Abstract
Replication of RNA viruses is regulated by cis-acting RNA elements, including promoters, replication silencers, and replication enhancers (REN). To dissect the function of an REN element involved in plus-strand RNA synthesis, we developed an in vitro trans-replication assay for tombusviruses, which are small plus-strand RNA viruses. In this assay, two RNA strands were tethered together via short complementary regions with the REN present in the nontemplate RNA, whereas the promoter was located in the template RNA. We found that the template activity of the tombusvirus replicase preparation was stimulated in trans by the REN, suggesting that the REN is a functional enhancer when located in the vicinity of the promoter. In addition, this study revealed that the REN has dual function during RNA synthesis. (i) It binds to the viral replicase. (ii) It interacts with the core plus-strand initiation promoter via a long-distance RNA-RNA interaction, which leads to stimulation of initiation of plus-strand RNA synthesis by the replicase in vitro. We also observed that this RNA-RNA interaction increased the in vivo accumulation and competitiveness of defective interfering RNA, a model template. We propose that REN is important for asymmetrical viral RNA replication that leads to more abundant plus-strand RNA progeny than the minus-strand intermediate, a hallmark of replication of plus-strand RNA viruses.
Replication of plus-strand RNA viruses of eukaryotes, which include important pathogens of humans, animals, and plants, such as severe acute respiratory syndrome coronavirus, West Nile virus, and hepatitis C virus, leads to production of large amounts of viral progeny in infected cells. Various cis-acting elements are present in both plus- and minus-strand (which is an intermediate template) RNAs to achieve selective yet robust synthesis of virus RNA progeny by the virus-coded RNA-dependent RNA polymerase (RdRp) (1, 3, 10). These elements include the essential promoter (initiation) elements, as well as regulatory RNA elements such as template recruitment elements (31), RNA replication enhancers (REN) that up-regulate (16, 17, 27), and RNA replication silencer that down-regulates (23, 34) RNA synthesis. While considerable effort has been made to dissect the role of promoter elements that are involved in initiation of RNA synthesis for many plus-strand [(+)]RNA viruses (reviewed in references 3, 5, and 6), current understanding of the roles of REN elements, which are involved in plus-strand RNA synthesis, is incomplete (10).
Tombusviruses, including Tomato bushy stunt virus (TBSV) and Cucumber necrosis virus (CNV), have ∼4.8-kb, single-component (+)RNA genomes that encode two replication proteins, p33 and p92 (Fig. 1) (32). Previous work with partially purified CNV and TBSV replicases revealed that two separate short promoters, termed gPR and cPR (Fig. 1A), are essential for minus- and plus-strand synthesis, respectively (20). Additional work led to the identification of a replication silencer element, which inhibited the level of minus-strand [(−)]RNA synthesis by ∼7 fold in vitro (23), and two replication enhancer elements, which stimulated (+)RNA synthesis by ∼2 to 10 fold in vitro (Fig. 1A) (17, 19, 27). The strongest of the two RENs is termed RIII(−) (Fig. 1A), which was highly active both in vivo and in vitro (17, 27). Deletion analysis revealed that either of two stem-loops, SL1-III(−) and SL2-III(−) (Fig. 1A) together with a single-stranded region between the stem-loops in RIII(−) played important roles in (+)RNA synthesis in the in vitro replicase assay (17).
FIG. 1.
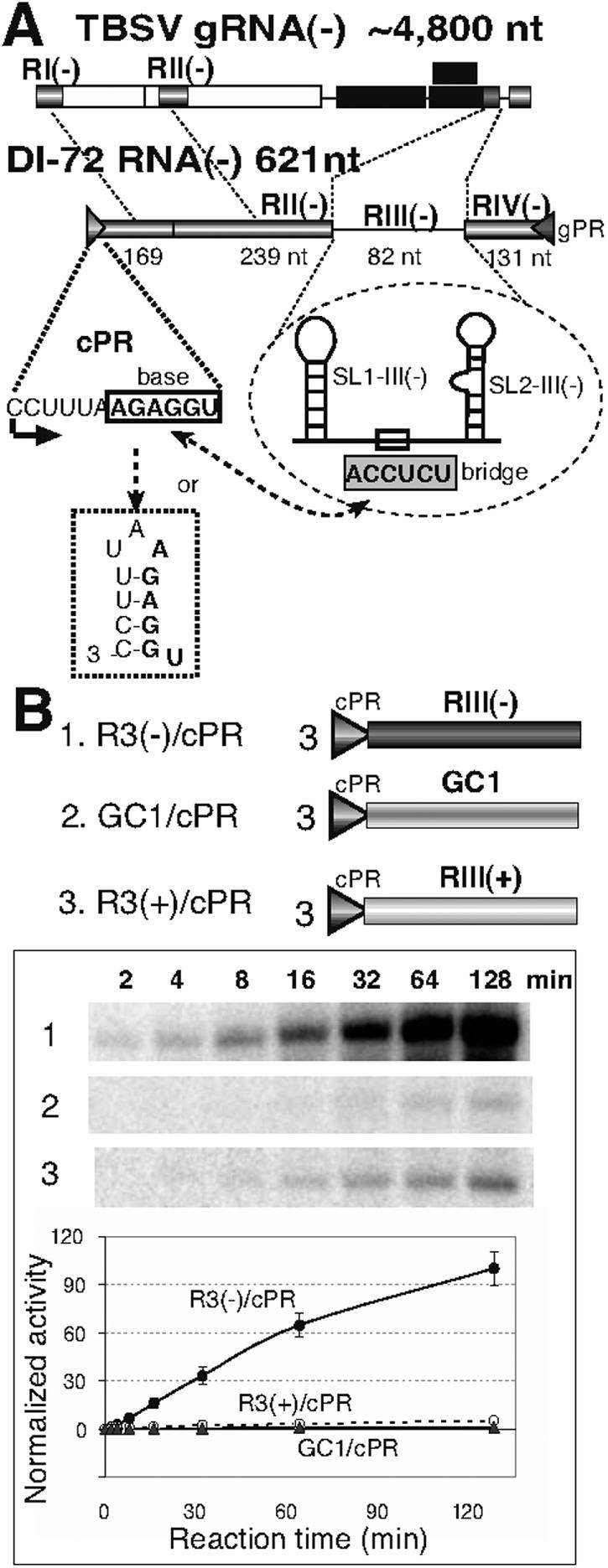
(A) A schematic presentation of a typical tombusvirus genome (TBSV; the minus strand is shown in a 3′-to-5′ orientation) and a prototypical DI RNA (DI-72). The four noncontiguous regions that are present in the DI-72 RNA are indicated with gray boxes. RIII(−) bears the replication enhancer function, whereas the site of plus-strand initiation is indicated by a solid arrow. The minimal plus-strand initiation promoter (termed cPR, indicated by a triangle), which is located at the 3′ end of RI(−), contains the base sequence (boxed). The predicted long-distance RNA-RNA base pairing (indicated by a two-headed arrow) takes place between the bridge and base sequences, which is predicted to disrupt the hairpin formed in cPR (shown at the bottom). The stem-loop structures SL1-III(−) and SL2-III(−) in RIII(−) are also shown schematically. (B) Time course of RNA synthesis by CNV replicase in vitro, based on denaturing gel analysis. The RNA templates were used in equal molar amounts in the in vitro assays. The 32P-labeled replicase products were normalized, based on the number of templated urilydates. The templates used contain the cPR promoter and the similar-sized RIII(−) REN, an artificial GC sequence, and RIII(+), respectively. The length of the incubation period during the in vitro assay is shown below the gel. Graphic representation of the quantified and normalized replicase products obtained from the experiments shown in panel B is also shown.
