Abstract
Chinese hamster ovary (CHO) cells are traditionally regarded as nonpermissive cells for herpes simplex virus type 1 (HSV-1) infection as they lack the specific entry receptors, and modified CHO cells have been instrumental in the identification of HSV-1 receptors in numerous studies. In this report we demonstrate that the HSV-1 strain 17+ variant HSV1716 is able to infect unmodified CHO cells but only if the virus is propagated in baby hamster kidney (BHK) cells. Infection of CHO cells by BHK-propagated HSV1716 results in expression of immediate-early, early, and late viral genes, and infectious progeny virions are produced. In normally cultured CHO cells, up to a maximum of 50% of cells were permissive for BHK-propagated HSV1716 infection, with 24 h of serum starvation increasing this to 100% of CHO cells, suggesting that the mechanism used by BHK-propagated virus to infect CHO cells was cell cycle dependent. The altered tropism of HSV1716 was also evident in another nonpermissive mouse melanoma cell line and is an exclusive property resulting from propagation of the virus using BHK cells, as viruses propagated on Vero, C8161 (a human melanoma cell line), or indeed, CHO cells were completely unable to infect either CHO or mouse melanoma cells.
Herpes simplex virus type 1 (HSV-1), an alphaherpesvirus, is a ubiquitous eukaryotic pathogen, and following infection of permissive cells, the lytic cycle of viral gene expression can be divided into three temporal stages (reviewed in reference 5). The virion tegument protein VP16 (Vmw65) initiates transcription of viral immediate-early (IE) genes whose protein products, Vmw175 (ICP4), Vmw63 (ICP27), Vmw110 (ICP0), and Vmw68, are principally involved in the orchestration of early and late gene expression. ICP27 and ICP4 are essential proteins, and deletion of their respective genes prevents viral replication at early stages of infection in tissue culture. ICP4 transactivates viral gene expression through DNA binding, and ICP27 is a multifunctional protein involved in the export, 3′ processing, and poly(A) usage of viral RNAs. Early gene products are detectable by 4 to 5 h postinfection and are mostly enzymes involved in DNA synthesis and replication. Late genes are efficiently expressed after 6 to 7 h postinfection and mostly encode structural capsid proteins and membrane glycoproteins.
Initiation of infection requires cells to display the appropriate receptors to permit HSV-1 entry, a process requiring the complex interplay of a number of cellular and viral membrane components (reviewed in reference 19). Four virus membrane glycoproteins, gB, gD, and the heterodimer comprising gH and gL (gH/gL), have been shown to be necessary and sufficient for HSV-1 entry into cells. Initial contact with the cell is between gB and cellular heparan sulfate; gD then interacts specifically with the cellular receptors for HSV-1 entry, which include herpesvirus entry mediator (HVEM), nectins 1 and 2, and 3-O-sulfated heparan sulfate. Membrane fusion then requires the concerted activities of gB and gH/gL. An additional receptor, nectin 3, has been reported to mediate the entry of the HSV-1 mutant JMP into nonpermissive J cells (3).
Identification of cellular receptors for HSV-1 has relied upon the ability of nonpermissive cell lines, such as Chinese hamster ovary (CHO), to be rendered permissive by transfection of these cells with plasmids that express putative receptor proteins. Model systems have used HSV-1 to infect nonpermissive CHO cells transfected with plasmids that express either HVEM (12), a member of the tumor necrosis factor receptor family, nectin 1 (7, 11), or nectin 2 (10, 21), members of the immunoglobulin superfamily. Infection is monitored with reporter genes, such as β-galactosidase, either encoded by the virus or by stably transformed cell lines in which expression is driven by an HSV-1 IE promoter. HSV-1 entry by endocytosis into CHO cells has been reported, but infection requires that the cells express HVEM, nectin 1, or nectin 2 (13, 14). Thus, although HSV-1 is endocytosed by CHO cells, evidence of infection, such as viral gene expression, is detected only in those cells modified to display an HSV-1 entry mediator, such as nectin 1. In normal CHO cells, the endocytosed HSV-1 is destroyed and this process is prevented by the cellular presence of the HSV-1 receptor and the virion glycoproteins gB, gD, and gH/gL (14). Interestingly, CHO cells are more permissive for HSV-2 infection, and potentially, this is due to differences in the HSV-2 gD sequence that allow it, but not HSV-1 gD, to interact with the Chinese hamster homologue of nectin 2 (23).
Other experimental systems used to elucidate the requirements for HSV-1 entry mediating structural glycoproteins and cellular receptors have again capitalized on nonpermissive CHO cells cotransfected with plasmids expressing cellular receptors and the viral glycoproteins gB, gD, and gH/gL. Coexpression of a receptor plus the four viral glycoproteins essential for entry results in cell fusion and syncytium formation (2, 9, 15, 20).
A principal underpinning all of these studies is the ability to manipulate normally nonpermissive cells such that they allow HSV-1 entry and gene expression, and CHO cells are central to many of these experiments. However, we have previously noted that HSV-1 strain 17+ is able to infect CHO cells, and we have detected expression of viral IE genes (ICP27, ICP0, and large subunit of viral ribonucleotide reductase [R1]), early genes (small subunit of viral ribonucleotide reductase and 65-k DNA binding protein), and late genes (gB, gC, VP16, and ICP34.5) (J. Conner, unpublished data). Further, we have previously reported activation of the stress-activated protein kinases p38 and JNK in CHO cells infected with HSV-1 strain 17+, an activation that requires entry of infectious virus into cells (22). This apparent ability of CHO cells to support HSV-1 strain 17+ entry and infection contrasts with the observations of others (7, 12) who report poor infection of CHO cells by HSV-1 strain 17+ unless the cells expressed an HSV-1 receptor. Therefore, we investigated these apparently contradictory results using a variant of HSV-1 strain 17+, HSV1716, that expresses green fluorescent protein (HSV1716gfp).
MATERIALS AND METHODS
Cells and viruses.
BHK cells were grown in Glasgow modified Eagle medium (Invitrogen, Paisley, Scotland) with 10% tryptose phosphate broth and 10% newborn calf serum (NCS; Invitrogen). CHO-K1, Vero, HeLa, and C8161 (a human melanoma cell line) cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% NCS. B16-F10 mouse melanoma cells were also cultured in Dulbecco's modified Eagle's medium supplemented with 10% NCS and 4.2 mg/liter glucose. For this study, fresh aliquots of CHO-K1 and mouse melanoma cells were obtained from the American Type Culture Collection/LGCpromochem, London, United Kingdom. SK-N-SH cells were cultured in Dulbecco's modified Eagle's medium supplemented with nonessential amino acids and 10% NCS.
