Abstract
Though first described as a lymphotropic virus, human herpesvirus 6 (HHV-6) is highly neuropathogenic. Two viral variants are known: HHV-6A and HHV-6B. Both variants can infect glial cells and have been differentially associated with central nervous system diseases, suggesting an HHV-6 variant-specific tropism for glial cell subtypes. We have performed infections with both viral variants in human progenitor-derived astrocytes (HPDA) and monitored infected cell cultures for cytopathic effect (CPE), intra- and extracellular viral DNA load, the presence of viral particles by electronic microscopy, mRNA transcription, and viral protein expression. HHV-6A established a productive infection with CPE, visible intracellular virions, and high virus DNA loads. HHV-6B-infected HPDA showed no morphological changes, intracellular viral particles, and decreasing intra- and extracellular viral DNA over time. After long-term passage, HHV-6B-infected HPDA had stable but low levels of intracellular viral DNA load with no detectable viral mRNA. Our results demonstrate that HHV-6A and HHV-6B have differential tropisms and patterns of infection for HPDA in vitro, where HHV-6A results in a productive lytic infection. In contrast, HHV-6B was associated with a nonproductive infection. These findings suggest that HHV-6 variants might be responsible for specific infection patterns in glial cells in vivo. Astrocytes may be an important reservoir for this virus in which differential tropism of HHV-6A and HHV-6B may be associated with different disease outcomes.
Human herpesvirus-6 (HHV-6), a ubiquitous β-herpesvirus commonly acquired during childhood and infancy (29), is being actively investigated as an underrecognized pathogen of human neurologic disease. The occurrence of encephalitis and febrile or afebrile seizures as complications of primary infection has been described previously (41, 43), suggesting that HHV-6 has neuropathogenic potential. In addition, HHV-6 has also been demonstrated to be the causative agent of encephalitis in adult immunosuppressed patients such as bone marrow transplant recipients (40). These findings, possibly due to local viral reactivation, would suggest that not only does HHV-6 spread to the central nervous system (CNS), but it can also establish latency in the CNS, as confirmed by the detection of HHV-6 DNA in autopsy samples from healthy individuals (9).
Consistent evidence supporting the possible association of HHV-6 in multiple sclerosis (MS) would suggest that HHV-6 is also involved in this inflammatory disease of the CNS (33). Recent studies on autopsy brain samples from patients with MS have reported a higher frequency of HHV-6 DNA in MS plaques as opposed to normal-appearing white matter of the same patients and to unaffected brain specimens (7, 17), and a recent investigation has documented significantly higher levels of HHV-6 gene transcripts in both lesional and normal-appearing white matter in autopsy brain samples from MS patients than in those from normal controls (27).
However, since CNS disorders such as MS and encephalitis are associated with inflammation, the detection of HHV-6 in these diseases may be an epiphenomenon related to the presence of inflammatory cells. Arguments against this hypothesis have been suggested by a recent report demonstrating the presence of active HHV-6 infection in the absence of inflammation in brain samples from a subset of patients with mesial temporal lobe epilepsy (14). Importantly, this study demonstrated the presence of HHV-6-infected astrocytes, supporting the assumption that resident glial cells and not only peripheral lymphocytes leaking through the blood brain barrier might harbor the virus. The capacity of astrocytes to be infected with HHV-6 is consistent with a number of reports (10, 20, 42). The role of astrocytes in CNS function is increasingly being studied and is challenging the view of glia as mere ancillary cells in the CNS circuitry. Rather, evidence has suggested that astrocytes can play an important role in neural functions that actively modulate synaptic transmission (4). Therefore, it is possible that the clinical manifestations of HHV-6 infection in the CNS may be related to the capacity of this virus to replicate in resident glial cells.
HHV-6 has been subtyped into two variants, A and B (1), based on genetic polymorphisms and clinical and biological characteristics of these two variants. The sequences of the U1102 strain for the A variant and of the Z29 strain for B variant are known (13). Although HHV-6A and HHV-6B share an average sequence homology of 90%, the two variants have unique properties and features so as to fulfill the criteria for classification into two distinct viral species (5).