To advance our understanding of REN-mediated enhancement of RNA synthesis, we demonstrate in this report that the two stem-loops in RIII(−) are the binding sites for the tombusvirus replicase. In addition, we also show that a 6-nucleotide (6-nt) single-stranded stretch within RIII(−), termed the bridge sequence, likely interacts with the cPR promoter sequence via a long-distance RNA-RNA interaction, which alters the structure of the promoter and that this interaction leads to ∼2-fold increased initiation from cPR. Overall, our results suggest that the REN may have a dual function by binding to the replicase and interacting with the cPR promoter, which might facilitate correct positioning of the promoter for initiation of RNA synthesis.
MATERIALS AND METHODS
Plant inoculation and CNV replicase preparation.
Nicotiana benthamiana plants were inoculated with CNV genomic RNA transcripts obtained by standard T7 RNA transcription by using a SmaI-cut clone of pK2/M5p20STOP for CNV (28). CNV replicase preparations were obtained from systemically infected leaves as previously described (14).
Replicase assay.
For the in vitro experiments, RNA templates were obtained by in vitro transcription reaction with T7 RNA polymerase using PCR-amplified DNA templates (17). To make the partly double-stranded constructs for the trans-replication assay, we annealed the heat-denatured RNA transcripts (94°C for 2 min) in STE buffer (10 mM TRIS [pH 8.0], 1 mM EDTA, and 100 mM NaCl) and then slowly (over 30 min) cooled them to 25°C. The annealed RNAs were loaded onto a 5% nondenaturing 15-cm-long polyacrylamide gel. After electrophoresis, the gels were stained with ethidium bromide, and the annealed RNA bands were excised. The RNAs were eluted into 0.6 M ammonium acetate, followed by phenol-chloroform extraction and ethanol precipitation. Replicase reaction mixtures (each, 50 μl) were carried out as previously described (14). Each replicase reaction mixture contained 0.2 μg of template RNA. In the template competition experiments, the amount of template RNA (100 ng/μl) was the same in each reaction, while the molar amount of the competitor was one, three, and ninefold higher than that of the template. The replicase products were analyzed on a 20-cm-long denaturing 5% polyacrylamide-8 M urea gels, followed by analysis with a phosphorimager as previously described (14).
Gel mobility shift assay.
The gel shift assay was performed with a 10-μl volume containing 1× replicase buffer (14), 10% glycerol, 3 U of RNasin, 50 ng of recombinant p92 and p33C, and 1.0 μg of yeast tRNA (Sigma, Inc.) as a nonspecific competitor (∼500-fold excess over labeled RNA). Recombinant CNV p92 and p33C were produced as a fusion protein with the maltose binding protein as previously described (21). After 10 min of preincubation at room temperature, the 32P-labeled RNA probe (0.5 pmol; 5′ end label) was added, and incubation was continued for 15 more minutes at room temperature. The reaction products were loaded on a native 5% polyacrylamide gel and ran for 1 h at 1 V/cm at 4°C. The gel was dried and then exposed to a phosphorimager.
In vivo experiments with defective interfering (DI) RNAs.
The prototypical DI-72 and DI-72Δbr RNAs and CNV helper (pK2/M5p20STOP) (28), 4 μg each, were mixed together and rub inoculated onto N. benthamiana leaves. The inoculated and uninoculated leaves were harvested 7 and 10 days after inoculation. One microgram of total RNA was used for reverse transcription-PCR (RT-PCR) with primers #313 (5′-CCCAACAAGAGTAACCTGTATGCTATGCCA-3′) and #131 (5′-GTAATACGACTCACTATAGGAGAACCTTCGTAAAAGCAGA-3′). The RT-PCR products obtained were separated on an 8% polyacrylamide gel. The protoplast experiments and Northern blot analysis were performed according to reference 21. Briefly, T7-transcribed DI RNA transcripts and helper CNV RNA (1 μg each) were gel isolated prior to electroporation into the primary-infected protoplasts. Five micrograms of total RNA extracts obtained from the primary infected protoplasts (24 h incubation after electroporation) was used for electroporation to a new batch of protoplasts (first passage).
Expression of DI-72 RNA and its mutated derivatives in the presence of p33 and p92 in yeast was done as described in reference 18. Total RNA extraction from yeast, followed by Northern blotting, was performed as described in reference 18.
RESULTS
To dissect the mechanism of REN-mediated enhancement of RNA synthesis by the tombusvirus replicase, first we tested the dynamics of RNA synthesis in time course experiments by an in vitro CNV replicase assay (14, 20). We found that R3(−)/cPR template carrying both RIII(−) REN and the 3′-located cPR promoter stimulated RNA synthesis from cPR by up to ∼20 fold (Fig. 1A and B) (17) when compared to similar-sized control templates carrying other viral (e.g., the complementary RIII(+) in construct R3(+)/cPR) or artificial (e.g., construct GC1/cPR) sequences (Fig. 1B). The stability of the secondary structures in RIII(+) (27) and GC1 (13) templates are similar to that in RIII(−) (not shown). The increasing gap in RNA synthesis between the RIII(−)-containing and control templates is consistent with the role of RIII(−) in (i) stimulation of initiation of RNA synthesis and/or (ii) rapid recycling (binding) of the replicase for sequential rounds of RNA synthesis.