The principal virus used in this study was HSV1716gfp, an ICP34.5-null mutant of HSV-1 strain 17+ with an expression cassette comprising the enhanced green fluorescent protein gene driven by the human cytomegalovirus IE promoter inserted in the UL43 gene. In some instances, HSV17+gfp, an HSV-1 strain 17+ variant that expresses an ICP34.5/enhanced green fluorescent protein fusion protein, was used. The construction of both of these viruses was described by Harland et al. (8). For production of virus stocks, confluent monolayers of BHK, Vero, and C8161 cells in T175 flasks were infected with 1 × 105 PFU HSV1716gfp, and after 4 days of infection, total virus was harvested and titrated on BHK cells.
Fluorescence microscopy.
Twenty-four hours prior to infection, the various cell lines, in appropriate media, were plated out on 13-mm glass coverslips in 24-well plates and incubated at 37°C in 5% CO2. Cells were between 60 to 90% confluent prior to infection. Cells, between 0.6 × 106 and 1 × 106/well, were infected with 1 × 106, 5 × 106, or 1 × 107 PFU of virus, incubated at 37°C in 5% CO2 for 8 h, washed once in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde in PBS for 30 min, and then washed once again in PBS. Glass coverslips were then removed from the wells and mounted on microscope slides with a drop of Vectashield (Vector Laboratories Ltd., Peterborough, United Kingdom), and green fluorescent protein was observed by fluorescence microscopy. Numbers of green fluorescent cells were counted per field of view at ×200 magnification, with three fields of view counted per microscope slide. The number of cells for each cell type per field of view was determined using visible light.
Western blotting.
Prior to infection, the various cell lines, in appropriate media, were plated out in 35-mm plates and incubated for 24 h at 37°C in 5% CO2. Confluent monolayers of cells, approximately 2 × 106/well, were then infected with 1 × 107 PFU of virus and incubated for 8 h at 37°C in 5% CO2. Cells were then washed once with 1 ml PBS, and whole-cell extracts were harvested by the direct addition of 0.2 ml sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transfer to nitrocellulose membranes, blots were probed with monoclonal antibodies to gC and gB (both from Abcam, Cambridge, United Kingdom), ICP27, ICP4, and ICP0 (all from Europa Bioproducts Ltd, Cambridge, United Kingdom), and VP16 (a kind gift from Tony Minson, University of Cambridge, United Kingdom) and a polyclonal antibody to the R1 subunit of the viral ribonucleotide reductase (4).
Transmission electron microscopy.
CHO cells were plated out in six-well dishes, and after 24 h in culture, confluent monolayers of cells were infected with 1 × 107 PFU HSV1716gfp. The infection was allowed to proceed for 24 h at 37°C in 5% CO2, and the cells were then harvested for electron microscopy. After removal of the growth medium, cells were washed once with PBS, scraped from the monolayer in 1 ml of PBS, and pelleted by centrifugation in BEEM tubes at 1,000 rpm for 4 min in a microcentrifuge. Cells were fixed for at least 1 h by careful removal of the PBS and replacement with 0.5 ml cold 2.5% glutaraldehyde in PBS. The pellets were rinsed with PBS and incubated at room temperature for 1 h in 1% osmium tetroxide. The pellet was again rinsed in PBS and then dehydrated through a graded alcohol series before being infiltrated with Epon 812 resin (TAAB) and placed in an embedding oven at 65°C for 2 days to harden. Embedded samples were sectioned using a Reichart Jung Ultracut E microtome and stained with uranyl acetate and lead citrate before being examined using a JEOL 100S electron microscope.
RESULTS
HSV1716gfp propagated on BHK but not Vero cells can infect CHO and mouse melanoma cells.
HSV1716gfp grown in BHK cells (HSV1716gfp/BHK) was used to infect a variety of different permissive (BHK, HeLa, SK-N-SH, and C8161) and nonpermissive (CHO-K1 and B16-F10 mouse melanoma) cell lines. Twenty-four hours prior to infection, the various cell lines, in appropriate media, were plated out on 13-mm glass coverslips in 24-well plates and incubated at 37°C in 5% CO2. Cells were infected with 1 × 106, 5 × 106, or 1 × 107 PFU of virus, incubated at 37°C in 5% CO2 for 8 h, washed, fixed, and washed again. Glass coverslips were then mounted, and green fluorescent protein was observed by fluorescence microscopy. At 1 × 106 PFU (data not shown), 5 × 106 PFU (Fig. 1a to d), and 1 × 107 PFU (data not shown), 90 to 100% of the permissive BHK (Fig. 1a), C8161 (Fig. 1b), HeLa (Fig. 1c), and SK-N-SH (Fig. 1d) cells were infected. Surprisingly, cells displaying green fluorescent protein expression were observed with the nonpermissive CHO and mouse melanoma cell lines, indicating that HSV1716gfp/BHK was also able to infect these cell lines (Fig. 1e to h). At 1 × 106 PFU, approximately 5 to 10% of CHO cells were infected (Fig. 1e), at 5 × 106 PFU, approximately 15 to 20% of CHO cells were infected (Fig. 1f), and at 1 × 107 PFU, approximately 30 to 40% of CHO cells were infected (Fig. 1g). Numbers of green fluorescent cells were lower for mouse melanoma cells with approximately 1%, 3 to 5%, or 5 to 10% of cells infected at 1 × 106 (data not shown), 5 × 106 (data not shown), or 1 × 107 PFU (Fig. 1h), respectively.
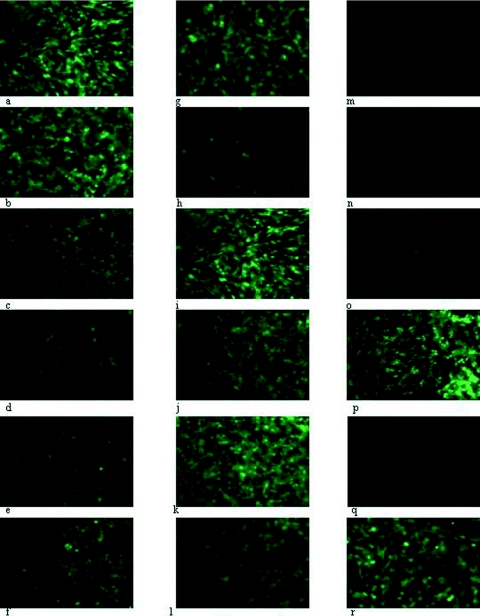
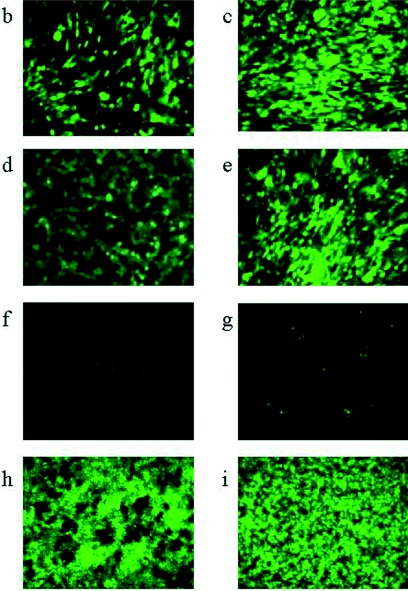
FIG. 1.
