Abstract
A putative deoxyuridine triphosphatase (dUTPase) gene from chlorella virus PBCV-1 was cloned, and the recombinant protein was expressed in Escherichia coli. The recombinant protein has dUTPase activity and requires Mg2+ for optimal activity, while it retains some activity in the presence of other divalent cations. Kinetic studies of the enzyme revealed a Km of 11.7 μM, a turnover kcat of 6.8 s−1, and a catalytic efficiency of kcat/Km = 5.8 × 105 M−1 s−1. dUTPase genes were cloned and expressed from two other chlorella viruses IL-3A and SH-6A. The two dUTPases have similar properties to PBCV-1 dUTPase except that IL-3A dUTPase has a lower temperature optimum (37°C) than PBCV-1 dUTPase (50°C). The IL-3A dUTPase differs from the PBCV-1 enzyme by nine amino acids, including two amino acid substitutions, Glu81→Ser81 and Thr84→Arg84, in the highly conserved motif III of the proteins. To investigate the difference in temperature optima between the two enzymes, homology modeling and docking simulations were conducted. The results of the simulation and comparisons of amino acid sequence suggest that adjacent amino acids are important in the temperature optima. To confirm this suggestion, three site-directed amino acid substitutions were made in the IL-3A enzyme: Thr84→Arg84, Glu81→Ser81, and Glu81→Ser81 plus Thr84→Arg84. The single substitutions affected the optimal temperature for enzyme activity. The temperature optimum increased from 37 to 55°C for the enzyme containing the two amino acid substitutions. We postulate that the change in temperature optimum is due to reduction in charge and balkiness in the active cavity that allows more movement of the ligand and protein before the enzyme and substrate complex is formed.
The gene encoding deoxyuridine triphosphatase (dUTPase), which catalyzes the hydrolysis of dUTP to dUMP and PPi, is ubiquitous in prokaryotic and eukaryotic organisms and is also present in many viruses. The enzyme serves two critical biological roles. First, dUTPase provides the substrate for thymidylate synthase, the major biosynthesis pathway to deoxythymidine triphosphate (dTTP). Second, it maintains low dUTP levels in the cell that prevents misincorporation of uracil into DNA (10, 25). Incorporation of uracil into DNA induces excision repair, DNA fragmentation and cell death (13, 14).
The wide distribution of the dUTPase gene (dut) among DNA and RNA viruses, such as poxviruses, adenoviruses, herpesviruses, asfarviruses, and a subset of retroviruses indicates that strict control of dUTP concentrations is important for virus replication (1). Misincorporation of uracil into retroviral DNA affects the specificity of plus-strand synthesis initiation during lentivirus reverse transcription, thereby disrupting the viral life cycle (24). Disruption of the varicella-zoster virus dut gene results in a virus that grows to lower titers and is impaired in syncytium formation (36). Mutations in the dut gene from herpes simplex virus type 1 and pseudorabies virus significantly attenuate virus production in animals (22, 35). Therefore, dUTPase activity is one of the key sites for intervention in both chemotherapy and antiretroviral therapy (19).
Sequence analysis of the 330-kb chlorella virus PBCV-1 (family Phycodnaviridae) genome revealed an open reading frame, A551L, which encodes a protein similar to dUTPases from many organisms (27). PBCV-1 encodes genes for 12 additional putative enzymes involved in nucleotide metabolism (41). This is probably not surprising because the total DNA in virus-infected cells increases 4- to 10-fold by 4 h postinfection (p.i.) (42). To begin characterizing these nucleotide-metabolizing enzymes, we report here that the PBCV-1-encoded dUTPase, as well as homologs from chlorella virus IL-3A and SH-6A, have properties similar to those of dUTPases from eukaryotes, bacteria, and other viruses. Surprisingly, however, the temperature optimum for activity from IL-3A dUTPase is lower than for the other two enzymes. To assess the key amino acids related to the temperature differences between the enzymes, we modeled the three-dimensional structures of chlorella virus dUTPases and investigated docking kinetics by computer simulation. The model predicts that adjacent residues are important to determine the optimal temperature. To confirm this prediction, we generated three mutations in IL-3A dUTPase motif III and characterized the mutants with respect to predicted protein structure and kinetic properties.
MATERIALS AND METHODS
Viruses and host strains.
The growth of PBCV-1 host Chlorella NC64A on MBBM medium and Chlorella Pbi on FES medium, the production and purification of the viruses, and the isolation of virus DNAs have been described (43, 44). E. coli strains DH5αMCR (E. coli Genetic Stock Center, New Haven, CT) and BL21(DE3)/pLysS (Novagen, Madison, WI) were grown in Luria-Bertani (LB) medium (37).
Cloning, expression, and purification of dUTPase.
PBCV-1 a551l was cloned from PCR-amplified viral DNA with the following oligonucleotide primers: 5′ primer 5′-GGAATTCATGCTTCTCTCCTCGTA-3′ and 3′ primer 5′-TTACTCGAGGATTCCCGTACTTCCAAA-3′. The 5′ primer contained an EcoRI restriction site, and the 3′ primer contained an XhoI restriction site. The a551l gene was amplified with KOD hot start DNA polymerase (Novagen) in 50-μl reactions which contained 100 ng of virus DNA; 15 pM concentrations of each primer; 0.2 mM concentrations each of dATP, dGTP, dCTP, and dTTP; and 1 mM MgSO4 in 35 cycles of heating and cooling as follows: 15 s at 94°C for denaturing, 30 s at 60°C for annealing, and 1 min at 68°C for elongation. The PCR products were purified from 1.2% agarose gels by using a Qiaex II gel extraction kit (QIAGEN, Valencia, CA), digested with EcoRI and XhoI, and inserted into the EcoRI/XhoI sites of the pET23a+ expression vector (Novagen). This process produced a His6 tag at the C terminus of the target protein. The construction of the recombinant expression plasmid, named pET-DUT, was confirmed by DNA sequencing. Expression of the dUTPase protein was carried out in E. coli BL21(DE3)/pLysS, which contains the T7 RNA polymerase gene under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacUV5 promoter and a plasmid constitutively expressing T7 lysozyme that is the inhibitor of T7 RNA polymerase. Cells were transformed with plasmid pET-DUT and grown overnight in LB medium at 37°C. Flasks containing 400 ml of LB medium, which contain 100 μg of ampicillin/ml and 37 μg of chloramphenicol/ml, were inoculated with 1/40 volume of the overnight culture and incubated at 37°C until the absorbance at 595 nm reached 0.6 to 0.8. IPTG (Sigma, St. Louis, MO) was added to a final concentration of 0.5 mM, and incubation was continued for 2 h at 30°C. One liter of cells was harvested by centrifugation at 5,000 × g for 5 min and resuspended in 50 ml of NPI-10 buffer (50 mM phosphate buffer [pH 8.0], 300 mM NaCl, 10 mM imidazole). The cells were disrupted on ice by sonication for 4 min by using a Tekmar sonic disruptor at 100% amplitude in 5-s pulses. The samples were centrifuged at 12,000 × g for 10 min to separate soluble and insoluble fractions. Then, Ni-nitrilotriacetic acid (NTA) agarose beads (QIAGEN, Hilden, Germany) equilibrated with NPI-10 buffer were added to the soluble fraction. After mixing for 1 h at 4°C, the resin was loaded in a column and washed with 20 column volumes of NPI-20 buffer (50 mM phosphate buffer [pH 8.0], 300 mM NaCl, 20 mM imidazole). The recombinant dUTPase protein was eluted from the column by NPI-250 buffer (50 mM phosphate buffer [pH 8.0], 300 mM NaCl, 250 mM imidazole). Glycerol was added to the sample to a final concentration of 50%, and the enzyme was stored at −20°C.