HHV-6 variant B is widely recognized as the agent associated with primary infection and its complications, while no well-defined syndrome has been definitively linked to HHV-6A, although an association of variant A in MS has been reported (22, 34). In vitro studies suggest that the two variants of HHV-6 have different patterns of infection and tropism for glial cell subtypes. Though both variants can grow in glial precursor cells (12), human primary glial cells (3, 20), and glia cell lines (10, 42), infection with HHV-6A has been reported to be associated with CPE and viral DNA detection (10), consistent with a number of clinical observations suggesting a greater neurotropism for this viral variant (19, 22, 34).
The wide cellular host range of HHV-6 may be attributed to the ubiquity of its cellular receptor, CD46, which is shared by both variants (31). CD46 is a member of a family of glycoproteins that function as regulators of complement activation to prevent spontaneous activation of complement on autologous cells. Other viral and bacterial agents use CD46 as a receptor for entry or fusion through the use of different extracellular domains (18, 21) consisting of a region rich in serine, threonine, and proline (STP site) linked to four short consensus repeats (SCR1 to SCR4). Though necessary, CD46 might not be sufficient as the HHV-6 receptor, as suggested by the lack of HHV-6 replication and cell-to-cell viral-mediated fusion in a number of T-cell lines that are CD46 positive (31). The molecular basis of interaction between CD46 and HHV-6 is not yet fully understood; however, recent in vitro studies have suggested that virus-cell fusion required for entry and cell-cell fusion of infected cells are not dependent on the same cellular mechanisms (24).
To understand and define possible HHV-6 variant-specific tropisms and pathogenicity in glial cells, the outcomes of infection by HHV-6A and HHV-6B in human progenitor-derived astrocytes (HPDA) were evaluated. Cellular toxicity, protein expression, ultrastructural analysis, DNA quantitation, and RNA detection were used to evaluate the biological effect and efficacy of infection by either variant. The results obtained showed remarkable differences in the capacity of the two HHV-6 variants to replicate in human astrocytes. HHV-6A was associated with active virus growth and replication, while HHV-6B infection was nonproductive. Additionally, HHV-6A active infection of HPDA could be inhibited using a monoclonal antibody raised against a specific CD46 SCR. These results highlight further differences in HHV-6 tropism for human astrocytes and provide a framework to better understand the role of HHV-6 variants in CNS disease.
MATERIALS AND METHODS
Cell cultures and HPDA differentiation.
HPDA were obtained from human fetal brain tissue and characterized as previously described (23). Selective differentiation was achieved by culturing adherent cells in Eagle minimal essential medium (HyClone, Logan, UT) supplemented with 10% FCS, 25 mM l-glutamine, 100 IU/ml penicillin/streptomycin (Biowhittaker, Walkersville, MD), and 50 μg/ml gentamicin in poly-d-lysine (Sigma, St. Louis, MO)-treated flasks. Cells were passaged at 80 to 90% confluence at a split ratio of 1:2 and plated at a density of 5 × 105 cells/75-cm2 flask. After 20 days of culture, cells were considered differentiated when they appeared >90% positive for the astrocytic marker glial fibrillary acidic protein (GFAP) expression using an immunofluorescence assay (described below) and were ready for use. The lymphocytic cell lines SupT-1 and Jjhan were maintained in RPMI 1640 medium (Gibco Invitrogen Corp., Grand Island, NY) containing 10% (for noninfected cells) or 5% (infected cells) fetal calf serum (HyClone), 25 mM l-glutamine, and 100 IU/ml penicillin/streptomycin.
Virus infections.
Cell-free viral inocula were obtained by culturing HHV-6A (U1102 and GS strains, obtained from C. Cermelli, Modena University, Italy, and D. V. Ablashi, Advanced Biotechnologies Inc, Columbia, MD, respectively) and HHV-6B (Z29 strain, a kind gift from P. Secchiero, Institute of Human Virology, Baltimore, MD), respectively, on Jjhan, HSB2, and SupT-1 cells. When CPE was >80%, the cell suspensions were centrifuged for 5 min at 200 × g. The supernatants were harvested and centrifuged again at 1,800 × g for 5 min. The cell-free supernatants were stored frozen at −70°C. A frozen aliquot was tested for viral DNA quantitation using a real-time TaqMan PCR (described below). All the frozen cell-free supernatants used as inocula were thawed not more than once. Virus-free supernatants from uninfected SupT-1 and Jjhan cells were obtained by applying the same procedure and used as mock infection inoculum.