Unprocessed images (magnification, ×184) from fluorescent microscopy of various cell lines after 8 h of infection with HSV1716gfp propagated either in BHK (a to h, o to r) or Vero (i to n) cells. Permissive BHK (a, i), C8161 (b, j), HeLa (c, k), or SK-N-SH (d, l) cells were infected with 5 × 106 PFU HSV1716gfp. Nonpermissive CHO cells were infected with 1 × 106 PFU, 5 × 106 PFU, or 1 × 107 PFU HSV1716gfp/BHK (e, f, g, respectively) or with 1 × 107 PFU HSV1716gfp/Vero (m). Nonpermissive mouse melanoma cells were infected with 1 × 107 PFU HSV1716gfp/BHK (h) or HSV1716gfp/Vero (n). Infection of BHK (o, p) or CHO (q, r) cells was completely neutralized using a sheep anti-HSV-1 antiserum (o and q, respectively) but was unaffected by normal sheep serum (p and r, respectively).
The above experiment was repeated using HSV1716gfp that had been propagated in Vero cells (HSV1716gfp/Vero). At 1 × 106 PFU (data not shown), 5 × 106 PFU (Fig. 1i to l), and 1 × 107 PFU (data not shown), 90 to 100% of the permissive BHK (Fig. 1i), C8161 (Fig. 1j), HeLa (Fig. 1k), and SK-N-SH (Fig. 1l) cell lines were infected. Significantly, neither the nonpermissive CHO nor the mouse melanoma cells displayed any evidence of HSV1716gfp/Vero infection (Fig. 1m and n). Unlike HSV1716gfp/BHK-infected CHO and mouse melanoma cells at 1 × 107 PFU (Fig. 1g and 1h, respectively), no green fluorescent CHO (Fig. 1m) or mouse melanoma (Fig. 1n) cells were visible using 1 × 107 PFU HSV1716gfp/Vero. Even at 2 × 107 PFU, HSV1716gfp/Vero was unable to infect either CHO or mouse melanoma cells (data not shown), suggesting that growth of HSV1716gfp on BHK cells confers on the virus the ability to infect normally nonpermissive CHO and mouse melanoma cells.
Sheep anti-HSV-1 antiserum was generated by four consecutive monthly injections of 1 × 106 PFU HSV1716. Preincubation for 18 h at 4°C of 1 × 107 PFU HSV1716gfp/BHK with a 100-fold dilution of antiserum obtained after the fourth injection completely neutralized viral infectivity, as assessed by the total lack of green fluorescent protein expression 8 h after the addition of the neutralized virus to BHK cells (Fig. 1o, 0% cells infected with antiserum-incubated virus), whereas incubation of 1 × 107 PFU HSV1716gfp/BHK with normal sheep serum at a similar dilution had no effect on viral infectivity (Fig. 1p, 100% of cells infected by serum-incubated virus). Similarly, all HSV1716gfp/BHK infectivity of CHO cells was neutralized by the sheep anti-HSV-1 antiserum at 1:100 (Fig. 1q), but infectivity was unaffected by normal sheep serum at 1:100 (Fig. 1r, ca. 50% of cells infected with serum-incubated virus). These results suggest that HSV1716gfp/BHK infection of CHO cells, like HSV1716gfp/BHK infection of BHK cells, is probably receptor mediated.
Using three T175 flasks per cell line, confluent monolayers of BHK, Vero, and C8161 cells were infected with 1 × 105 PFU HSV1716gfp and total virus was harvested and titrated on BHK cells. The yields of virus from each flask for each cell line were almost identical (Table 1). Six different cell lines (BHK, HeLa, SK-N-SH, C8161, CHO, and mouse melanoma) were infected with 5 × 106 PFU of each of the preparations of HSV1716gfp/BHK, HSV1716gfp/Vero, or HSV1716gfp/C8161,and after 8 h of infection, green fluorescent protein expression was assessed by fluorescence microscopy. For each virus preparation, the number of green fluorescent cells per field of view was recorded as a percentage of the total number of cells for three separate fields of view at ×400 magnification, and the mean values are shown in Table 1. All three preparations of each virus infected 100% of the BHK, HeLa, SK-N-SH, and C8161 cells, but only the viruses produced in BHK cells were able to infect CHO or mouse melanoma cells (Table 1). Between 12 and 15% of CHO cells and between 4 and 6% of mouse melanoma cells were infected by HSV1716gfp/BHK, whereas no infected CHO or mouse melanoma cells were visible with either HSV1716gfp/Vero or HSV1716gfp/C8161. The efficiency with which each virus preparation was able to infect BHK, HeLa, SK-N-SH, and C8161 cells was assessed by adding the amount of virus required to infect approximately 50% of cells and then counting the number of green fluorescent cells after 8 h of infection. Each of the HSV1716gfp/BHK, HSV1716gfp/Vero, and HSV1716gfp/C8161 preparations infected BHK, HeLa, SK-N-SH, and C8161 cells with approximately equal efficiencies (data not shown).
TABLE 1.