The dut genes from viruses IL-3A and SH-6A were cloned, expressed, and purified with the same primers and protocols used for PBCV-1 dut.
Enzyme assays.
Unless otherwise indicated, dUTPase activity was determined in a 20-μl reaction mixture containing 50 mM Tris-HCl (pH 8.5), 0.05% bovine serum albumin (BSA), 5 mM MgCl2, 2 mM dithiothreitol, 50 μM [5-3H]dUTP (Amersham, Piscataway, NJ), and 1 ng of enzyme. Kinetic studies were performed under standard assay conditions with various substrate concentrations ranging from 1 to 50 μM dUTP and 1 ng of enzyme. The determination of optimal temperature was performed at 25, 30, 37, 42, 50, 55, 60, and 65°C. A 10 mM buffer mixture of PIPES, HEPES, Tris, and Capso (pH 6.0 to 11.0), and 50 mM Tris-maleate buffer (pH 2.0-8.0) was used to determine the optimal pH for dUTPase activity. Divalent cation experiments were performed in the same reaction system by substituting 5 mM MnCl2, CaCl2, CuCl2, CoCl2, NiCl2, or ZnSO4 for MgCl2. Competition experiments were assessed in the same reaction mixture with the addition of unlabeled deoxynucleoside triphosphates at 10 to 50 μM.
Northern and dot blot s analyses.
Chlorella cells (109 cells) were collected at various times after PBCV-1 infection, frozen in liquid nitrogen, and stored at −80°C. RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA), denatured with formaldehyde, separated on a 1.2% agarose denaturing gel, and transferred to a nylon membrane (Micron Separations, Inc., Westborough, MA) as described previously (15). The membrane was subsequently photographed under UV illumination to visualize transferred RNA. The RNA was hybridized with a double-stranded 32P-dut probe at 65°C in 50 mM NaPO4, 1% BSA, and 2% sodium dodecyl sulfate (SDS). The probe was labeled with 32P by using a random primers DNA labeling kit (Invitrogen). After hybridization, radioactivity bound to the membranes was detected and quantified with a Storm 840 PhosphorImager and ImageQuant software (Molecular Dynamics, Inc., Sunnyvale, CA). To monitor possible loading differences between samples, the relative amounts of the 3.6-kb rRNA in each lane were determined by converting the photographs of stained membranes to digital images with a Hewlett-Packard ScanJet 4C scanner and analyzing the images with ImageQuant software.
Viral DNAs used for dot blots were denatured, applied to nylon membranes fixed by UV, and hybridized with the same probes used for the Northern analysis.
SDS-PAGE and Western blots.
Chlorella cells, prepared as described above, were suspended in 200 μl of cell lysis buffer (10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3], 140 mM NaCl, 2.7 mM KCl) with 200 mg of 0.10-mm glass beads and vortexed at high speed for 5 min. Then, 5 μl of protease inhibitor cocktail (Sigma) was added to each sample. Disrupted cells were centrifuged at 13,000 × g for 10 min to remove particulate material. Protein concentrations were measured by the Bradford method with a Coomassie Plus protein assay kit (Pierce, Rockford, IL). Fifty micrograms of protein from each sample was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) using a 4 to 20% gradient (Cambrex, Rockland, ME) at 200 V for 1 h. Protein gels were stained with Coomassie brilliant blue R or transferred to a nitrocellulose membrane and reacted with PBCV-1 dUTPase antiserum produced in mice. The polyclonal antiserum was preincubated with an acetone powder of host chlorella protein to reduce extraneous reacting protein (18).
Site-directed mutagenesis.
Site-directed mutants were generated by two-step PCR with virus IL-3A dut DNA as a template. The PCRs were conducted with the same enzyme and procedure described above. The first-step PCR included two PCRs that used 5′ primer of the PBCV-1 dut gene and mutagenesis primer 2 in reaction 1, and 3′ primer of PBCV-1 dut gene and mutagenesis primer 1 in reaction 2. The products of reactions 1 and 2 were used as a template together, and 5′ and 3′ primers of PBCV-1 dut gene were used for the second-step PCR. The products from the second-step PCR, which contained the mutant site, were digested with EcoRI and XhoI and inserted into the EcoRI/XhoI sites of the pET23a+ expression vector. Mutations in the IL-3A dut gene were confirmed by DNA sequencing. The mutant proteins were expressed and purified by the same procedures described above. Oligonucleotide sequences of the mutagenic primers are listed in Table 1.
TABLE 1.
DNA sequences of the mutagenic primers used for the site-directed mutagenesis of the IL-3A dut genea
| Primer | Sequence |
|---|---|
| Mu-2 | |
| Primer 1 | 5′-GATGAAGATTATAGAGGTGAA-3′ |
| Primer 2 | 5′-TTCACCTCTATAATCTTCATC-3′ |
| Mu-8 | |
| Primer 1 | 5′-GATTCAGATTATACCGGTGAA-3′ |
| Primer 2 | 5′-TTCACCGGTATAATCTGAATC-3′ |
| Mu-22 | |
| Primer 1 | 5′-GATTCAGATTATAGAGGTGAA-3′ |
| Primer 2 | 5′-TTCACCTCTATAATCTGAATC-3′ |
Codons for the changed amino acids are underlined.
Molecular modeling and analysis.