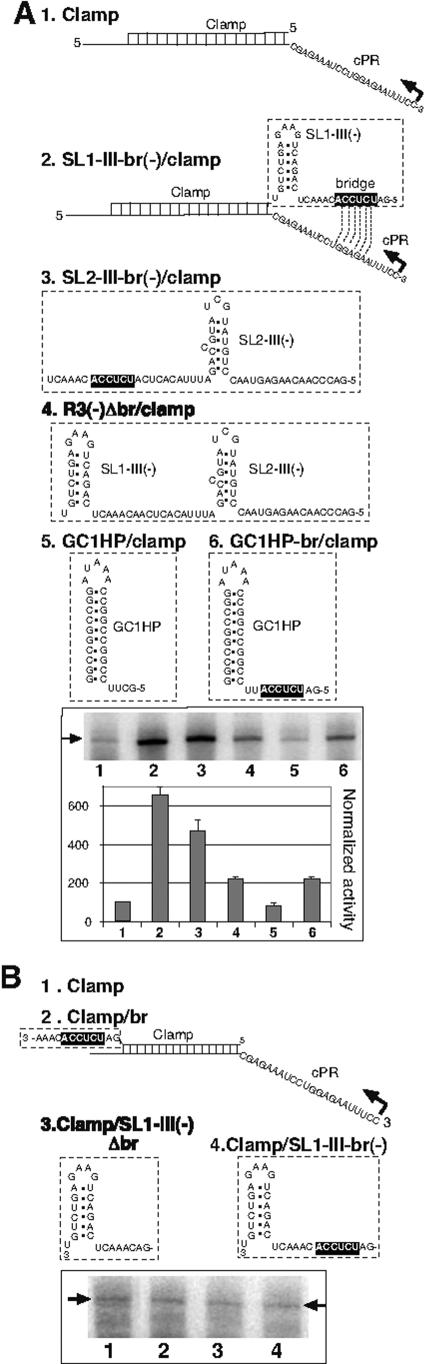
Development of a trans-replication assay to test the role of RIII(−) in stimulation of RNA synthesis.
Based on the previous experiments in which RIII(−) and cPR were present in the same template (cis function) (Fig. 1B) (17), we could not discriminate if RIII(−) enhanced the efficiency of initiation or termination. This is because more-efficient termination of RNA synthesis could also enhance the efficiency of initiation by facilitating recycling of the replicase for new rounds of RNA synthesis. Therefore, we developed a trans-replication assay by placing an RIII(−)-derived sequence on a nontemplate RNA, whereas the template RNA contained the cPR promoter. However, the nontemplate and the template RNAs were tethered together via complementary regions (termed a clamp), which positioned the REN and cPR in close proximity (3′-trans position) [shown as template SL1-III-br(−)/clamp in Fig. 2A ]. Note that we used the minimal REN element that included only the SL1-III hairpin and the short bridge sequence from RIII(−), which showed levels of stimulation on RNA synthesis in vitro comparable to that of the entire RIII(−) (17). Testing the template activity of SL1-III-br(−)/clamp (Fig. 2A, lane 2) in the trans-replication assay revealed that it supported an ∼7-fold-higher level of RNA synthesis from the cPR promoter than the control template (i.e., Fig. 2A, clamp and lane 1), which included the cPR promoter and the clamp region, but it lacked the minimal REN. This observation suggests that the REN can function in trans (i.e., when present in the nontemplate RNA) during RNA synthesis if it is positioned proximally to the cPR promoter. Altogether, these data (i) strongly support a role for REN in initiation and (ii) indicate that REN is unlikely to affect the efficiency of termination. This is because the nontemplate RNA carrying the REN should be separated from the template RNA by the progressing CNV replicase after initiation in the trans-replication assay.
FIG.2.
Stimulation of RNA synthesis by REN in trans. (A, top) Schematic representation of the in vitro trans-replication system. Each of the shown templates consists of two RNA strands. The template strand contains the cPR promoter, which is fused to RII(+) of DI-72 (Fig. 1A). The nontemplate strand carrying RII(−) can base pair with the template RNA, as indicated by the label “clamp” (shown schematically by a ladder). Further details can be found in Fig. 1A. Note that the template strand is the same in these experiments and that each nontemplate strand can form identical clamp structures with the template strand. Only the boxed sequences are different among the RNA constructs. The bridge sequence, which can base pair with cPR, is boxed. A representative denaturing gel of radiolabeled replicase products synthesized by in vitro transcription with CNV replicase is shown at the bottom. The gel-isolated, annealed RNAs were used in equal molar amounts. The template-sized tombusvirus replicase products are marked by an arrow. (B) The effect of REN location on trans-replication activity. Note that the presence of REN sequences (lanes 3 and 4) reduced template activity by ∼50%, likely due to competition between the distant cPR and REN for replicase binding (Fig. 3A). See further details in panel A.
To test if the minimal REN sequence can also function when positioned distantly from the cPR sequence (5′ trans position) on the nontemplate RNA, we constructed template SL1-III-br(−)/clamp (Fig. 2B, lane 4). An in vitro CNV replicase assay revealed that the REN was ineffective in stimulating RNA synthesis initiated from cPR in this arrangement (Fig. 2B, compare lanes 1 and 4), suggesting the REN cannot stimulate RNA synthesis when positioned distantly from the cPR promoter. Overall, the above data suggest that the REN can only stimulate RNA synthesis from cPR when it is located in a proximal position to cPR, which is in agreement with its proposed role in stimulation of initiation of RNA synthesis.
RNA-RNA interaction between RIII(−) replication enhancer and the promoter.