For infection, 6 × 105 HPDA were seeded onto a 75-cm2 flask or 1 × 105 cells were seeded onto 2-well chamber slides (Nunc, Rochester, NY). After an overnight incubation, which allowed the cells to adhere, cultures were washed twice with phosphate-buffered saline (PBS) and infected with freshly thawed cell-free supernatant containing 108 DNA viral copies/106 HPDA. After a 3-hour incubation at 37°C in 5% CO2, cultures were washed at least three times with PBS, and fresh medium was added to the cultures. Since viral stocks may range in their infectivity, and the viral DNA copies titrated in the samples might not correspond entirely to viable virus, the viability of either inoculum was confirmed by infecting SupT-1 cells, which are permissive to both variants, in parallel with the same inoculum following the same procedures as those for infection of HPDA. Procedures for mock infections and viral infections were performed, respectively, in two separate laminar flow hoods.
Cell cultures were checked for CPE every day after infection. At every time point (3 h, day 1, day 3, day 5, and day 7) after the infection, cell cultures were centrifuged and pelleted after two washes in PBS, with adherent HPDA being previously detached using trypsin (Sigma, St. Louis, MO) (5- to 10-min incubation at 37°C). Two aliquots of cell pellet were obtained and stored at −70°C either as dry pellet for DNA or resuspended in 350 μl of RTL buffer (QIAGEN Inc., Valencia, CA) for RNA isolation. The supernatants of infected and control SupT-1 and HPDA cultures obtained after a double centrifugation were frozen, while an aliquot of 200 μl underwent treatment for DNA extraction. Infection of HPDA was performed at least five independent times with HHV-6A and six times using HHV-6B.
For HHV-6 entry-blocking experiments, HPDA were seeded onto 6- and 2-well chamber slides. The infection, performed for 3 hours with HHV-6A (U1102) supernatant from Jjhan cultures, was preceded by a 20-min incubation at 37°C with monoclonal antibodies (10 μg/ml) M177, M160 (generously donated by Tsukasa Seya, Osaka Medical Center, Osaka, Japan), and J4.48 (Immunotech, Marseille, France), respectively, for anti-CD46 SCR-2, SCR-3, and SCR-1 (18) or using an antibody against HLA-DR (BD Biosciences, San Jose, CA). No antibody was used in the controls.
Immunofluorescence assay (IFA).
HPDA were cultured directly on poly-d-lysine-coated 2-chamber glass slides, and the infection was performed as described below. At each time point, cells were fixed in an ice-cold acetone/methanol (50:50, vol/vol) solution for 10 min at −20°C, rinsed with PBS, and blocked with 3% bovine serum albumin solution at room temperature for 10 min. PBS antibody solution containing 1/100 mouse monoclonal anti-HHV-6 gp116/54/64 (Advanced Biotechnologies Inc., Columbia, MD) and 1/100 rabbit anti-GFAP (DAKO Corp., Carpinteria, CA) was distributed on slides for 30 min at 37°C. After three rinses in PBS and two rinses in distilled water, the secondary antibody-PBS solution containing a 1/1,000 dilution of Alexa Fluor 488 (green fluorescence)-labeled anti-rabbit immunoglobulin G and a 1/1,000 dilution of Alexa Fluor 594 (red fluorescence)-labeled anti-mouse immunoglobulin G2b (Molecular Probes, Eugene OR) was distributed on the slides and incubated for 30 min at 37°C. Experiments on fixed SupT-1 cells infected with either variant were performed using a 1/100-diluted fluorescein isothiocyanate-labeled anti-mouse antibody (Sigma, St. Louis, MO) as a secondary antibody. Slides were rinsed as described above. Coverslips were mounted using Vectashield Hard Set mounting medium (Vector Laboratories, Burlingame, CA), with DAPI (4′,6′-diamidino-2-phenylindole). Samples were examined using a fluorescent microscope (Carl Zeiss Microimaging Inc., Thornwood, NY) at ×20 or ×32 magnification, with exposure times of 30 ms for the DAPI filter and 400 ms for both fluorescein isothiocyanate and rhodamine filters.
Electron microscopic analysis.