Percent infection of BHK, C8161, HeLa, SK-N-SH, CHO, or mouse melanoma cells with 5 PFU/cell HSV1716gfp/BHK, HSV1716gfp/Vero, or HSV1716gfp/C8161
| Virus | Titre (PFU/ml) | % Infection of cell typea:
|
|||||
|---|---|---|---|---|---|---|---|
| BHKc | C8161 | HeLa | SK-N-SH | CHO | MMb | ||
| BHK1c | 1.0 × 109 | 100 | 100 | 100 | 100 | 12 | 4 |
| BHK2 | 1.0 × 109 | 100 | 100 | 100 | 100 | 15 | 4 |
| BHK3 | 1.2 × 109 | 100 | 100 | 100 | 100 | 12 | 6 |
| Vero1 | 0.9 × 109 | 100 | 100 | 100 | 100 | 0 | 0 |
| Vero2 | 1.0 × 109 | 100 | 100 | 100 | 100 | 0 | 0 |
| Vero3 | 1.1 × 109 | 100 | 100 | 100 | 100 | 0 | 0 |
| C8161/1 | 0.9 × 109 | 100 | 100 | 100 | 100 | 0 | 0 |
| C8161/2 | 0.9 × 109 | 100 | 100 | 100 | 100 | 0 | 0 |
| C8161/3 | 1.0 × 109 | 100 | 100 | 100 | 100 | 0 | 0 |
Mean percentages of green fluorescent cells from three fields of view at a magnification of ×400.
Mouse melanoma cells.
HSV1716gfp/BHK.
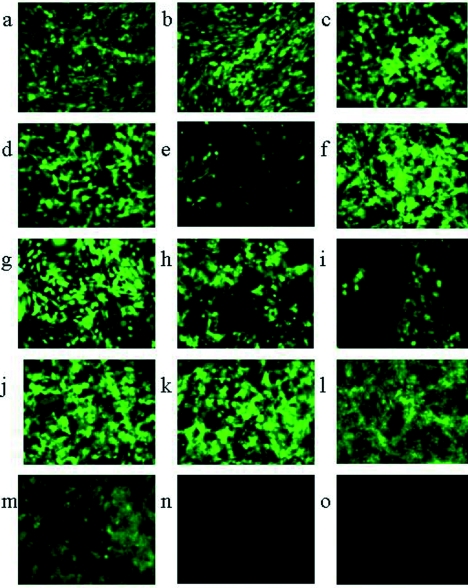
HSV1716gfp/BHK was used to infect Vero cells in a T175 flask, and 1 × 105 PFU of harvested virus was used to infect a T175 flask of BHK cells. After harvest, 1 × 105 PFU of virus was used to infect a T175 flask of Vero cells followed by a subsequent propagation of 1 × 105 PFU of harvested virus in BHK cells. These alternate passages in BHK and Vero cells were repeated starting with HSV1716gfp/Vero, and 1 × 107 PFU of each of the resulting virus preparations was used to infect CHO cells. Fluorescence microscopy indicated that only when the virus was propagated in BHK cells was it able to infect CHO cells (Fig. 2a to h). Strikingly, a single passage in BHK cells was sufficient to confer on HSV1716gfp the ability to infect CHO cells with approximately 30 to 40% of cells displaying green fluorescent protein (GFP) expression (Fig. 2b, d, e, and g), whereas this CHO infectivity was lost completely following a single passage in Vero cells (Fig. 2a, c, f, and h, 0% cells infected). Similarly, HSV17+gfp propagated on BHK cells was able to infect CHO cells (Fig. 2i, ca. 20% of CHO cells infected), whereas CHO infectivity was lost by a single passage of HSV17+gfp in Vero cells (Fig. 2j, 0% CHO cells infected). HSV17+gfp expresses an ICP34.5-GFP fusion protein (8) and was used to confirm that the lack of ICP34.5 in HSV1716 did not affect CHO infectivity.
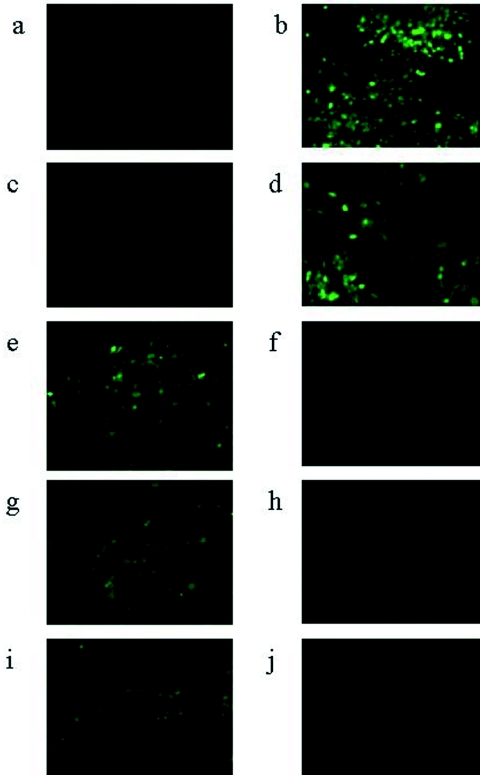
FIG. 2.
Unprocessed images (magnification, ×184) from fluorescent microscopy of CHO cells after 8 h of infection with either 5 PFU/cell HSV1716gfp (a to h) or HSV17+gfp (i, j) propagated alternatively in either BHK (b, d, e, g, i) or Vero (a, c, f, h, j) cells.
Infection of CHO cells by HSV1716gfp is productive.
Western blotting confirmed that HSV1716gfp/BHK was able to infect CHO and mouse melanoma cells and that both of these cell lines were resistant to HSV1716gfp/Vero infection (Fig. 3a). Four 35-mm dishes each of Vero, CHO, and mouse melanoma cells were infected with two separate preparations of either HSV1716gfp/BHK or HSV1716gfp/Vero at 5 PFU/cell. After 8 h of infection, whole-cell extracts were harvested directly and samples were Western blotted and probed with antiserum 106 directed against the R1 subunit of the viral ribonucleotide reductase (Fig. 3a). R1 was detected in Vero cells infected with both preparations of either HSV1716gfp/Vero and HSV1716gfp/BHK (Fig. 3a, lanes 1 and 2 and 7 and 8, respectively). Although weaker in intensity, R1 was detected in extracts of CHO and mouse melanoma cells infected with both preparations of HSV1716gfp/BHK (Fig. 3a, lanes 9 and 10 and 11 and 12, respectively), but R1 was not detected in CHO or mouse melanoma extracts prepared from cells infected with both preparations of HSV1716gfp/Vero (Fig. 3a, lanes 3 and 4 and 5 and 6, respectively).
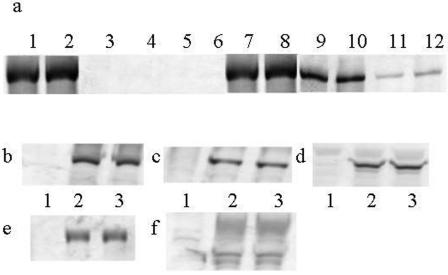
FIG. 3.