Previous work revealed that the single-stranded sequence between SL1-III(−) and SL2-III(−) hairpins was also important in stimulation of RNA synthesis in the in vitro replicase assay (17). Interestingly, there is a 6-nt-long sequence termed the bridge sequence between SL1-III(−) and SL2-III(−), which is predicted to interact with the base sequence present in cPR via a long-distance base pairing (Fig. 1A). To test if this interaction is important for stimulation of RNA synthesis, we compared its effect on RNA synthesis using our trans-replication assay (Fig. 2A). Deletion of the bridge sequence [construct R3(−)Δbr/clamp] (Fig. 2A, lane 4) resulted in ∼3-fold reduction in RNA synthesis when compared to a template carrying the bridge sequence in combination with one of the hairpins [constructs SL1-III-br(−)/clamp and SL2-III-br(−)/clamp] (Fig. 2A, lanes 2 to 3). We also found that the bridge sequence by itself, i.e., in the absence of SL1-III(−) and SL2-III(−), increased the efficiency of RNA synthesis by ∼2-fold (compare construct GC1HP-br/clamp with construct GC1HP/clamp) (Fig. 2A, lanes 6 and 5). These data confirmed that the bridge sequence affects RNA synthesis initiated from the cPR promoter in vitro.
The replication enhancer promotes binding of the tombusvirus replication proteins to the template in vitro.
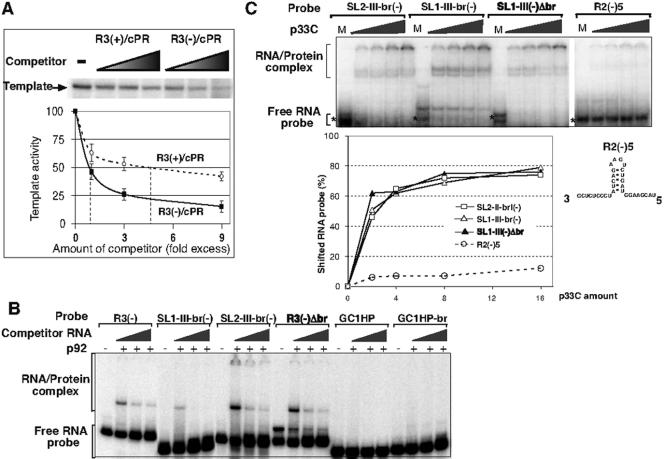
Based on the above data, we predicted that RIII(−) REN-driven enhancement of RNA synthesis depends on efficient binding of RIII(−) to the tombusvirus replicase. To test the involvement of RIII(−) in replicase binding, we performed template competition and in vitro binding experiments. In the template competition experiments, RIII(−) was present in a competitor RNA [i.e., R3(−)/cPR] (Fig. 1B), whereas the template RNA lacked RIII(−) but contained cPR to facilitate initiation by the CNV replicase. We found that the competitor R3(−)/cPR RNA carrying RIII(−) decreased the activity of the CNV replicase on the template RNA fivefold-more efficiently than a second competitor RNA [R3(+)/cPR] (Fig. 1B) carrying RIII(+) did (Fig. 3A). These data suggest that the presence of RIII(−) makes the RNA more competitive for the CNV replicase than RNAs lacking this sequence. Overall, the enhanced competitiveness of RIII(−)-containing RNA is consistent with the model that RIII(−) is involved in binding to the replicase.
FIG. 3.
In vitro binding of CNV replicase proteins to REN. (A) An RIII(−)-containing RNA is an efficient competitor for the CNV replicase. The template used was MDV(−)/cPR (20), which contains, in addition to the 3′-terminal cPR sequence (Fig. 1A), a 221-nt heterologous sequence derived from the minus-stranded satellite RNA of Qβ bacteriophage. The competitor RNAs were R3(+)/cPR and R3(−)/cPR (Fig. 1B). Lane −, sample lacking a competitor RNA in the replicase reaction. Filled triangles show the increasing amounts of competitor RNAs (from 0.1 μg to 1.0 μg). For graphic representations of data, the template activity of MDV(−)/cPR without competitor is given as 100%. The 50% inhibitory concentration for each competitor is shown by a dotted line. (B) Purified recombinant p92 binds to REN in a gel mobility shift assay. Each probe (as shown at the top) was labeled with [32P]UTP and was used in four lanes. The left lane represents the free probe (no p92 was added), while the other three lanes have a combination of the free probe and the CNV p92 expressed and purified from E. coli. The nonspecific RNA competitor was used in increasing amounts (500, 1,000, and 2,000-fold excess over the probe). The migration of the free probe is indicated. Protein-RNA complexes are indicated by a bracket on the left. The RNA probes are boxed in Fig. 2A. (C) Efficient binding of recombinant p33 to REN in vitro. The same amounts of labeled probes (as shown on the top) were used in the presence of increasing amounts of p33 (note that p33C, a functional truncated form of the p33 protein, was used). In this native polyacrylamide gel, some of the RNA probes ran as multiple bands, likely due to the formation of alternative secondary structures. The relative amounts of the shifted probes were quantified and shown as percentages of the probe without p33 added (labeled as M). The sequence of R2(−)5′ derived from RII(−) of DI-72 (Fig. 1A) is shown on the right. Note that the presence of more than one shifted bands is likely due to the binding of different number of p33 molecules to the same RNA molecule.
To test if RIII(−) could directly bind to the tombusvirus replicase proteins, we performed gel mobility shift analyses between 32P-labeled RNA probes and the affinity-purified recombinant CNV replicase proteins expressed in Escherichia coli (Fig. 3B and -C) (21). First, we demonstrated that the recombinant p92, which contains the RdRp motifs, bound to the RNA probes that contained full or partial RIII(−) sequences [construct R3(−), SL1-III-br(−), and SL2-III-br(−)] (Fig. 3B). Interestingly, the RNA construct [R3(−)Δbr] (Fig. 3B) that carried the two hairpins but lacked the bridge sequence bound to p92 as efficiently as R3(−), suggesting that the bridge sequence does not contain the binding site for p92. This conclusion was further supported by the lack of p92 binding to an artificial GC-rich sequence with or without the bridge sequence (constructs GC1HP and GC1HP-br) (Fig. 3B). The overall binding of the RIII(−)-containing RNA probes to the recombinant p92 was weak, resulting in only partial shift of the probes (Fig. 3B). This is in agreement with previous data that p92 binds less efficiently to viral RNA than p33 replication protein (24).