HPDA grown on poly-d-lysine-coated 2-well glass chambers were fixed for 3 h with 4% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at room temperature. The cells were then washed in 0.1 M cacodylate buffer, mordanted en bloc with 0.25% uranyl acetate overnight, washed and dehydrated through a graded series of ethanol, and finally embedded in epoxy resin. Thin sections were thereafter counterstained with uranyl acetate and lead citrate and examined under a JEOL 1200 transmission electron microscope operated at 60 kV. Images were captured with a digital camera system (XR-100 from AMT, Danvers, MA).
DNA extraction and quantitative real-time PCR.
DNA extractions were carried out using the Qiamp blood kit or the Viral RNA kit (QIAGEN), respectively, for extraction from cells or from supernatants according to the manufacturer's instructions. The final elution volume of extracted DNA samples was 100 μl for cells and 70 μl for supernatants. DNA concentrations were adjusted to 10 ng/μl and applied to a 96-well TaqMan plate in triplicates. Quantitative real-time PCR specific for each of the two viral variants was performed as described previously (14) using reagents from Life Technologies and a Sequence Detector system (model 7700 [TaqMan]; Perkin Elmer). The primers and probes used (Applied Biosystems, Foster City, CA) were specific for the immediate-early region of variants A and B (25). Samples were also quantitatively titrated for human genomic β-actin DNA on a third real-time PCR assay, and final viral DNA load per 106 cells was calculated by the following formula: (HHV-6 DNA copy number/β-actin DNA copy number/2) × 106 = HHV-6 DNA copy number/106 cells (14). Results were plotted and sorted using the Sequence Detector System program (Perkin-Elmer, Wellesley, MA).
Long-term passage of HHV-6B-infected HPDA.
Six different Z29 infections of HPDA were performed in which three cultures were grown for up to six passages. HPDA were infected with 108 viral copies/106 cells (Z29 strain) and were passaged every 6 days or at 80% confluence. Intracellular viral DNA and RNA transcription were monitored at each passage. At passage 6, each flask was split in two, and one was incubated in culture medium containing 50 nmol/ml phorbol myristate acetate (PMA) (Sigma). The remaining flask was treated in parallel with PMA-free medium. After 24 h, the medium was removed, the cultures were washed twice with PBS, and fresh medium was added. Three days after PMA treatment, samples for DNA and RNA analysis were taken from all cultures for viral determination.
RNA extraction and retrotranscription-PCR (RT-PCR).
RNA extractions were performed on frozen pelleted cells using the RNeasy kit (QIAGEN) according to the manufacturer's instructions. The final elution volume was 30 μl. In order to remove all viral DNA, up to 1 μg of RNA samples was digested with DNase I (Invitrogen) for 20 min at room temperature. The enzyme was then inactivated at 65°C for 10 min. The retrotranscription with random primers was performed using ImProm-II Reverse Transcription system (Promega, Madison, WI) according to the manufacturer's instructions. cDNA samples were stored at −20°C. One microgram of cDNA underwent a 35-cycle primary PCR for each primer pair, with annealing temperatures of 57°C (U33 and U12), 54°C (U39), 52°C (U27), 59°C (U94), and 59°C (human β-actin). In the case of U12, U27, U33, and U94, a 35-cycle nested PCR was run on 5 μl of amplified template, with annealing temperatures of 57°C (U12 and U33), 59°C (U94), and 50°C (U27). All primers were derived from GenBank sequences and designed using Primer 3.0 (Whitehead Institute [http://www.genome.wi.mit.edu/genome]). All cDNA samples were first tested for the human β-actin fragment in a 35-cycle primary PCR specific for a spliced mRNA region. Primary and nested PCRs were performed using QIAGEN Mastermix in a 50-μl final volume containing 25 pmol of each primer pair. Ten microliters of the amplification product was electrophoresed in agarose gel and stained with ethidium bromide as described previously (2).
U12 primers were designed to detect mRNA and not viral DNA, since they amplify a spliced region, while for all other primers, viral DNA removal was confirmed by the absence of any PCR signal at PCR for each of the RNA viral primers on nonretrotranscribed RNA samples. Each set of primers was tested on sample cDNA, on nonretrotranscribed RNA as a negative control, and on a positive control cDNA obtained from SupT-1 cultures infected with either viral variants.
RESULTS
HHV-6 variant-specific patterns of infection in HPDA.