(a) Western blots developed with anti HSV-1 R1 antiserum 106. In duplicate, whole-cell extracts were prepared from Vero (lanes 1, 2, 7, 8), CHO (lanes 3, 4, 9, 10) or mouse melanoma (lanes 5, 6, 11, 12) cells infected with 5 PFU/cell HSV1716gfp/Vero (lanes 1 to 6) or HSV1716gfp/BHK (lanes 7 to 12). For panels b to f, Western blots developed with monoclonal antibodies against ICP4 (b), ICP0 (c), VP16 (d), gB (e), and gC (f) are shown. Whole-cell extracts were prepared from CHO (lanes 1 and 2) or BHK (lane 3) cells infected with 10 or 5 PFU/cell HSV1716gfp/Vero (lane 1 and 3, respectively) or 10 PFU/cell HSV1716gfp/BHK (lanes 2).
In a separate experiment, CHO cells were infected with either HSV1716gfp/BHK or HSV1716gfp/Vero at 10 PFU/cell and BHK cells were infected with HSV1716gfp/Vero at 5 PFU/cell. After 8 h, whole-cell extracts were prepared and the expression of ICP4 (Fig. 3b), ICP0 (Fig. 3c), VP16 (Fig. 3d), gB (Fig. 3e), and gC (Fig. 3f) in the two cell types was assessed by Western blotting. To compensate for the reduced level of infection in CHO cells, 20 μl of whole-cell extract was loaded compared to 5 μl for the BHK whole-cell extract. There was little difference in the pattern of expression of ICP4 (Fig. 3b), ICP0 (Fig. 3c), VP16 (Fig. 3d), gB (Fig. 3e), and gC (Fig. 3f) in CHO cells infected with 10 PFU/cell HSV1716gfp/BHK (Fig. 3b to f, lanes 2) compared to BHK cells infected with 5 PFU/cell HSV1716gfp/Vero (Fig. 3b to f, lanes 3). In contrast, none of the viral proteins was detected by Western blotting in CHO cells infected with 10 PFU/cell HSV1716gfp/Vero (Fig. 3b to f, lanes 1). Similar results were obtained using antibodies to R1 and ICP27 (data not shown). When the experiment was repeated using mouse melanoma cells, expression of ICP0, ICP4, ICP27, R1, VP16, gB, and gC was detected in cells infected with HSV1716gfp/BHK but not in cells infected with HSV1716gfp/Vero. However, since only ca. 5% of cells were infected, band intensities were much lower than in similarly infected CHO cells (data not shown).
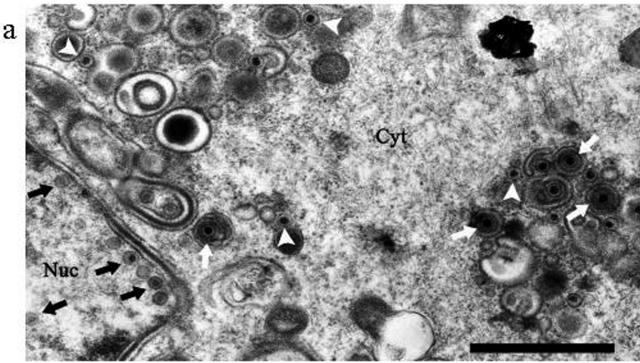
Initially, we were unable to detect infectious virions from the supernatant of CHO cells infected with HSV1716gfp/BHK. Thus, 2.5 × 106 CHO cells in a 60-mm dish were infected with 1 × 105 PFU HSV1716gfp/BHK. After 72 h, the supernatant was removed and 2 ml was used to infect BHK cells on glass coverslips in 24-well plates, but no GFP-positive cells were observed. In contrast, when either Vero or C8161 cells were infected similarly and their supernatants tested, 100% of BHK cells were GFP positive following addition of 0.1 ml supernatant from the infected cells (data not shown). To determine the outcome of infection in CHO cells, cells were infected with 5 PFU/cell HSV1716gfp/BHK for 24 h and analyzed by electron microscopy (Fig. 4a), which clearly demonstrated that infection had proceeded normally, with filled nucleocapsids and enveloped virions clearly visible in the nucleus and cytoplasm, respectively. No evidence of any defects in maturation or egress was observed in any of our electron micrographs, and since apparently normal virions were seen in the electron microscopy samples, further experiments were carried out to look for the production of infectious virus. CHO cells (2.5 × 106) in a 60-mm dish were infected with 5 × 105 PFU HSV1716gfp/BHK. After 72 h, cells were scraped from the dish into the supernatant and sonicated for 2 min in a sonicator bath, and 1 ml was used to infect BHK, C8161, or Vero cells on glass coverslips in 24-well plates. Approximately 50% of the BHK (Fig. 4b), C8161 (Fig. 4d), or Vero (not shown) cells were GFP positive, indicating the presence of infectious virions produced by the CHO cells. From a 0.1-ml preparation of similarly infected and harvested Vero or C8161 cells, 100% of BHK cells were GFP positive (Fig. 4c and e, respectively). The titers of the viruses obtained from duplicate preparations of CHO, C8161, and Vero cells infected with 5 × 105 PFU HSV1716gfp/BHK were 2 × 106 and 3 × 106 PFU (CHO), 8 × 106 and 1 × 107 PFU (C8161), and 4 × 107 and 5 × 107 PFU (Vero), indicating 4- to 6-fold, 16- to 20-fold, and 80- to 100-fold yields of virus, respectively. Thus, both GFP expression studies and titration results clearly demonstrate that viable progeny are produced following infection of CHO cells by HSV1716gfp/BHK. Interestingly, the virus obtained from CHO cells was unable to infect CHO cells, as no GFP-positive cells were observed after the addition of 5 PFU/cell HSV1716gfp/CHO (Fig. 4f). This inability of HSV1716gfp/CHO to infect CHO cells was confirmed by adding 1 × 103 PFU HSV1716gfp/BHK to CHO cells in a 60-mm dish. After 5 days in culture, no plaques were visible and only individual, GFP-positive cells were observed (Fig. 4g). Plaques were clearly visible after 48 h on Vero or BHK cells infected with 1 × 103 PFU HSV1716gfp/BHK or HSV1716gfp/CHO, all cells were infected, and the monolayer destroyed after 5 days in culture. Figure 4h and i, respectively, shows BHK and Vero cells 5 days after infection with 1 × 103 PFU HSV1716gfp/CHO.
FIG. 4.