Because p33 is also part of the tombusvirus replicase complex (22), and its C-terminal domain (designated p33C) binds better to RNA than p92 (25), we tested the ability of p33C to bind to RNA templates in a gel mobility shift experiment (Fig. 3C). We found that p33C bound to SL1-III-br(−), SL2-III-br(−), SL1-III(−)Δbr efficiently, while it bound poorly to a DI-72 RNA sequence representing portion of RII(−), which is comparable in length to SL1-III-br(−) (Fig. 3C). In summary, we found that both p92 and p33 replicase proteins bound better to RNAs carrying RIII(−) REN sequences than to RNAs lacking RIII(−) sequences, supporting the model that REN is important in binding to the tombusvirus replicase.
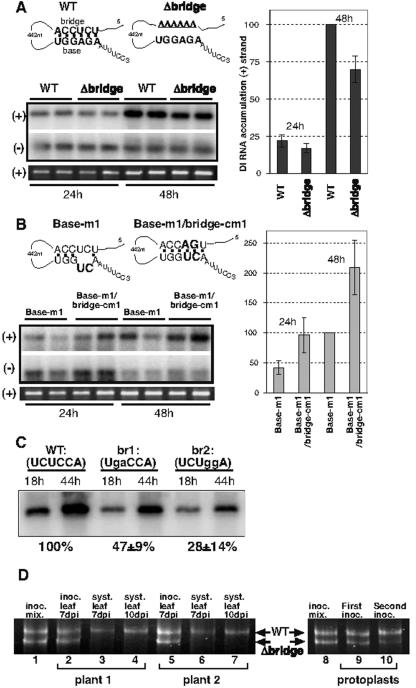
Interaction between the bridge and base sequences affects replication and competitiveness of DI RNAs in plant cells, whole plants, and yeast.
To test for the significance of the long-distance RNA-RNA interaction between the base and the bridge sequences, we inoculated N. benthamiana protoplasts with the TBSV-derived DI-72 RNAs (a model template) carrying either the wild-type (wt) bridge sequence or lacking this sequence (constructs wt and Δbridge) (Fig. 4A) in the presence of CNV helper virus. Northern blot analysis of the total RNA extracts obtained from N. benthamiana protoplasts showed that Δbridge DI RNA supported a 30% reduced level of (+)DI RNA accumulation when compared with wt DI-72 RNA after 48 h of incubation (Fig. 4A), whereas the levels of (−)RNA for these constructs were comparable (Fig. 4A). For further testing of the effect of base pairing between the base and the bridge sequences on DI RNA replication, we generated (i) a mutant carrying a 2-nt change within cPR (Base-m1) (Fig. 4B) and (ii) a “compensatory” mutant that had a second mutation within the bridge sequence, which restored the ability to form base pairs between the two elements (Base-m1/bridge-cm1) (Fig. 4B). Northern blot analysis showed that the (+)RNA for Base-m1/bridge-cm1 accumulated to a twofold-higher level than that of Base-m1 (Fig. 4B), supporting the model that base-bridge interaction between the promoter and the replication enhancer is important in plus-strand synthesis.
FIG. 4.
Effect of mutations within the RIII bridge sequence on DI RNA accumulation in N. benthamiana. (A and B) A schematic representation of the predicted base pairing between the base and bridge sequences in wt (Fig. 1A) and mutated DI-72 RNAs. Deleted nucleotides are shown by Δ, while the mutated nucleotides are in boldface type. See further details in Fig. 1A. Northern blot analysis shows DI-72 accumulation over time in N. benthamiana protoplasts. The membrane was probed to detect either positive strands (top) or negative strands (middle). The ethidium bromide-stained gel is shown as a loading control at the bottom. The levels of plus-stranded RNA accumulation obtained with the mutated DI RNAs are compared to wt DI-72 RNA after 48 h incubation (100%) (A) or to Base-m1 (B), based on three independent experiments. Note that Base-m1 accumulates only to 10% of wt DI-72 RNA (not shown), suggesting that these mutations affected the function of either cPR or that of the 5′ end of the plus-strand RNA. (C) Accumulation of wt and mutated DI-72 RNA in yeast. Mutations within the bridge sequence are shown with small letters. The level of DI RNA accumulation was estimated based on Northern blots. (D) In vivo competition studies between the wt DI-72 RNA and the bridge deletion mutant (A). RT-PCR analysis of total RNA extracts obtained from N. benthamiana plants or from protoplasts (lanes 9 to 10) inoculated with an equal mixture of WT DI-72 and Δbridge RNAs (see panel A), along with the transcripts of CNV helper by using high-resolution polyacrylamide gels. The RT-PCR was performed either from the inoculation mixture (mixed transcripts) (lanes 1, 8), from inoculated leaf 7 days post inoculation (dpi) (lanes 2 and 5), from noninoculated leaves 7 and 10 dpi (lanes 2, 3 and 6, 7), or from protoplasts (first and second inoculations) (lanes 9 and 10).
The role of the base-bridge interaction in DI-72 RNA accumulation was also tested in yeast cells, which coexpressed p33 and p92 proteins together with DI-72 RNAs, resulting in robust viral RNA replication (18, 22). Mutations of 2 nt within the bridge sequence (constructs br1 and br2) (Fig. 4C) in DI-72 RNA reduced its accumulation by two- to fourfold, suggesting that the bridge sequence affects viral RNA accumulation in plant and yeast host cells as well.
Since the viral gRNA and DI RNA are likely to compete with one another for limited amounts of replicase molecules in the infected cells, it is possible that the proposed long-distance base-bridge interaction may influence the ability of the replicating DI RNAs to compete with other templates. This was tested by coinoculating the wt and the Δbridge DI-72 RNAs in a 1:1 ratio onto N. benthamiana plants in the presence of the CNV helper. RT-PCR analyses of total RNA extracts obtained from the systemically infected leaves (Fig. 4D, lanes 3 to 4 and 6 to 7) revealed the presence of the wt DI-72 RNA, while the Δbridge DI RNA was not detected in these samples. We conclude that the wt DI-72 RNA is more competitive in plants than the Δbridge DI RNA. This observation was also supported by protoplast experiments, which showed that the wt DI-72 RNA became more abundant than the Δbridge DI RNA over time (∼30% and 60% decrease in Δbridge DI RNA levels) (Fig. 4D, lanes 9 and 10). Since previous protoplast experiments demonstrated that RIII did not affect the stability of the DI RNA in protoplast (27), we suggest that the base-bridge interaction is important under competitive conditions.