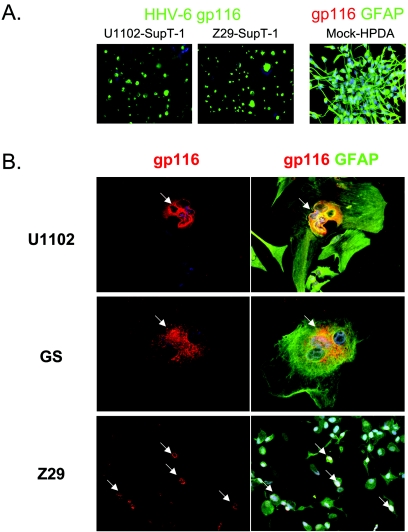
HPDA cultures uniformly stained positively for GFAP and morphologically resembled astrocytes as described previously (23). Cell-free supernatants containing either HHV-6 variant A or B were used to infect these cells at comparable levels of HHV-6 DNA (1 × 108 copies/106 cells) as determined by quantitative real-time PCR. At 3 h and 1, 3, 5, and 7 days postinfection, cells were fixed for IFA and stained for HHV-6 expression with a monoclonal antibody specific for the HHV-6 gp116/54/64 surface glycoprotein and antibodies specific for GFAP. HHV-6A (U1102)-infected HPDA showed formation of syncytia (Fig. 1B, top), and by day 5 postinfection, more than 60% of infected cells showed a marked CPE, with marked cellular swelling and cell-cell fusion. Anti-HHV-6 gp116/54/64 reactivity was clearly localized perinuclearly in the cytoplasm of most multinucleate giant cells which were intensely reactive for GFAP, demonstrating that the infected cells retained their astrocytic phenotype (Fig. 1B). The number of adherent cells decreased over time, indicating a high rate of cell death. By day 14, the number of adherent cells in the culture had decreased by 80%, and it was not possible to maintain a population of surviving cells in culture. The occurrence of CPE and anti-HHV-6 gp116/54/64 reactivity was confirmed on HPDA infected with the same concentration of another HHV-6A strain, GS (Fig. 1B, center panels). CPE and virus protein expression were demonstrated in control cultures of HHV-6A-infected T-cell line SupT-1 (Fig. 1A). Moreover, cell-free supernatants from HHV-6A-infected HPDA were used to inoculate a known permissive naive T-cell line (Jjhan) that demonstrated productive infections with CPE, cell death, and high intracellular HHV-6A viral load at all time points analyzed, peaking between 107 and 109 copies/106 cells at day 5 (data not shown).
FIG. 1.
Detection of HHV-6 gp116/54/64 glycoproteins in human progenitor-derived astrocytes and in T lymphocytes. (A) SupT-1 cells (left images) infected with either HHV-6A (U1102) or with HHV-6B (Z29) show bright expression of viral glycoproteins gp116/54/64 (green) in the cytosol. In contrast, mock-infected HPDA (right image) stain positively with an antibody against GFAP (green) and show no unspecific staining for HHV-6 glycoproteins (lack of red signal). (B) Double IFA for viral glycoproteins (red, arrows) and GFAP (green) on HPDA at 3 days postinfection with HHV-6. HPDA form syncytia following infection with the HHV-6A strains U1102 or GS but not with HHV-6B strain Z29. Magnifications, ×20 (A), ×32 (B, top and center panels), and ×20 (B, bottom panels).
In marked contrast to these observations in HHV-6A-infected HPDA, cells infected with the same amount of viral copies of cell-free HHV-6B (strain Z29) showed no CPE with continued growth and replication. A small percentage of HHV-6B-infected cells (5 to 15% depending on the experiments) showed weak specific perinuclear staining for HHV-6 gp116/54/64 beginning at day 3 to day 7 postinfection (Fig. 1B, bottom). Though viral genomes could be detected in the supernatant (see Fig. 3C), no infectious virus was released, since cell-free supernatants from HHV-6B-infected HPDA failed to infect control SupT-1 cells (data not shown). To demonstrate that this lack of infectivity for HHV-6B was specific for HPDA, a known susceptible T-cell line was infected with the same HHV-6B inoculum. SupT-1 cells infected with the Z29 HHV-6B variant showed a marked CPE and a strong immunoreactivity for HHV-6 gp116/54/64, thus confirming that the supernatants used for the infection contained viable virus (Fig. 1A). Control mock-infected HPDA cultures tested at the same time points resembled uninfected HPDA and showed no morphological change or anti-HHV-6 gp116/54/64 reactivity while remaining GFAP positive (Fig. 1A).