Homology models of the chlorella virus dUTPases were constructed by using the human dUTPase structure (28) (PDB ID 1Q5H) by SWISS-MODEL (38), 3D-JIGSAW (2), and CCP4 (9). The visual presentation was done with PyMOL (11). To locate the dUTP ligand binding site, dUTP coordinates in human dUTPase were used as the initial structure and then refined. The binding free energy of dUTP against the dUTPases was calculated by AutoDock (30).
Other procedures.
DNA and putative protein sequences were analyzed with the University of Wisconsin Computer Group package of programs (version 10.1; Genetics Computer Group). The GenBank accession number for the PBCV-1 open reading frame (ORF) A551L is U42580. GenBank numbers for the dUTPase genes from viruses IL-3A and SH-6A are AY857869 and AY857870, respectively.
RESULTS
PBCV-1 ORF A551L is a dUTPase.
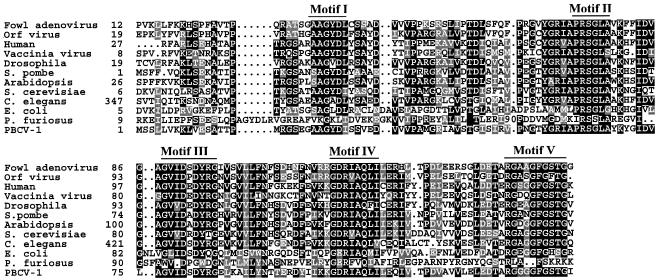
Chlorella virus PBCV-1 encodes a 141-codon ORF (A551L) that has 53 to 62% amino acid identity with dUTPases from other organisms. The dUTPase has an inferred molecular mass of 14.9 kDa that is among the smallest dUTPases in the public databases. Amino acid alignment of the enzyme with dUTPases from several sources indicates that A551L contains the five conserved motifs that comprise the dUTPase active site (6, 10, 26, 28, 34) (Fig. 1). Five gaps of one to six amino acids are scattered throughout the alignment that are shared by at least some of the other dUTPases.
FIG. 1.
Amino acid alignment of dUTPases. The amino acid sequences from fowl adenovirus (S26429), Orf virus (Q9YYS0), human (G02777), vaccinia virus (P21035), Drosophila melanogaster (JC7565), Schizosaccharomyces pombe (Q9P6Q5), Arabidopsis (T12954), Saccharomyces cerevisiae (P33317), Caenorhabditis elegans (T43673), Escherichia coli (P06968), Pyrococcus furiosus (AAL47572), and PBCV-1 were aligned with the Wisconsin GCG PILEUP program. The five conserved motifs of dUTPases are labeled.
The G+C content of a551l is 48%, which is higher than the 40% G+C content of the PBCV-1 genome and less than the 67% G+C content of Chlorella NC64A DNA (45). This observation suggests the gene might have arisen from horizontal gene transfer (1). Typically, the 50 nucleotides preceding PBCV-1 translational start codons are at least 70% A+T (41). The a551l gene fulfills this requirement, with 70% A+T. Three possible TATAAT-like sequences are located within the 15 nucleotides prior to the UTG translation start codon (results not shown). Thus, we expected a551l to encode a functional protein.
Expression and purification of PBCV-1 dUTPase.
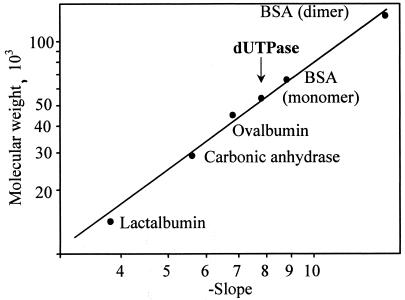
Plasmid pET-DUT, containing the PBCV-1 dut gene, was constructed, and the protein was expressed as a 17.6-kDa fusion protein containing a phage T7 tag at the N terminus and a His6 tag at the C terminus by using E. coli strain BL21(DE3)-pLysS. The His tag allowed the recombinant protein to be purified by Ni-NTA affinity chromatography. About 20 mg of soluble recombinant protein was obtained per liter of E. coli culture. dUTPases from various sources function as either monomers (3, 4), dimers (20), or trimers (6, 8, 26, 28, 32). Recently, a bifunctional enzyme with dUTPase activity from Methanococcus jannaschii was discovered that functions as a hexamer (21). The molecular size of PBCV-1 recombinant dUTPase on native polyacrylamide gels is ca. 54 kDa, suggesting the enzyme is a homotrimer (Fig. 2). Preliminary analysis of the crystal structure of IL-3A dUTPase also indicates it is a homotrimer (K. Homma and H. Moriyama, unpublished results).
FIG. 2.
The molecular mass of the recombinant dUTPase (∼54 kDa) was determined by nondenaturing electrophoresis on 7, 8, 9, and 10% polyacrylamide gels as described in a kit (Sigma) for measuring the molecular mass of nondenatured proteins. Lactalbumin, carbonic anhydrase, ovalbumin, and BSA were used as standards.
Characterization of PBCV-1 dUTPase.
Purified dUTPase is active over a wide pH range from 2.0 to 11.0, with an optimum pH at 8.5 (Table 2). The enzyme also functions over a wide temperature range from 25 to 65°C, with an optimal temperature of 50°C (Table 2). This temperature is much higher than the 25°C optimum temperature for growing the host and the virus.
TABLE 2.
Optimum reaction conditions for wild-type and mutant dUTPases
| dUTPase | Temp (°C) | pH | Cation |
|---|---|---|---|
| PBCV-1 | 50 | 8.5 | Mg2+ |
| SH-6A | 50 | 8.5 | Mg2+ |
| IL-3A | 37 | 8.5 | Mg2+ |
| Mu-2 | 50 | 8.5 | Mg2+ |
| Mu-8 | 42 | 8.5 | Mg2+ |
| Mu-22 | 55 | 8.5 | Mg2+ |
Addition of 1 mM EDTA completely inhibits dUTPase activity, indicating the enzyme requires a divalent cation for activity. To assess whether enzyme activity requires a specific divalent cation, the ability of several divalent cations to restore enzyme activity in the presence of EDTA was tested. Preincubation of the enzyme with 1 mM EDTA for 5 min, followed by addition of 5 mM divalent cations, established that several divalent cations restore enzyme activity. The enzyme prefers Mg2+ but exhibits activity in the present of Mn2+, Co2+, Ni2+, Zn2+, Ca2+, and Cu2+ (Table 3). However, like dUTPases from feline immunodeficiency virus (48), vaccinia virus (5) and African swine fever virus (32), the recombinant PBCV-1 enzyme is active without added divalent cations (Table 3). This result implies that divalent cations bind tightly to the protein and remain attached to the protein after purification. Competition experiments with unlabeled deoxynucleotide triphosphates established that only dUTP competed with [3H]dUTP, indicating a high preference for dUTP (Table 4).