DISCUSSION
The emerging picture in RNA virus replication is that viral RNA plays multiple roles, in addition to being used as a template (5). These roles include (i) facilitating the assembly of the functional viral replicase complex (22, 31), (ii) promoter (initiation) functions for genomic minus- and plus-strand synthesis (6, 20) and subgenomic RNA synthesis (11), and (iii) additional regulatory functions, such as up- and down-regulation of complementary strand synthesis via REN and replication silencer elements, respectively (9, 17, 19, 23, 27, 34), or as riboregulators (29). The regulatory RNA elements present in the minus-stranded intermediate templates are currently poorly characterized for plus-stranded RNA viruses, except for the minimal promoter sequences (6). However, regulatory elements are predicted to play significant roles in the synthesis of the plus-stranded RNA progeny, because the minus-strand templates are used up to 100-fold-more efficiently during replication than plus-stranded templates are (1, 3). Among the identified regulatory elements that are present in the minus-stranded RNA templates are REN elements, including TBSV RIII(−) and M1H of turnip crinkle virus-associated satC, which were found to stimulate plus-strand synthesis 10 to-20 fold in vitro using replicase preparations (16, 17). The TBSV (−)RNA contains an additional REN, termed the promoter proximal element, which stimulated RNA synthesis by ∼3 fold by in vitro replicase assays (19).
Increased template binding by the tombusvirus replication proteins in the presence of RIII(−) REN.
To gain insight into the mechanism of REN-mediated stimulation of RNA synthesis in tombusvirus replication, we show evidence in this paper that REN can enhance binding of the template to both p92 and p33 replicase proteins (Fig. 3B and C). A more detailed analysis of RIII(−) sequences that are important for binding revealed that the stem-loop structures in RIII(−) bound to the recombinant replicase proteins with the highest efficiency (Fig. 3). Interestingly, p33 was also shown to bind in vitro to an unrelated stem-loop structure present in RII(+), which is important for template selection and recruitment (12, 24). The relevance of the observed binding of RIII(−) to p33 and p92 proteins is also supported by both kinetics and template competition experiments, which demonstrated that a template carrying the RIII(−) sequence produced ∼20-fold-more RNA products (Fig. 1B) and that it was more competitive than a template carrying the RIII(+) sequence (Fig. 3A). In summary, we conclude that RIII(−) can facilitate the binding of the minus-stranded template to the viral replicase, which is predicted to lead to more efficient use of the minus-stranded RNAs as templates, resulting in asymmetrical RNA synthesis (more plus than minus-stand progeny RNAs).
Stimulation of RNA synthesis by RIII(−) REN from the cPR promoter in vitro.
An in vitro trans-replication assay, based on a minimal REN tethered to the template RNA via a complementary clamp region to bring REN and cPR in close proximity, was used to demonstrate that the REN could enhance RNA synthesis in trans (when not present in the template RNA) (Fig. 2A). On the contrary, placing REN at a distal position relative to cPR in the tethered RNAs nullified the effect of REN on RNA synthesis (Fig. 2B), suggesting that the REN should be in the vicinity of the cPR promoter for stimulation of RNA synthesis in vitro. However, unlike the inflexible double-stranded clamp located between the cPR and REN, the full-length TBSV (−)RNA or DI-72 (−)RNA might be flexible enough to allow proximal positioning of these elements during replication (see also below).
Because in the trans-replication system the REN is not copied by the replicase and the REN-containing RNA is being removed (due to unwinding of the “clamped” RNA region) during the ongoing RNA synthesis on the template strand, we propose that the primary function of the REN is to stimulate the initiation step rather then the termination step in RNA synthesis. However, the role of the REN in stimulating strand elongation by the replicase cannot be excluded, based on the data presented.
Stimulatory role of a long-range RNA-RNA interaction between RIII(−) REN and the cPR promoter on viral RNA replication and competitiveness.
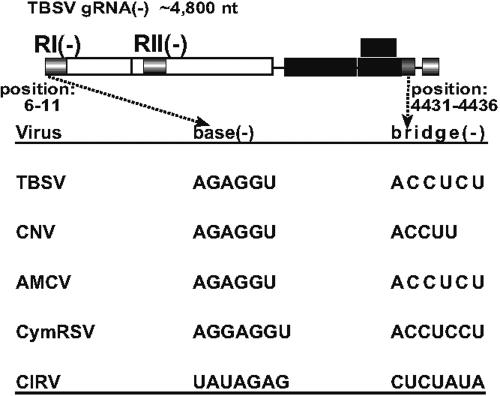
In vitro experiments showed that that REN performs an additional function via its 6-nt bridge sequence, which can interact with the base sequence in cPR. This interaction alone enhanced RNA synthesis from cPR by ∼2 fold (Fig. 2A). The existence of the bridge-base interaction is based on the following (i) Phylogenetic analysis shows that tombusviruses and their DI RNAs carry conserved sequences that could form long-distance base pairing (Fig. 5). (ii) Solution structure probing indicated that the bridge sequence, albeit present within a predicted single-stranded region between SL1-III(−) and SL2-III(−) hairpins in RIII(−), was susceptible to cleavage by double-strand-specific V1 nuclease (27), a finding that is consistent with its proposed role of interaction with the base sequence. (iii) Deletion or mutagenesis of the bridge sequence reduced the level of RNA synthesis by two- to threefold in vitro (Fig. 2) and reduced the accumulation of plus-stranded DI RNA by ∼30% in protoplasts (Fig. 4A) and by 50 to 70% in yeast (Fig. 4C). (iv) The proposed base-bridge interaction also affected the competitiveness of the DI RNAs in planta or in protoplasts (Fig. 4D), arguing that DI RNAs capable of this interaction have selective advantage in vivo over DI RNAs lacking this interaction. Based on these observations, we propose that the bridge-base interaction may play a role in facilitating the presentation of the cPR sequence to the replicase bound to RIII(−). One such role of the bridge-base interaction may be the facilitation of formation of a secondary structure within cPR that results in a 6-nt double-stranded region and a 4-nt-long 3′ single-stranded tail. Interestingly, this structure is remarkably similar to the hairpin structure predicted to exist for the minimal minus-strand initiation promoter termed gPR (20). Although the cPR promoter is functional in the absence of the bridge sequence (20), it is a less active promoter without the bridge sequence (Fig. 2A). This could be due to the formation of a stable hairpin with a 3-nt loop in cPR (Fig. 1A) that might inhibit initiation from cPR. The base-bridge interaction is predicted to disrupt this hairpin structure (Fig. 1A), thus facilitating the formation of single-stranded tail necessary to fit into the RdRp template channel. Accordingly, the formation of an accessible single-stranded 3′ initiation sequence of 4 to 6 nt is apparently required for efficient RNA synthesis by the hepatitis C virus and brome mosaic virus RdRp (7, 26). Altogether, the bridge sequence is likely involved in “switching” the core cPR promoter to a more active form for plus-stranded RNA synthesis.