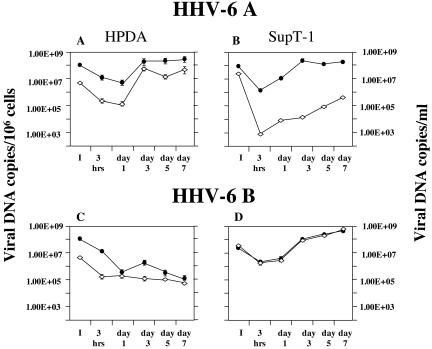
FIG. 3.
Quantitative detection of intracellular (filled circles) and extracellular (empty diamonds) HHV-6 DNA at 3 h and 1, 3, 5, and 7 days postinfection of HPDA and SupT-1 cells. While both viral variants (U1102 and Z29) establish a productive infection in SupT-1 cells, only infection with U1102 is productive in HPDA. Intracellular DNA is expressed as viral copies/106 cells; extracellular DNA is expressed as viral copies/milliliter ± standard error of the mean of triplicate values. I, inoculum.
Ultrastructural analysis of HHV-6A-infected HPDA.
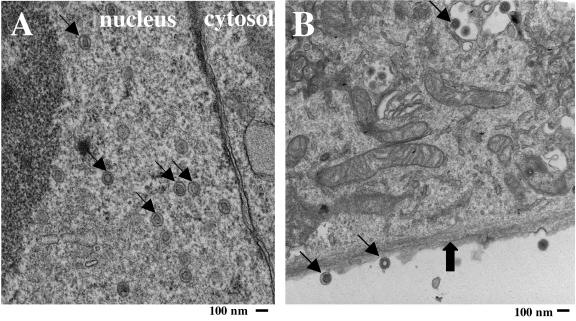
To confirm further that HHV-6A was replication competent in HPDA, electron microscopic analyses were performed on HHV-6A-infected HPDA at 5 and 7 days postinfection. High numbers of HHV-6 viral nucleocapsids and virus with tegument could be visualized in the cell nucleus (Fig. 2A, arrows). In the cytoplasm, particles with tegument and virions forming envelopes could be observed (Fig. 2B, thin arrows). In addition, virus particles were also visualized outside the cell (Fig. 2B, thin arrows). HPDA were ultrastructurally identified by their characteristic intracytoplasmic glial filaments (Fig. 2B, thick arrow) (32). Each infected cell harbored more than 100 viral particles. In contrast, no viral particles could be visualized in HPDA infected with HHV-6B at any time point (data not shown). These data support the idea that only HHV-6A, but not HHV-6B, can establish a productive infection of HPDA.
FIG. 2.
Detection of viral particles in HHV-6A-infected HPDA by electron microscopy. Viral particles (thin arrows) are present in the nucleus (A) as well as in the cytosol at different stages of maturation (B) of infected HPDA, identified by characteristic glial filaments (thick arrow), at day 7 after infection. Viral particles are also visible extracellularly (B). Magnification, ×17,000.
Time course of HHV-6 infection in HPDA.
The quantification of viral load over time in HPDA infected with cell-free HHV-6 is illustrated in Fig. 3. HPDA infected with the HHV-6A variant U1102 showed an increase in viral DNA associated with active viral replication (Fig. 3A). The intracellular viral DNA load decreased in the first 24 h and then increased from day 1 to day 3 postinfection which then plateaued between 1.7 × 108 and 3 × 108 copies/106 cells for the remaining observation period (day 7). In parallel, detection of viral DNA in cell-free supernatants was similar, with a peak in viral load of 5.7 × 107 copies/ml at day 3 postinfection. These results were comparable to those obtained with HHV-6A-infected control SupT-1 cells in which productive HHV-6A virus could be demonstrated (Fig. 3B); interestingly, viral load in the supernatants was consistently higher in HPDA cultures.