TABLE 3.
Effect of added divalent cations on PBCV-1 dUTPase activity
| Added cation (concn [mM]) | Relative activitya (%) with:
|
|
|---|---|---|
| No EDTA | 1 mM EDTA | |
| None | 80 | 0 |
| Mg2+ (5) | 100 | 100 |
| Mn2+ (5) | 71 | 70 |
| Co2+ (5) | 61 | 59 |
| Zn2+ (5) | 52 | 49 |
| Ni2+ (5) | 47 | 30 |
| Ca2+ (5) | 38 | 31 |
| Cu2+ (5) | 32 | 28 |
The results are expressed as the percentage of enzyme activity relative to the activity obtained with Mg2+. The results are the average of three independent experiments.
TABLE 4.
Effect of unlabeled deoxynucleotides on PBCV-1 dUTPase activity
| Added deoxynucleotide | Relative activitya (%) |
|---|---|
| None | 100 |
| dTTP | 98 |
| dATP | 98 |
| dGTP | 99 |
| dCTP | 99 |
| dUTP | 52 |
The results are expressed as the percentage of enzyme activity relative to the activity (23.9 μmol min−1 mg−1) obtained without adding any other nucleotide. The results are the average of three independent experiments.
The PBCV-1 dUTPase has a Km for dUTP of 11.7 μM and a specific activity of 23.9 μM dUTP converted to dUMP min−1 mg of protein−1 at 50°C (Table 5). The turnover number (kcat) is 6.8 s−1. The kcat/Km of the PBCV-1 dUTPase is 5.8 × 105 M−1 s−1. The PBCV-1 recombinant enzyme was stored for over a year at −20°C without loss of activity.
TABLE 5.
Kinetic constants for wild-type and mutant dUTPases
| Enzyme | Km (μM) | kcat (s−1) | Vmax (μmol min−1 mg−1) | kcat/Km (105 M−1 s−1) |
|---|---|---|---|---|
| PBCV-1 | 11.7 | 6.8 | 23.9 | 5.8 |
| SH-6A | 11.9 | 4.9 | 16.6 | 4.1 |
| IL-3A | 11.7 | 9.2 | 31.4 | 7.9 |
| Mu-2 | 8.2 | 9.8 | 33.6 | 11.9 |
| Mu-8 | 7.6 | 4.1 | 14.0 | 5.4 |
| Mu-22 | 8.5 | 4.4 | 14.9 | 5.2 |
dut expression in PBCV-1-infected Chlorella cells.
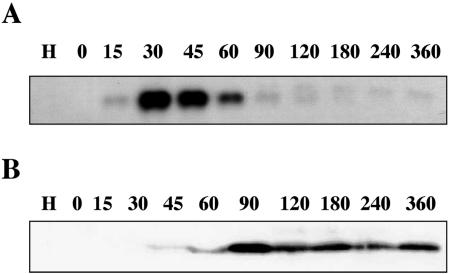
RNA was extracted from virus-infected cells at various times after infection and hybridized to an a551l probe to determine when the gene is transcribed during PBCV-1 replication. The probe hybridized to an ∼0.7-kb RNA transcript extracted from cells at 15 min p.i. The hybridization intensity increased until 45 min p.i., followed by a rapid decrease (Fig. 3A). This result indicates that the PBCV-1 dut gene is expressed as an early gene. The 0.7-kb RNA transcript is a reasonable size for a 141-amino-acid protein.
FIG. 3.
(A) Transcription of PBCV-1-encoded dUTPase gene a551l. Total RNAs were isolated from uninfected (H) and PBCV-infected Chlorella NC64A cells at the indicated times (in minutes) and hybridized with a 32P-labeled a551l probe. (B) Western blot of PBCV-1-encoded dUTPase. Total proteins were isolated from uninfected (H) and PBCV-1-infected Chlorella NC64A cells at indicated times in minutes. The proteins were reacted with dUTPase antiserum.
Antibody to the A551L protein reacted with an ∼15-kDa protein that first appeared at ∼45 min p.i. The protein increased to a maximum at 90 min p.i. and then remained at a relative constant level throughout virus replication (Fig. 3B).
dut occurrence in other chlorella viruses.
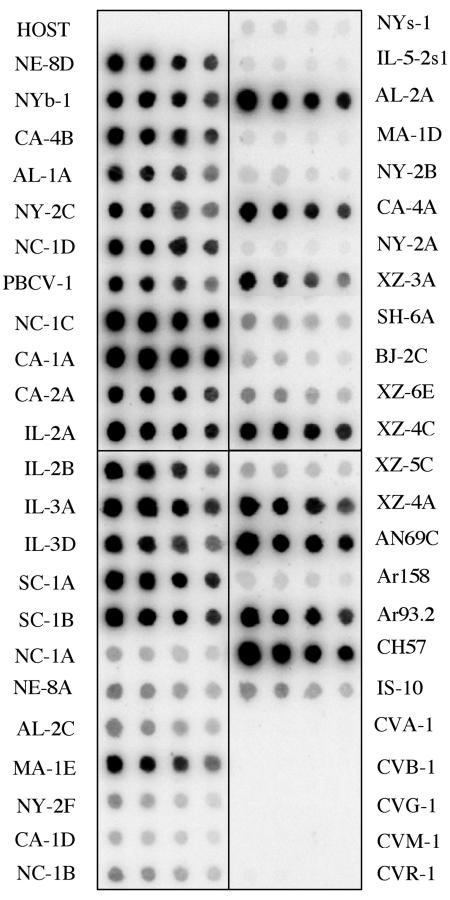
Genomic DNAs from 47 chlorella viruses, isolated from diverse geographical regions, were hybridized to the PBCV-1 a551l probe to determine how widely the dut gene was distributed among the viruses. These viruses infect either Chlorella NC64A (NC64A viruses) or Chlorella Pbi (Pbi viruses). The PBCV-1 a551l probe hybridized strongly to 24 DNAs and weakly to 11 DNAs from the 42 NC64A virus DNAs (Fig. 4). This latter finding was surprising because gene homologs from these 11 viruses, such as NE-8A, NY-2F, and SH-6A, typically hybridize strongly with PBCV-1 genes, e.g., the RNase III gene (49). The hybridizations with six other NC64A viruses—NYs-1, Il-5-2s1, MA-1D, NY-2B, NY-2A, and Ar158—were even weaker (Fig. 4). Typically, the nucleotide sequences of these six viruses differ significantly from PBCV-1 gene homologs and hybridization signals with PBCV-1 gene probes are either undetected or barely detected at moderate stringency. No hybridization was detected with DNA from the Chlorella NC64A host or DNAs from the five Pbi viruses. The nucleotide identity of gene homologs between the NC64A and Pbi viruses is typically 60 to 65% (41), and this probably explains the lack of a551l hybridization with the Pbi viruses.