FIG. 5.
Presence of conserved base and bridge sequences in tombusviruses. The complementary nucleotides are shown for the minus strands in a 3′-to-5′ orientation. TBSV, tomato bushy stunt virus; CNV, cucumber necrosis virus; AMCV, artichoke mottled crinkle virus; CymRSV, cymbidium ringspot virus; CIRV, carnation Italian ringspot virus. Note that CNV has an additional ACCUCU sequence at the 5′ end of RII(−).
Additional role of the bridge-base interaction might include bringing the REN and cPR into close proximity, which are located at the two opposite ends of the TBSV genomic or DI RNAs (Fig. 1A). Indeed, the proximity of REN and cPR was important for efficient initiation from cPR in the trans-replication assay (Fig. 2A and B). However, the 6-nt bridge-base interaction alone is unlikely to be enough to stabilize such RNA structure, because the bridge sequence, when positioned distantly from cPR, could not facilitate initiation in the trans-replication assay (Fig. 2B). We propose that self-folding of the minus-stranded viral RNA, including sequences outside of RIII(−), could facilitate the proximal positioning of RIII(−) and the cPR promoter (Fig. 6) in TBSV (−)RNA and DI (−)RNA; thus, this could promote the base-bridge interaction. It is also possible that additional RNA-protein and/or protein-protein interactions contribute to stabilizing the base-bridge interaction, as was proposed for bringing the 5′ and 3′ ends into proximity during poliovirus minus-strand synthesis (2). Altogether, the whole RIII(−) REN has been shown to be responsible for 10- to 20-fold stimulation of plus-strand synthesis (Fig. 1) (17), whereas the base-bridge interaction is shown to contribute two- to threefold enhancement. These in vitro data are in good correlation with in vivo data obtained with protoplasts and yeast cells (18, 27), which showed 10- to 20-fold inhibitory effect of the deletion of RIII and a 30 to 70% decrease for deletion or mutations within the bridge sequence (Fig. 4). These results strongly support that RIII(−) is the major REN element in TBSV RNA.
FIG. 6.
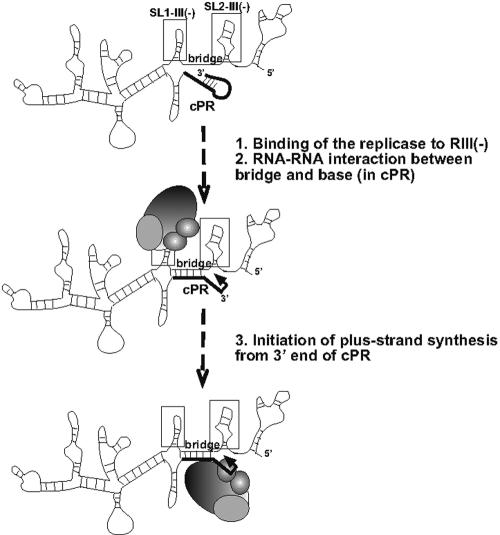
A model explaining the stimulation of RNA synthesis from the cPR plus-strand initiation promoter by the RIII(−) REN. The long-range RNA-RNA interaction between the bridge and the base sequences, possibly enhanced by the folding of the full-length RNA, is proposed to alter the structure of cPR. In addition, the formation of base-bridge interaction might also participate in bringing the REN and cPR into proximity. The tombusvirus replicase, containing p33, p92, and host factors, is proposed to bind to one of the two stem-loops, which then could result in positioning the active site of the replicase over the initiation site. This is predicted to lead to efficient initiation of plus-strand synthesis from cPR.
Although this paper is the first to give supporting evidence for the involvement of long range RNA-RNA interaction in replication enhancer-stimulated plus-strand RNA synthesis, there are a growing number of examples for long-range RNA-RNA interactions that have been indicated to play a role in replication of plus-strand viruses. For example, regulation of the subgenomic RNA synthesis in tombusviruses is thought to occur via long-distance RNA-RNA interaction between the distant activator domain and the complementary core sequence that involves 7 nt (4). The subgenomic RNA synthesis in RCNMV is controlled in trans by RNA-RNA interaction between two separate genomic RNAs (30). Also, bringing the replicase binding site and the initiation site for the minus-strand synthesis into proximity is mediated via an 8-nt RNA-RNA interaction in phage Qβ (8). The advantage for the long-distance interaction probably lies in fine-tuning regulative steps during replication, since proximal positioning of the regulatory elements was found to preserve or even increase the activity of certain replication elements (16, 26).
Model of replication stimulation by the RIII(−) replication enhancer.
Based on the above and previous data (17), we propose that RIII(−) replication enhancer in tombusviruses can facilitate plus-strand RNA synthesis by (i) binding to the replicase, (ii) changing the secondary structure of cPR via base-bridge interactions that may give rise to the formation of a hairpin-like structure with a 4-nt 3′ single-stranded tail, and (iii) promoting optimal positioning of the replicase on the minus-stranded template to facilitate de novo RNA initiation from the 3′-terminal cytidylate (Fig. 6). The above events may lead to efficient binding of the minus-stranded template to the replicase and efficient initiation of plus-strand synthesis. RIII(−) REN may also allow efficient rebinding of the replicase to the same template after termination of RNA synthesis (recycling model of replication). The role of REN in stimulation of plus-strand synthesis might be an important mechanism for tombusviruses to achieve asymmetric viral RNA replication, resulting far more plus-stranded than minus-stranded RNA progenies. Asymmetrical replication is thought to be important for tombusviruses, since (i) the large amount of plus strands can be used for virus encapsidation, cell-to-cell movement, protein synthesis, etc.; and (ii) the small amount of minus-strands may be helpful to avoid and/or delay the induction of gene silencing, a host resistance mechanism against double-stranded RNA molecules, which may form between the minus-strand and plus-strand RNAs during replication (1). Another advantage provided by REN is that it may speed up the replication process, which could allow the virus to race ahead of the gene-silencing process. Also, REN can increase the competitiveness of the viral RNA, which may be important during double-virus infections of hosts or during competition between genomic and defective RNAs.