In contrast, HPDA infected with a suspension of HHV-6B (Z29 strain) containing the same amount of viral DNA copies showed a gradual decrease in the viral DNA load over time (Fig. 3C). HPDA cells appeared to not support a productive HHV-6B infection, as demonstrated by a greater than 3-log decrease by day 7 in both the intracellular and extracellular viral load compared to the initial inoculum. The inability of HHV-6B to replicate in HPDA appeared to be cell specific, since HHV-6B infection was productive in SupT-1 cells (Fig. 3D). In these cultures, levels of HHV-6B were detected both intracellularly and in cell-free supernatants (1 × 109 copies/106 cells or/ml). These results further support the observations that HHV-6A is associated with productive viral infection in HPDA, while HHV-6B is not.
Infection of HPDA with HHV-6B is nonproductive and associated with mRNA transcription.
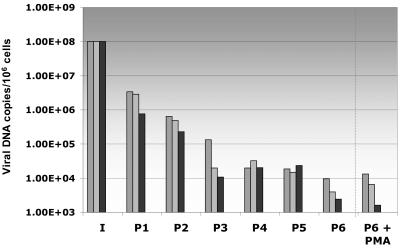
As HHV-6A-infected HPDA resulted in a productive viral infection associated with high viral loads and CPE, these cultures were difficult to maintain for greater than 7 days postinfection. However, the nonlytic infection of HPDA with HHV-6B afforded us the opportunity to propagate these cells for multiple passages. HHV-6B-infected HPDA were cultured for 6 weeks (cell cultures were split when confluent approximately once per week), and intra- and extracellular HHV-6B DNA load was measured at each passage using quantitative PCR. The results from three independent experiments are shown in Fig. 4, in which there was a consistent decrease in intracellular viral loads at each passage. HHV-6B DNA appeared to plateau after the fourth passage at a viral load of approximately 1 × 104 viral copies/106 cells. These results are consistent with the hypothesis that HPDA do not support productive HHV-6B infection.
FIG. 4.
Quantitative detection of intracellular viral DNA in HHV-6B-infected HPDA at subsequent passages and 3 days after PMA treatment. In these cultures, the viral load decreases with number of passages; treatment of infected cultures with PMA has no effect on the viral load. Each bar with the same filling represents one separate experiment.
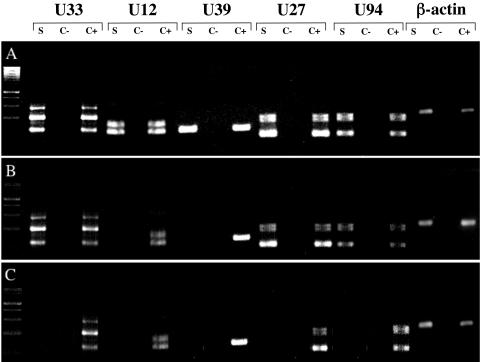
While low levels of HHV-6B DNA could be demonstrated in infected HPDA cultures (Fig. 4), it was of interest to determine if this was associated with the detection of HHV-6 mRNA for a variety of viral gene transcripts. Table 1 describes the reported function of five viral mRNA genes analyzed in this study (16). Immediate-early HHV-6 genes were represented by U94 transcripts: early genes were represented by U12, U27, and U33, and late genes were represented by U39. Of interest, the U94 gene has been reported to be associated with HHV-6 latency (28). HPDA and control SupT-1 cultures were infected with HHV-6A or HHV-6B and were tested for transcription using an RT-PCR assay. As shown in Fig. 5A (as a representative example of six separate experiments), all five HHV-6 transcripts were detectable in control HHV-6A-infected HPDA. In contrast, only U94, U27, and U33 were detected early (from day 1 to day 7 postinfection) in HHV-6B-infected HPDA (Fig. 5B). By the third passage (Fig. 4), no viral mRNA transcripts were detected in HPDA (Fig. 5C). In contrast, SupT-1 cells infected with either HHV-6A or HHV-6B were always positive for all five transcripts. The detection of viral DNA in cultures of HHV-6B-infected HPDA in the absence of detectable viral mRNA suggested the possibility that HHV-6B could establish a latent infection. However, attempts to reactivate HHV-6 using PMA (performed in three separate experiments) gave no appreciable increase in intracellular viral load at 3 days after PMA treatment (Fig. 4), and mRNA transcripts remained undetectable (data not shown).