FIG. 4.
Hybridization of the PBCV-1 dut gene to DNAs isolated from the host Chlorella NC64A, 42 NC64A viruses, and 5 Pbi viruses (CVA-1, CVB-1, CVG-1, CVM-1, and CVR-1). The DNAs were hybridized with a 32P-labeled a551l gene probe. The blots in each row contain 1, 0.5, 0.25, and 0.12 μg of DNA from left to right, respectively.
dut genes from other chlorella viruses.
The low hybridization of the PBCV-1 dut gene with some of the NC64A viruses prompted us to clone the gene from other chlorella viruses to examine sequence diversity. The dut genes from IL-3A, MA-1E, NC-1C, NE8A, NY2F, SH6A, and BJ-2C were successfully cloned by PCR. The G+C contents of these dut genes were 46.5 to 47%, which is similar to the 48% of the PBCV-1 gene. Collectively, 97 nucleotide substitutions occurred. Sixty-eight of these substitutions occurred in the third position of the codon (nonsynonymous substitution; results not shown). Based on the amino acid sequences, the dut genes from the chlorella viruses can be separated into three groups that are represented by PBCV-1, IL-3A, and SH-6A (Fig. 5).
FIG. 5.
Amino acid alignment of dUTPases from chlorella viruses PBCV-1, IL-3A, and SH-6A. Alignments were done with the Wisconsin GCG PILEUP program. The five most conserved motifs in dUTPase are labeled. (Note that virus IL-3A dUTPase differs by two amino acid residues in motif III from the other two dUTPases.)
The dut genes from IL-3A and SH-6A were expressed and the two new recombinant dUTPases had properties similar to those of the PBCV-1 enzyme. Surprisingly, however, the IL-3A dUTPase had a temperature optimum of 37°C rather than the 50°C for the PBCV-1 and SH-6A enzymes (Table 2). Furthermore, the larger turnover number (kcat) and catalytic efficiency (kcat/Km) (Table 5) indicate that IL-3A dUTPase is the most efficient enzyme of the three.
Protein modeling.
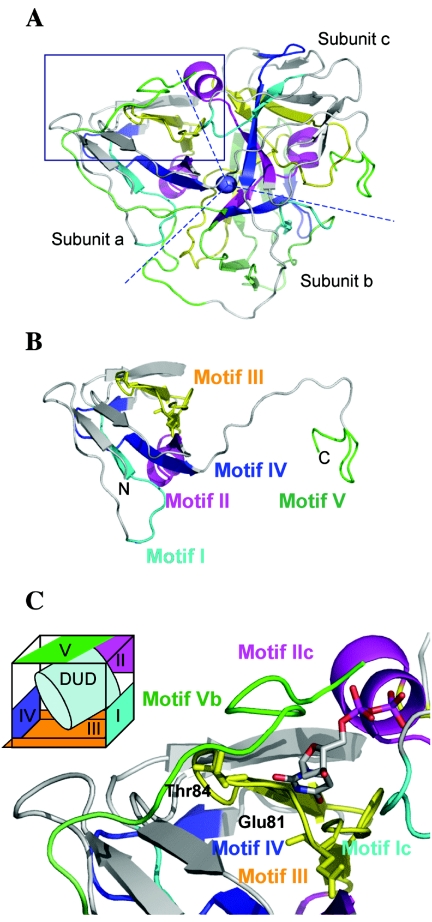
The difference in temperature optima between the dUTPases from PBCV-1 and IL-3A was unexpected. The IL-3A dUTPase differs by nine amino acids from the PBCV-1 enzyme, including two amino acid substitutions, Ser81→Glu81 and Arg84→Thr84, in the highly conserved motif III of the protein (Fig. 5). To determine whether these amino acid changes might affect temperature optima, homologous model building was undertaken. Since the primary amino acid sequence of IL-3A dUTPase is most similar to the human dUTPase (28) and both enzymes function as homotrimers, the enzyme was modeled by using the human dUTPase structure (PDB ID 1Q5U). The enzyme consists of three homo subunits: a, b, and c (Fig. 6A). Each subunit or monomer has two discreet structures, a core, and a tail (Fig. 6B). The core structure consists of motifs I through IV, and the tail structure consists of motif V. The monomers form a trimer that has threefold symmetry with a single magnesium atom located at the center of the structure (Fig. 6A). The active site (cavity) is cubic (Fig. 6C, inset); motifs III and IV of each subunit provide a base plate and a side plate. The other side plate and back wall are from motifs I and II of subunit c, respectively. Motif V of subunit b serves as a lid to the cubic structure. The substrate, dUTP, accesses the cavity via a hole in the back wall and lid.
FIG. 6.
Molecular modeling of chlorella virus IL-3A dUTPase. (A) Representative model of IL-3A dUTPase as a trimer. The boxed area represents an active cavity. Structural motifs are represented by colors. Each subunit has white, green, and gray-white motifs with the same color coding as in panel B. (B) Monomer view of the IL-3A dUTPase. (C) Enlarged view of the active site with dUDP (DUD) and schematic diagram of active site (an inset). Stick models were added at Glu81 and Thr84. (Note that dUDP is used to model the enzyme because it is a noncompetitive inhibitor that localizes in the active site.)
Amino acid residues 81 and 84 in motif III differ between PBCV-1 and IL-3A dUTPases. These two residues surround an aspartic acid and a tyrosine residue, which are essential for dUTPase activity (19). Motif V is highly flexible and may act after hydrolysis to eject reaction products from the active site (28). In human dUTPase the corresponding amino acids are Glu103 and Arg106 in the notation of the PDB entry 1Q5H. Both amino acids in PBCV-1 dUTPase interact with each other and, in addition, Arg84 has a polar contact with Gly132 in motif V (Table 6). Although the IL-3A dUTPase has human type Glu81, PBCV-1 dUTPase has a Ser at residue 81. In the high-temperature PBCV-1 and SH-6A dUTPases, the tail of Motif V is secured by polar contact between Arg84 and Gly132. The tail lid is then reinforced by an interaction between Ser81 and Arg84. However, low-temperature IL-3A dUTPase does not have a second interaction between Glu81 and Thr84. Instead, the Glu81 carbonyl oxygen OE3 interacts with the Gly55 nitrogen. From the modeling, we conclude that the interaction between the three amino acids in PBCV-1 dUTPase requires a high temperature to allow the lid to move, so that the substrate can approach the active site to form the ES complex. To test this idea, we did a docking experiment (17) between the virus dUTPase and dUTP at three potential active sites in the trimer. We placed a substrate molecule in the vicinity of an active site by using the identified binding site in human dUTPase as a guide. For each binding site, a best-fit conformation was determined by using the energy function in the AutoDock program. The results indicated that alternation of the 81st and/or 84th residue resulted in a change in binding between dUTPase and dUTP.