(a) Electron micrograph of CHO cells 24 h after infection with 5 PFU/cell HSV1716gfp/BHK. Examples of capsids in the nucleus (Nuc) are indicated by a black arrow, enveloped virions in the cytoplasm (Cyt) are indicated by a white arrow, and unenveloped capsids in the cytoplasm are indicated by a white arrowhead. Bar, 1 μm. For panels b to f, unprocessed images (magnification, ×200) from fluorescent microscopy are shown. (b) BHK cells after 8 h of infection with 5 PFU/cell HSV1716gfp propagated in CHO cells; (c) BHK cells after 8 h of infection with 5 PFU/cell HSV1716gfp propagated in Vero cells; (d) C8161 cells after 8 h of infection with 5 PFU/cell HSV1716gfp propagated in CHO cells; (e) BHK cells after 8 h of infection with 5 PFU/cell HSV1716gfp propagated in C8161 cells; (f) CHO cells after 8 h of infection with 5 PFU/cell HSV1716gfp propagated in CHO cells. For panels g, h, and i, unprocessed images (magnification, ×100) from fluorescent microscopy are shown for CHO (g), BHK (h), and Vero (i) cell lines 5 days after infection with 1 × 103 PFU HSV1716gfp/BHK (g, h) or HSV1716gfp/CHO (i).
Infection of CHO cells by HSV1716gfp/BHK is cell cycle dependent.
CHO, BHK, Vero, and C8161 cells were plated out on coverslips in 24-well dishes, and after 8 h, the medium was replaced with serum-free medium for 24 h. After this time, serum was reintroduced to the medium and the cells were infected with HSV1716gfp/BHK or HSV1716gfp/Vero either immediately or after 4, 8, or 16 h of incubation in serum-containing medium (Fig. 5). Serum deprivation of Vero (not shown), BHK (Fig. 5a), or C8161 (Fig. 5c) cells for 24 h had little effect on the ability of 5 PFU/cell of either HSV1716gfp/BHK (Fig. 5a and b) or HSV1716gfp/Vero (Fig. 5c and d) to infect these cell types compared to non-serum-starved Vero (not shown), BHK (Fig. 5b), or C8161 (Fig. 5d) cells, with 100% of cells infected in each case. Also, addition of virus after 4, 8, or 16 h following serum reintroduction did not affect the susceptibility of the cells to infection (data not shown). In marked contrast to this, serum starvation had a profound effect on the ability of HSV1716gfp/BHK to infect CHO cells. At 5 PFU/cell of HSV1716gfp/BHK, approximately 20% of normally cultured CHO cells were infected (Fig. 5e). After 24 h of serum starvation and immediate infection with 5 PFU/cell HSV1716gfp/BHK upon reintroduction of serum, 100% of CHO cells were infected, as shown in Fig. 5f. The addition of 5 PFU/cell of HSV1716gfp/BHK at 4, 8, and 16 h after serum reintroduction resulted in the infection of 90% (Fig. 5g), 60% (Fig. 5h), and 30% (Fig. 5i) of CHO cells, respectively. This experiment was repeated a number of times with different HSV1716gfp/BHK preparations, and 90 to 100% of CHO cells were always infected following simultaneous addition of serum and virus. Two other examples are shown in Fig. 5j and k. The addition of HSV17+gfp/BHK along with serum to 24-hour serum-starved CHO cells also resulted in infection of 100% of cells (Fig. 5l), and this had decreased to ca. 40% of CHO cells when 5 PFU/cell HSV1716gfp/BHK was added 16 h after reintroduction of serum (Fig. 5m). In contrast, HSV1716gfp/Vero was completely unable to infect CHO cells, even after 24 h of serum starvation (Fig. 5n and o). Virus yields obtained from duplicate preparations of serum-starved CHO, Vero, and C8161 cells immediately infected with 5 × 105 PFU HSV1716gfp/BHKfollowing reintroduction of serum were 6 × 106 and 8 × 106 PFU (CHO), 1 × 107 and 1 × 107 PFU (C8161), and 2 × 107 and 3 × 107 PFU (Vero), indicating 12- to 16-fold, 20-fold, and 40- to 60-fold yields of virus from CHO, C8161, and Vero cells, respectively.
FIG. 5.
Unprocessed images (magnification, ×196) from fluorescent microscopy of various cell lines after 8 h of infection with HSV1716gfp or HSV17+gfp propagated either in BHK or Vero cells. For panels a to d, serum-starved or non-serum-starved BHK (a, b) or C8161 (c, d) cells were infected with 5 PFU/cell HSV1716gfp/BHK (a, b) or 5 PFU/cell HSV1716gfp/Vero (c, d) added either to non-serum-starved cells (b, d) or to cells simultaneously with the reintroduction of serum (a, c). For panels e to k, serum-starved or non-serum-starved CHO cells were infected with 5 PFU/cell HSV1716gfp/BHK added either to non-serum-starved cells (e) or to serum-starved cells simultaneously with the reintroduction of serum (f, j, k) or 4 (g), 8 (h), or 16 (i) hours after serum addition. For panels l and m, 5 PFU/cell HSV17+gfp/BHK was added to serum-starved CHO cells either simultaneously with serum addition (l) or 16 h after the reintroduction of serum (m). For panels n and o, 5 PFU/cell HSV1716gfp/Vero was added to either non-serum-starved CHO cells (n) or to serum-starved CHO cells simultaneously with the reintroduction of serum (o).
DISCUSSION
The results of this study suggest the intriguing conclusion that the cell type used to propagate HSV-1 has an influence on the tropism of the progeny virions. The inability of HSV-1 to infect CHO cells is well documented (16), and the cell line has provided an excellent model system to identify receptors for HSV-1, with the normally nonpermissive CHO cells rendered permissive by the expression of an appropriate receptor (7, 12, 17). Montgomery et al. (12) report propagation of virus on either Hep-2 or Vero cells, whereas no details of virus propagation are given by Geraghty et al. (7) or Shukla et al. (17). When propagated in Vero or C8161 cells, HSV1716, a replication-competent mutant derived from HSV-1 strain 17+, completely fails to infect the nonpermissive CHO and mouse melanoma cell lines. This result is in keeping with the findings of others (7, 12) who reported that most strains of HSV-1, including strain 17+, failed to infect CHO cells unless the cells were modified to express a cellular receptor for virus entry. HSV-1 strain MP was able to infect CHO cells to a limited extent, and this was enhanced by cellular expression of HVEM or nectin 1. In contrast to these findings of others and to our results with HSV1716 propagated in Vero and C8161 cells, we have consistently observed HSV-1 strain 17+ infection of CHO cells (22;Conner, unpublished) and initiated this study to try to explain this discrepancy. Surprisingly, our results indicate that it is the cell line on which the virus is propagated which affects its ability to infect CHO cells. We regularly propagate HSV-1 strain 17+ and HSV1716 using BHK cells, and the resultant virus can always infect CHO cells. Our results indicate that, using a multiplicity of infection (MOI) of 10 PFU/cell, up to 50% of CHO cells in normal culture are infected by HSV1716gfp produced from BHK cells and, by serum starving the CHO cells for 24 h, up to 100% were infected. At similar or higher MOI and with or without 24 h of serum starvation, HSV1716gfp produced from Vero or C8161 cells was completely unable to infect CHO cells, as assessed by reporter gene or viral gene expression. Further, this difference in tropism between BHK- and Vero-propagated HSV1716 was not restricted to CHO cells, as a second nonpermissive cell line, B16-F10 mouse melanoma cells, was susceptible to HSV1716gfp/BHK infection but not to HVS1716gfp/Vero infection. Thus, the cell type used to propagate HSV-1 contributes to the tropism of the progeny virions, with production on BHK cells extending the permissive range of the virus. In view of this, caution should be exercised when using such viruses for studies on cellular receptors for HSV-1.
