a non-DNA-binding mechanism. J. Virol. 76:12044-12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 75.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78:4248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weger, S., E. Hammer, and R. Heilbronn. 2004. SUMO-1 modification regulates the protein stability of the large regulatory protein Rep78 of adeno associated virus type 2 (AAV-2). Virology 330:284-294. [DOI] [PubMed] [Google Scholar]
- 78.Wu, F. Y., H. Chen, S. E. Wang, C. M. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein alpha interacts with ZTA and mediates ZTA-induced p21(CIP-1) accumulation and G1 cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu, F. Y., J.-H. Ahn, D. J. Alecendor, W.-J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu, F. Y., Q. Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 99:10683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu, Y., J.-H. Ahn, M. Cheng, C. M. Rhys, C. J. Chiou, J. Zong, M. J. Matunis, and G. S. Hayward. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683-10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13:611-617. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]