TABLE 6.
Molecular contacts and substrate docking
| dUTPase | Residue 81 | Inter- action | Residue 84 | Interaction | dUTP binding (kcal M−1) |
|---|---|---|---|---|---|
| PBCV-1 | Ser | Arg84 | Arg | Ser81 Gly132 | −10.9 |
| IL-3A | Glu | Gly55 | Thr | Gly132 | −12.5 |
| Mu-2 | Glu | Arg84 | Arg | Glu81 Gly132 | −13.0 |
| Mu-8 | Ser | NAa | Thr | Gly132 | −13.3 |
| Mu-22 | Ser | Arg84 | Arg | Tyr83 Ser81 Ser127 Gly132 | −13.5 |
NA, not applicable.
Modulation of IL-3A dUTPase.
To investigate the effect of amino acid residues 81 and 84 on the temperature different between PBCV-1 and IL-3A dUTPases, IL-3A Glu81 and Thr84 were converted to Ser81 and Arg84 either singly or simultaneously. The three IL-3A dUTPase mutants are Mu-8 (Glu81→Ser81), Mu-2 (Thr84→Arg84), and Mu-22 (Glu81→Ser81 and Thr84→Arg84). The mutated dUTPases were isolated by the same procedure used for PBCV-1 dUTPase. Each mutant had properties similar to those of wild-type IL-3A dUTPase (Table 2). However, both amino acid substitutions affected the temperature optima of the enzyme. Substitution of Glu81→Ser81 (Mu-8) shifts the optimal temperature from 37 to 42°C, whereas substitution of Thr84→Arg84 (Mu-2) changed the optimal temperature from 37 to 50°C. Simultaneous substitutions (Mu-22) increased the optimal temperature from 37 to 55°C (Table 2). Kinetic analyses established that both substitutions (Glu81→Ser81 and Thr84→Arg84) slightly increased enzyme substrate (dUTP) binding. Interestingly, the change Thr84→Arg84 (Mu2) produced a larger kcat/Km value, which indicates the enzyme is more efficient than wild-type IL-3A dUTPase (Table 5).
DISCUSSION
Typically, cellular dUTPases are expressed at high levels in dividing, undifferentiated cells and at low levels in terminally differentiated, nondividing cells. Consequently, viruses with defective dut genes replicate poorly in nondividing host cells compared to wild-type viruses (7, 23, 39, 40). To guarantee a supply of deoxynucleotides in nonproliferating host cells, large DNA viruses often encode many nucleotide synthesis enzymes, including both subunits of ribonucleotide reductase and a dUTPase. Virus PBCV-1 follows this pattern and encodes at least 13 putative enzymes involved in nucleotide metabolism (41). The virus-encoded enzymes are important because the DNA concentration in a PBCV-1-infected cell increases 4- to 10-fold by 4 h p.i. (42). Consequently, PBCV-1 DNA synthesis requires large quantities of deoxynucleoside triphosphates that cannot be accounted for simply by recycling deoxynucleotides from degraded host DNA.
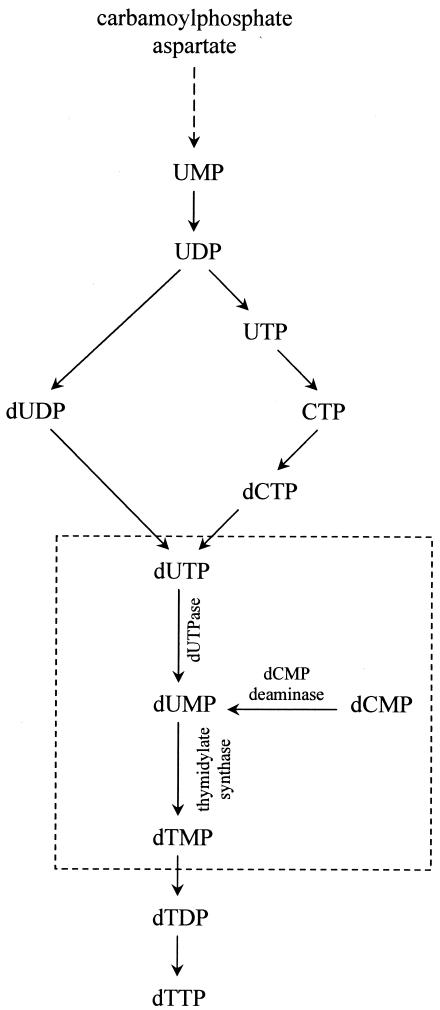
The PBCV-1 dUTPase has properties similar to those of dUTPases from other organisms. The product of the dUTPase reaction is dUMP that is the substrate for thymidylate synthase. In addition to dUTPase, PBCV-1 also encodes another enzyme, dCMP deaminase, which produces dUMP (Y. Zhang and J. L. Van Etten, unpublished results). Interestingly, PBCV-1 lacks a gene encoding the classical folate-dependent thymidylate synthase A. Instead, PBCV-1 encodes a flavin-dependent enzyme, called thymidylate synthase X, that synthesizes thymidylate (31). The recombinant PBCV-1 thymidylate synthase X is active (16), creating a PBCV-1 encoded pathway for dTMP synthesis that is part of dTTP biosynthesis (Fig. 7).
FIG. 7.
Biosynthesis of dTTP. PBCV-1-encoded enzymes—dUTPase, dCMP deaminase, and thymidylate synthase—form the pathway for dTMP synthesis that is highlighted in the dashed-line box.
Viruses also need dUTPases to prevent dUTP incorporation into viral DNA. Typically, the DNA repair enzyme uracil-DNA glycosylase (UNG), a member of the base excision-repair pathway, removes uracil from DNA. Unlike herpesviruses and poxviruses that encode UNG (7), PBCV-1 does not encode a UNG, and so it must rely on the host UNG to eliminate uracil from DNA. However, not all phycodnaviruses have dUTPase encoding genes. For example, virus EsV-1 that infects the filamentous brown alga, Ectocarpus siliculosus, lacks a dut gene (12), whereas the gene is present in a virus which infects a marine alga, Emiliania huxleyi (W. Wilson, unpublished data).