In spite of similar abilities of the known replication enhancers to up-regulate viral RNA synthesis, the role of RIII(−) REN in tombusviruses and possibly in carmoviruses (15, 16) is different from RENs found in the L-A virus of yeast as well as the M-site in Qβ phage (8, 33). This is because the RENs of L-A and Qβ are functional in the plus-stranded RNA templates and they are likely involved in minus-strand synthesis, in the assembly of the replicase, and/or in template recruitment into replication in infected cells (8, 33). In contrast, RIII(−) REN is present in the minus-stranded RNA template, and it is involved in plus-strand synthesis that takes place after the replicase assembly and template recruitment steps. Since plus-strand RNA synthesis is significantly more robust than minus-strand synthesis for all plus-strand RNA viruses (3, 10), it is possible that replication enhancers, similar to the TBSV RIII(−) described here, will also be found in other RNA viruses (10).
Acknowledgments
We thank Andy White, Judit Pogany, and John Shaw for the valuable comments and Zivile Panaviene for technical help with the protoplast experiments.
This work was supported by NSF (MCB0078152) and the University of Kentucky.
Footnotes
This study is publication no. 03-12-050 of the Kentucky Agricultural Experiment Station.
REFERENCES
- 1.Ahlquist, P. 2002. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270-1273. [DOI] [PubMed] [Google Scholar]
- 2.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, I. R., M. Ostrovsky, G. Zhang, and K. A. White. 2001. Regulatory activity of distal and core RNA elements in Tombusvirus subgenomic mRNA2 transcription. J. Biol. Chem. 276:41761-41768. [DOI] [PubMed] [Google Scholar]
- 5.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 6.Kao, C. C., P. Singh, and D. J. Ecker. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251-260. [DOI] [PubMed] [Google Scholar]
- 7.Kao, C. C., X. Yang, A. Kline, Q. M. Wang, D. Barket, and B. A. Heinz. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klovins, J., V. Berzins, and J. van Duin. 1998. A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA 4:948-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koev, G., S. Liu, R. Beckett, and W. A. Miller. 2002. The 3′-terminal structure required for replication of barley yellow dwarf virus RNA contains an embedded 3′ end. Virology 292:114-126. [DOI] [PubMed] [Google Scholar]
- 10.Lai, M. M. 1998. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology 244:1-12. [DOI] [PubMed] [Google Scholar]
- 11.Lin, H. X., and K. A. White. 2004. A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J. 23:3365-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monkewich, S., H.-X. Lin, M. R. Fabian, W. Xu, H. Na, D. Ray, O. A. Chernysheva, P. D. Nagy, and K. A. White. 2005. p92 polymerase coding region contains an internal RNA element required at an early step in tombusvirus genome replication. J. Virol. 79:4848-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagy, P. D., and J. J. Bujarski. 1998. Silencing homologous RNA recombination hot spots with GC-rich sequences in brome mosaic virus. J. Virol. 72:1122-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of cucumber necrosis virus and tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 15.Nagy, P. D., J. Pogany, and A. E. Simon. 2001. In vivo and in vitro characterization of an RNA replication enhancer in a satellite RNA associated with turnip crinkle virus. Virology 288:315-324. [DOI] [PubMed] [Google Scholar]
- 16.Nagy, P. D., J. Pogany, and A. E. Simon. 1999. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 18:5653-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panavas, T., and P. D. Nagy. 2003. The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol. 77:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panavas, T., and P. D. Nagy. 2003. Yeast as a model host to study replication and recombination of defective interfering RNA of tomato bushy stunt virus. Virology 314:315-325. [DOI] [PubMed] [Google Scholar]
- 19.Panavas, T., Z. Panaviene, J. Pogany, and P. D. Nagy. 2003. Enhancement of RNA synthesis by promoter duplication in tombusviruses. Virology 310:118-129. [DOI] [PubMed] [Google Scholar]
- 20.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 21.Panaviene, Z., J. M. Baker, and P. D. Nagy. 2003. The overlapping RNA-binding domains of p33 and p92 replicase proteins are essential for tombusvirus replication. Virology 308:191-205. [DOI] [PubMed] [Google Scholar]
- 22.Panaviene, Z., T. Panavas, S. Serva, and P. D. Nagy. 2004. Purification of the Cucumber necrosis virus replicase from yeast cells: role of coexpressed viral RNA in stimulation of replicase activity. J. Virol. 78:8254-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pogany, J., K. A. White, P. D. Nagy. 2005. Specific binding of the tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajendran, K. S., and P. D. Nagy. 2003. Characterization of the RNA-binding domains in the replicase proteins of tomato bushy stunt virus. J. Virol. 77:9244-9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjith-Kumar, C. T., X. Zhang, and C. C. Kao. 2003. Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol. 77:1830-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray, D., and K. A. White. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rochon, D. M. 1991. Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20-kDa nonstructural protein. Proc. Natl. Acad. Sci. USA 88:11153-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen, R., and W. A. Miller. 2004. Subgenomic RNA as a riboregulator: negative regulation of RNA replication by barley yellow dwarf virus subgenomic RNA 2. Virology 327:196-205. [DOI] [PubMed] [Google Scholar]
- 30.Sit, T. L., A. A. Vaewhongs, and S. A. Lommel. 1998. RNA-mediated trans-activation of transcription from a viral RNA. Science 281:829-832. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White, K. A., and P. D. Nagy. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187-226. [DOI] [PubMed] [Google Scholar]
- 33.Wickner, R. B. 1996. Prions and RNA viruses of Saccharomyces cerevisiae. Annu. Rev. Genet. 30:109-139. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, G., J. Zhang, and A. E. Simon. 2004. Repression and derepression of minus-strand synthesis in a plus-strand RNA virus replicon. J. Virol. 78:7619-7633. [DOI] [PMC free article] [PubMed] [Google Scholar]