Although in a preliminary analysis, we initially failed to detect infectious viruses in the supernatant of CHO cells infected with HSV1716gfp/BHK, the pattern and levels of viral protein production were comparable with those of infected BHK or Vero cells and electron microscopy indicated that infection in CHO cells proceeded normally with filled nucleocapsids and enveloped virions in the infected cell. When cells and supernatants were harvested together, infectious virions were readily detected, indicating that infection of CHO cells by HSV1716gfp/BHK was productive and suggesting that perhaps the progeny virions from CHO infection were not released as efficiently by the CHO cells compared to either Vero or BHK cells. Alternatively, since not all (20 to 50%) of CHO cells were infected by HSV1716gfp/BHK, the amount of virus released into supernatants in our initial experiment was too low to be detected and, in support of this, yields from infected CHO cells were much lower than from either Vero or C8161 cells. Additionally, the virions released by CHO cells were unable to spread to adjacent cells, as evidenced by the failure of both 5 PFU/cell of HSV1716gfp/CHO to infect CHO cells and a low MOI of HSV1716gfp/BHK to form plaques on CHO cells. Therefore, infection was limited to a single round of replication in the population of CHO cells susceptible to HSV1716gfp/BHK, thus further reducing viral yields and indicating that the factor responsible for the altered tropism of HSV1716 is not present in CHO cells. The acquisition of an infectivity factor from a round of replication in BHK cells, which is then lost following a round of replication in another cell line, is similar to the observation that gD-negative HSV-1 progeny virions produced in a gD-complementing cell line are infectious, as they acquire gD from the complementing cell line. However, a subsequent round of replication in a noncomplementing cell line results in noninfectious progeny, as they lack gD (for an example, see reference 24). The inability of HSV1716gfp/CHO to infect cells was restricted to the nonpermissive CHO and mouse melanoma lines, as the virus efficiently infected BHK, Vero, C8161, SK-N-SH (data not shown), and HeLa (data not shown) cells. However, even after serum starvation, with 100% of CHO cells infected, virus yields were lower than for C8161 or Vero cells, and some other mechanism, such as inefficient assembly or egress may contribute to lower productivity in CHO cells. Interestingly, Montgomery et al. (12) reported variation in progeny production in CHO-K1 cells that express HVEM, with different strains, HSV-1(KOS) and HSV-1(F), producing 10,000-fold and 100-fold more progeny virus, respectively, than control cells, suggesting that various strains of HSV-1 display different efficiencies of replication in CHO cells. Montgomery et al. (12) also reported 10- to 100-fold-lower yields from HSV-1 infection of CHO cells rendered permissive by expression of the HVEM receptor than those from permissive HeLa cells.
The ability of HSV1716gfp/BHK to infect CHO cells was dependent on the cell cycle status, as 24 h of serum starvation rendered 100% of the CHO cells permissive for infection upon simultaneous introduction of serum and virus. The addition of virus at various times after reintroduction of serum resulted in decreased permissiveness of the CHO cells such that only 30% were infected 16 h after cessation of serum starvation. Serum starvation had no apparent effects on the susceptibility of either BHK or Vero cells to HSV1716gfp/BHK, and serum starvation did not alter the nonpermissiveness of CHO cells to HSV1716gfp/Vero. Serum starvation arrests CHO cells in the G1 stage of the cell cycle, and presumably, all cells are susceptible to HSV1716gfp/BHK infection at this stage. The addition of serum reestablishes cell cycling, and the number of CHO cells susceptible to HSV1716gfp/BHK infection decreases as cells move out of the G1 phase. Therefore, in normal CHO cultures, only those cells in the G1 phase are infected by HSV1716gfp/BHK, and presumably, cell cycle variation is responsible for the inability to infect more than 50% of cells in asynchronously growing cultures. HSV-1 infection per se has been shown to arrest cells in the G1 phase of the cell cycle (6, 18), but to our knowledge, this is the first demonstration that the cell cycle affects the susceptibility of cells to HSV-1 infection.
In Vero cells, HSV-1 penetrates the plasma membrane after specific interactions with its receptors and the nucleocapsid is delivered directly into the cytosol after membrane fusion. In contrast, CHO cells endocytose the HSV-1 virions and the fate of the endocytosed virus is governed by the presence or absence of an HSV-1 gD receptor. Electron microscopy studies by Nicola and Strauss (14) clearly show that, regardless of whether or not the CHO cells express an HSV-1 entry receptor, the virus particles are internalized by endocytosis. In normal CHO cells, the endocytosed virions are subjected to lysosomal degradation and no infection results, whereas in CHO cells that express specific HSV-1 receptors, such as nectin 1, the endocytosed virions follow an alternative route that escapes lysosomal degradation and infection ensues (14). The precise nature of the mechanism that allows alternative trafficking of HSV-1-containing endocytotic vesicles in the presence of an HSV-1 gD receptor is unknown, but as in plasma membrane fusion, it requires gB, gD, and gH/gL (14) and the two processes possibly share some common mechanistic features. Thus, when HSV1716gfp/Vero, HSV1716gfp/C8161, or HSV1716gfp/CHO cells are endocytosed, the vesicles are most likely routed for lysosomal degradation and infection does not occur. In contrast, HSV1716gfp/BHK virions, even in the absence of an HSV-1 gD receptor, must enter the cells through a route that avoids lysosomal degradation, allowing infection to occur, and based on the electron microscopy studies of others (14), this most likely involves alternative trafficking of endocytosed virions. We suggest that some factor acquired as a component of HSV1716gfp/BHK virions during replication in BHK cells compensates for the lack of a specific gD receptor and allows alternative trafficking with subsequent infection to occur.