Accumulating evidence indicates that the chlorella viruses probably have a long evolutionary history (41). Phylogenic analysis of DNA polymerases places the phycodnavirus enzymes near the root of all eukaryotic δ DNA polymerases (46, 47). Likewise, phylogenetic analyses of other PBCV-1-encoded proteins often position them near the root of their eukaryotic counterparts, e.g., the potassium ion channel protein Kcv (33), ornithine decarboxylase (29), and GDP-mannose 4,6-dehydratase (G. Duncan and J. L. Van Etten, unpublished results). Finally, despite the fact that PBCV-1 encodes a mixture of prokaryote- and eukaryote-like proteins, the G+C content of the PBCV-1 genome (40%) (45) is relatively uniform throughout its genome. This pattern suggests that the genes have existed together in PBCV-1 for a long time. Therefore, it was surprising to discover that the G+C content of the PBCV-1 dut gene is significantly higher (48%) than the rest of the virus genome. This finding suggests that PBCV-1 might have acquired the dut gene after the other genes. A recent comprehensive analysis of dUTPase amino acid sequence relationships from viruses and their hosts is consistent with this hypothesis (1). The authors of that study concluded that PBCV-1 dut probably resulted from horizontal gene transfer from a eukaryotic organism, possibly from its host.
Another surprise was that the nucleotide sequence in dut genes from some of the viruses that infect Chlorella NC64A differed substantially from the PBCV-1 homolog. Whereas significant differences in the dut gene were expected for NC64A viruses NY-2A, NYs-1, IL-5-2s1, MA-1D, NY-2B, and Ar158, gene homologs from viruses NC-1A, NE-8A, Al-2C, NY-2F, CA-1D, NC-1B, SH-6A, BJ-2C, XZ-6E, XZ-5C, IS-10, MA-1E, and IL-3A are usually similar to their PBCV-1 counterparts. This finding suggests that the dut gene either evolves faster than the other chlorella virus genes or the dut gene was acquired on more than one occasion. If the gene was acquired more than once, the gene might have different locations in the virus genomes. It will be interesting to determine the genes that flank dut in some of these viruses.
The finding that the temperature optimum for the IL-3A dUTPase (37°C) was significantly different than the PBCV-1 enzyme (50°C) was also surprising. The two enzymes differ by nine amino acids (Fig. 5). Interestingly, two of the amino acid substitutions at positions 81 (Ser to Glu) and 84 (Arg to Thr) occur in the highly conserved motif III that is part of the enzyme catalytic site. Both computer modeling predicted and amino acid substitution experiments established that these two amino acids were at least partially, if not totally, responsible for the difference in temperature optima.
The SH-6A dUTPase differs by 12 amino acids from the PBCV-1 enzyme (Fig. 5). However, none of these amino acids are located in the five conserved motifs of the enzyme. Since these two enzymes have similar properties, the amino acid changes in these nonconserved regions do not appear to alter the structure of the activity center of the enzyme.
Thus, we determined the following from the present study. (i) Chlorella viruses encode a functional dUTPase with properties similar to those of dUTPases from other organisms. (ii) The native structure of the dUTPase is a trimer. (iii) Transcription of the dut gene begins 15 min after PBCV-1 infection; the dUTPase protein appears at 45 min p.i. and remains until host cell lysis. (iv) The dut gene may have been acquired by the chlorella viruses later than the other genes and on more than one occasion. (v) Finally, amino acid residues Glu81 and Thr84 of IL-3A dUTPase have key roles in determining the optimal temperature of the enzyme for activity.
Acknowledgments
We thank Benjamin Bixenmann for technical help.
This investigation was supported in part by Public Health Service grant GM32441 (J.L.V.E.), NIH grant P20RR15635 from the COBRE program of the National Center for Research Resources (J.L.V.E.), and a Layman award from the University of Nebraska-Lincoln (H.M.).
REFERENCES
- 1.Baldo, A. M., and M. A. McClure. 1999. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J. Virol. 73:7710-7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, P. A., L. A. Kelley, R. M. MacCallum, and M. J. Sternberg. 2001. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 2001(Suppl. 5):39-46. [DOI] [PubMed] [Google Scholar]
- 3.Bergman, A. C., P. O. Nyman, and G. Larsson. 1998. Kinetic properties and stereospecificity of the monomeric dUTPase from herpes simplex virus type 1. FEBS Lett. 441:327-330. [DOI] [PubMed] [Google Scholar]
- 4.Bjornberg, O., A. C. Bergman, A. M. Rosengren, R. Persson, I. R. Lehman, and P. O. Nyman. 1993. dUTPase from herpes simplex virus type 1: purification from infected green monkey kidney (Vero) cells and from an overproducing Escherichia coli strain. Protein Expr. Purif. 4:149-159. [DOI] [PubMed] [Google Scholar]
- 5.Broyles, S. S. 1993. Vaccinia virus encodes a functional dUTPase. Virology 195:863-865. [DOI] [PubMed] [Google Scholar]
- 6.Cedergren-Zeppezauer, E. S., G. Larsson, P. O. Nyman, Z. Dauter, and K. S. Wilson. 1992. Crystal structure of a dUTPase. Nature 355:740-743. [DOI] [PubMed] [Google Scholar]
- 7.Chen, R., H. Wang, and L. M. Mansky. 2002. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 83:2339-2345. [DOI] [PubMed] [Google Scholar]
- 8.Climie, S., T. Lutz, J. Radul, M. Sumner-Smith, E. Vandenberg, and E. McIntosh. 1994. Expression of trimeric human dUTP pyrophosphatase in Escherichia coli and purification of the enzyme. Protein Expr. Purif. 5:252-258. [DOI] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project, N. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.Dauter, Z., R. Persson, A. M. Rosengren, P. O. Nyman, K. S. Wilson, and E. S. Cedergren-Zeppezauer. 1999. Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex. J. Mol. Biol. 285:655-673. [DOI] [PubMed] [Google Scholar]
- 11.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, Calif.
- 12.Delaroque, N., D. G. Muller, G. Bothe, T. Pohl, R. Knippers, and W. Boland. 2001. The complete DNA sequence of the Ectocarpus siliculosus virus EsV-1 genome. Virology 287:112-132. [DOI] [PubMed] [Google Scholar]
- 13.el-Hajj, H. H., H. Zhang, and B. Weiss. 1988. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 170:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadsden, M. H., E. M. McIntosh, J. C. Game, P. J. Wilson, and R. H. Haynes. 1993. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 12:4425-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graves, M. V., C. T. Bernadt, R. Cerny, and J. L. Van Etten. 2001. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 285:332-345. [DOI] [PubMed] [Google Scholar]
- 16.Graziani, S., Y. Xia, J. R. Gurnon, J. L. Van Etten, D. Leduc, S. Skouloubris, H. Myllykallio, and U. Liebl. 2004. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria chlorella virus-1. J. Biol. Chem. 279:54340-54347. [DOI] [PubMed] [Google Scholar]
- 17.Halperin, I., B. Ma, H. Wolfson, and R. Nussinov. 2002. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins 47:409-443. [DOI] [PubMed] [Google Scholar]
- 18.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 19.Harris, J. M., E. M. McIntosh, and G. E. Muscat. 1999. Structure/function analysis of a dUTPase: catalytic mechanism of a potential chemotherapeutic target. J. Mol. Biol. 288:275-287. [DOI] [PubMed] [Google Scholar]
- 20.Hokari, S., I. Koyama, K. Shioda, and Y. Sakagishi. 1994. Deoxyuridine triphosphatase in human hepatoma. Int. J. Biochem. 26:487-490. [DOI] [PubMed] [Google Scholar]
- 21.Huffman, J. L., H. Li, R. H. White, and J. A. Tainer. 2003. Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii. J. Mol. Biol. 331:885-896. [DOI] [PubMed] [Google Scholar]
- 22.Jons, A., V. Gerdts, E. Lange, V. Kaden, and T. C. Mettenleiter. 1997. Attenuation of dUTPase-deficient pseudorabies virus for the natural host. Vet. Microbiol. 56:47-54. [DOI] [PubMed] [Google Scholar]
- 23.Jons, A., and T. C. Mettenleiter. 1996. Identification and characterization of pseudorabies virus dUTPase. J. Virol. 70:1242-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klarmann, G. J., X. Chen, T. W. North, and B. D. Preston. 2003. Incorporation of uracil into minus strand DNA affects the specificity of plus strand synthesis initiation during lentiviral reverse transcription. J. Biol. Chem. 278:7902-7909. [DOI] [PubMed] [Google Scholar]
- 25.Kornberg, A., and T. Baker. 1991. DNA replication, 2nd ed. W. H. Freeman Co., New York, N.Y.
- 26.Larsson, G., L. A. Svensson, and P. O. Nyman. 1996. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUDP). Nat. Struct. Biol. 3:532-538. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., Z. Lu, L. Sun, S. Ropp, G. F. Kutish, D. L. Rock, and J. L. Van Etten. 1997. Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology 237:360-377. [DOI] [PubMed] [Google Scholar]
- 28.Mol, C. D., J. M. Harris, E. M. McIntosh, and J. A. Tainer. 1996. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure 4:1077-1092. [DOI] [PubMed] [Google Scholar]
- 29.Morehead, T. A., J. R. Gurnon, B. Adams, K. W. Nickerson, L. A. Fitzgerald, and J. L. Van Etten. 2002. Ornithine decarboxylase encoded by chlorella virus PBCV-1. Virology 301:165-175. [DOI] [PubMed] [Google Scholar]
- 30.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. O. Belew, and A. J. lson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 31.Myllykallio, H., G. Lipowski, D. Leduc, J. Filee, P. Forterre, and U. Liebl. 2002. An alternative flavin-dependent mechanism for thymidylate synthesis. Science 297:105-107. [DOI] [PubMed] [Google Scholar]
- 32.Oliveros, M., R. Garcia-Escudero, A. Alejo, E. Vinuela, M. L. Salas, and J. Salas. 1999. African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. J. Virol. 73:8934-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plugge, B., S. Gazzarrini, M. Nelson, R. Cerana, J. L. Van Etten, C. Derst, D. DiFrancesco, A. Moroni, and G. Thiel. 2000. A potassium channel protein encoded by chlorella virus PBCV-1. Science 287:1641-1644. [DOI] [PubMed] [Google Scholar]
- 34.Prasad, G. S., E. A. Stura, D. E. McRee, G. S. Laco, C. Hasselkus-Light, J. H. Elder, and C. D. Stout. 1996. Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus. Protein Sci. 5:2429-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyles, R. B., N. M. Sawtell, and R. L. Thompson. 1992. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J. Virol. 66:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234:186-195. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turelli, P., F. Guiguen, J. F. Mornex, R. Vigne, and G. Querat. 1997. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J. Virol. 71:4522-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turelli, P., G. Petursson, F. Guiguen, J. F. Mornex, R. Vigne, and G. Querat. 1996. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J. Virol. 70:1213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Etten, J. L. 2003. Unusual lifestyle of giant chlorella viruses. Annu. Rev. Genet. 37:153-195. [DOI] [PubMed] [Google Scholar]
- 42.Van Etten, J. L., D. E. Burbank, J. Joshi, and R. H. Meints. 1984. DNA synthesis in a chlorella-like alga following infection with the virus PBCV-1. Virology 134:443-449. [DOI] [PubMed] [Google Scholar]
- 43.Van Etten, J. L., D. E. Burbank, Y. Xia, and R. H. Meints. 1983. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126:117-125. [DOI] [PubMed] [Google Scholar]
- 44.Van Etten, J. L., R. H. Meints, D. E. Burbank, D. Kuczmarski, D. A. Cuppels, and L. C. Lane. 1981. Isolation and characterization of a virus from the intracellular green algae symbiotic with Hydra viridis. Virology 113:704-711. [DOI] [PubMed] [Google Scholar]
- 45.Van Etten, J. L., A. M. Schuster, L. Girton, D. E. Burbank, D. Swinton, and S. Hattman. 1985. DNA methylation of viruses infecting a eukaryotic chlorella-like green alga. Nucleic Acids Res. 13:3471-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villarreal, L. P. 1999. DNA viruses: their influence on host evolution, p. 391-420. In E. Domingo, R. Webster, J. J. Holland, and T. Pickett (ed.), Origin and evolution of viruses. Academic Press, Inc., New York, N.Y.
- 47.Villarreal, L. P., and V. R. DeFilippis. 2000. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Virol. 74:7079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagaman, P. C., C. S. Hasselkus-Light, M. Henson, D. L. Lerner, T. R. Phillips, and J. H. Elder. 1993. Molecular cloning and characterization of deoxyuridine triphosphatase from feline immunodeficiency virus (FIV). Virology 196:451-457. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., I. Calin-Jageman, J. R. Gurnon, T. J. Choi, B. Adams, A. W. Nicholson, and J. L. Van Etten. 2003. Characterization of a chlorella virus PBCV-1 encoded ribonuclease III. Virology 317:73-83. [DOI] [PubMed] [Google Scholar]