Interestingly, the ability of HSV1716gfp to infect CHO cells was lost or gained by a single round of replication in Vero or BHK cells, respectively. Thus, the progeny from a single round of HSV1716gfp/Vero replication in BHK cells acquired the ability to infect CHO cells, whereas this ability was lost following a single passage of HSV1716gfp/BHK in Vero cells. The relatively straightforward ease with which CHO cell infectivity was gained or lost by a single passage in BHK or Vero cells suggests that the mechanism responsible for this switch is most likely to be through the acquisition (or loss) of a factor during replication in BHK cells. Since this factor directs alternative trafficking of endocytosed virions by CHO cells, it seems most probable that it is a membrane component that circumvents the requirement for a specific HSV-1 gD receptor. Thus, we can envisage several mechanisms through which HSV1716 grown on BHK but not Vero or C8161 cells might be able to infect normally nonpermissive CHO cells. HSV-1 binds to heparan sulfate on the surface of CHO cells (16) and is endocytosed, but as the cells lack the entry receptors (HVEM or nectin 1 or 2) for the specific entry interaction with HSV-1 gD, the endocytosed virus is targeted for destruction by the cell (14). CHO cells are permissive for HSV-2 infection (12), with this difference possibly residing in the N-terminal amino acids of gD, which allows HSV-2 gD, but not HSV-1 gD, to interact with a receptor, possibly the Chinese hamster homologue of nectin 2 (23). As with HSV-1 infection of CHO cells rendered permissive by expression of an HSV-1 receptor, HSV-2 infection of CHO cells is productive (12), and presumably, the resultant progeny virions can infect other CHO cells possessing the appropriate receptor. Possibly, posttranslational processing of gD in BHK cells is different from that which occurs in Vero or C8161 cells and the resultant gD can interact with an alternative receptor on CHO and, to a lesser extent, mouse melanoma cells. However, differences in gD processing seem unlikely, as the efficiency with which HSV1716gfp/BHK infected permissive cell types compared to HSV1716gfp/Vero or HSV1716gfp/C8161 was unaffected.
An alternative and perhaps more intriguing explanation stems from the basic observation that HSV-1 propagated on rodent cells is better able to infect rodent cells than an identical virus propagated on monkey or human cells. Subtle differences in viral membrane composition, including the possible incorporation of a rodent cell component, may result from replication in BHK compared to primate Vero or human C8161 cells and allow the rodent cell-derived virus access to an alternative entry route present on CHO cells and, to a lesser extent, on mouse melanoma cells. Previously, infection of cells that transiently express the vesicular stomatitis virus glycoprotein gG (VSV-gG) with a gD-negative HSV-1 results in the formation of a pseudotype HSV-1 with VSV-gG receptor binding, as the VSV-gG is acquired by progeny virions (1). We conclude, therefore, that the tropism of HSV-1 is derived not only genetically but also from the cells used for propagation and it will be interesting to compare the tropisms of HSV-1 derived from human, monkey, and rodent cell lines on apparently nonpermissive human and monkey cells.
REFERENCES
- 1.Anderson, D. B., S. Laquerre, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2000. Pseudotyping of glycoprotein D-deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein G enables virus attachment and entry. J. Virol. 74:2481-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 3.Cocchi, F., L. Menotti, V. D. Ninni, M. Lopez, and G. Campadelli-Fiume. 2004. The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J. Virol. 78:4720-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conner, J., J. Furlong, J. Murray, M. Meighan, A. Cross, H. Marsden, and J. B. Clements. 1993. Herpes simplex virus type 1 ribonucleotide reductase large subunit: regions of the protein essential for subunit interaction and dimerization. Biochemistry 32:13673-13680. [DOI] [PubMed] [Google Scholar]
- 5.Davison, A. J., and J. B. Clements. 1997. Herpesviruses: general properties, p. 309. In B. W. J. Mahy and L. H. Collier (ed.), Topley and Wilsons' principles of bacteriology virology and immunology. Edward Arnold, London, United Kingdom.
- 6.Ehmann, G. L., T. I. McLean, and S. L. Bachenheimer. 2000. Herpes simplex virus type 1 infection imposes a G1/S block in asynchronously growing cells and prevents G1 entry in quiescent cells. Virology 267:335-349. [DOI] [PubMed] [Google Scholar]
- 7.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 8.Harland, J., P. Dunn, E. Cameron, J. Conner, and S. M. Brown. 2003. The herpes simplex virus (HSV) protein ICP34.5 is a virion component that forms a DNA-binding complex with proliferating cell nuclear antigen and HSV replication proteins. J. Neurovirol. 9:477-488. [DOI] [PubMed] [Google Scholar]
- 9.Jones, N. A., and R. J. Geraghty. 2004. Fusion activity of lipid-anchored envelope glycoproteins of herpes simplex virus type 1. Virology 324:213-228. [DOI] [PubMed] [Google Scholar]
- 10.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menotti, L., M. Lopez, E. Avitabile, A. Stefan, F. Cocchi, J. Adelaide, E. Lecocq, P. Dubreuil, and G. Campadelli-Fiume. 2000. The murine homolog of human nectin1δ serves as a species nonspecific mediator for the entry of human and animal αherpesviruses in a pathway independent of a detectable binding to gD. Proc. Natl. Acad. Sci. USA 97:4867-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 13.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicola, A. V., and S. E. Strauss. 2004. Cellular and viral requirements for rapid entry of herpes simplex virus. J. Virol. 78:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion by herpes simplex virus glycoprotein gB, gD and gH/gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 16.Shieh, M.-T., D. WuDunn, R. I. Montgomery, J. D. Esko, and P. G. Spear. 1992. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 116:1273-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 18.Song, B., K.-C. Yeh, J. Liu, and D. M. Knipe. 2001. Herpes simplex virus gene products required for viral inhibition of expression of G1-phase functions. Virology 290:320-328. [DOI] [PubMed] [Google Scholar]
- 19.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2 and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 22.Zachos, G., B. Clements, and J. Conner. 1999. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J. Biol. Chem. 274:5097-5103. [DOI] [PubMed] [Google Scholar]
- 23.Zago, A., and P. G. Spear. 2003. Differences in the N termini of herpes simplex virus type 1 and 2 gDs that influence functional interactions with the human entry receptor nectin-2 and an entry receptor expressed in Chinese hamster ovary cells. J. Virol. 77:9695-9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]