TABLE 1.
Primers used in RT-PCRa
| Coding ORF | Functionb | Primers | cDNA length (bp) | DNA length (bp) |
|---|---|---|---|---|
| U33 | Capsid protein | Primary F, GACAGCGCTGACACTGAAAA | 174 | 174 |
| Primary R, CGGTGATCGGAACAAATTCT | ||||
| Nested F, CGGTCACGTTGTAGAGATGC | 117 | 117 | ||
| Nested R, CGGTGATCGGAACAAATTCT | ||||
| U12 | G-protein-coupled receptor | Primary F, CACTGTCATTGAGCTGTCCAA | 238 | 327 |
| Primary R, ACCACATGAGCACAAAATCG | ||||
| Nested F, CACTGTCATTGAGCTGTCCAA | 113 | 202 | ||
| Nested R, TGAGCGTGATTCCGGTAACT | ||||
| U39 | Glycoprotein B | Nested F, CCATACCCTCCTCCTTTTCC | 129 | 129 |
| Nested R, CTTTCCTTATGCCACCAATCC | ||||
| U27 | DNA polymerase processivity factor | Primary F, CGGGAACATAGAGAAACGAGAG | 209 | 209 |
| Primary R, GACAAAAACAAACATTCCGCC | ||||
| Nested F, CTACAGTGACTTTCACGCC | 100 | 100 | ||
| Nested R, GACAAAAACAAACATTCCGCC | ||||
| U94 | Transactivation, AAV-2 | Primary F, GCATACGTGCACCAATCATC | 168 | 168 |
| Primary R, ACGCTCAAGCGGAGAATAAA | ||||
| Nested F, GCATACGTGCACCAATCATC | 100 | 100 | ||
| Nested R, GCGCAACGATAGGAAAACAC | ||||
| Human β-actin | Primary F, CAAGAGATGGCCACGGCTGCT | 275 | 340 | |
| Primary R, TCCTTCTGCATCCTGTCGGCA |
ORF, open reading frame; AAV-2, adeno-associated virus type 2.
See reference 16.
FIG. 5.
Detection of viral transcripts by RT-PCR in HPDA infected with HHV-6A or HHV-6B. RT-PCR for U33, U12, U39, U27, and U94 at day 3 postinfection is shown. Each set of primers was tested on HPDA cDNA (S), nonretrotranscribed RNA as a negative control (C−), and a positive control of cDNA obtained from SupT-1 cultures infected with either viral variant (C+). (A) HPDA infected with U1102. (B) HPDA at day 3 postinfection with HHV-6B strain Z29. (C) After the third passage, no viral transcripts were detected in HPDA infected with Z29. β-actin was used as an internal control.
HHV-6A uses the SCR-2 domain of CD46 as its receptor in HPDA.
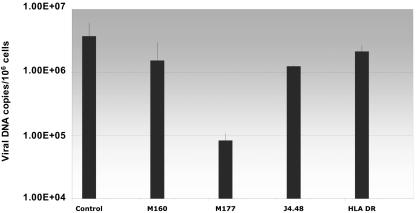
The ubiquitous cell surface molecule CD46 has been demonstrated to be a receptor for a variety of microbial organisms including HHV-6 (6). However, different agents may use different extracellular domains of CD46 (18). It was therefore of interest to determine which SCR domain of CD46 is used by HHV-6A in infected HPDA. SCR-specific anti-CD46 monoclonal antibodies were used to demonstrate the requirement of this surface protein for HHV-6 entry. HPDA were infected with HHV-6A in the presence of the different blocking monoclonal antibodies. As shown in Fig. 6, incubation with a monoclonal antibody that blocks the SCR-2 domain (M177) resulted in a greater than 1-log reduction in viral load compared to that of HHV-6A-infected HPDA (no antibody) or virus-infected cells incubated with an irrelevant antibody (HLA-DR). In addition, incubation with the anti-SCR-2 monoclonal antibody also resulted in inhibition of CPE (data not shown). Incubation with monoclonal antibodies to the SCR-3 domain (M160) and SCR-4 (J4.48) had no effects on HHV-6A virus loads (Fig. 6) and did not interfere with HHV-6A CPE in infected HPDA. These results suggest that HHV-6A utilizes the SCR-2 domain of CD46 as a cellular receptor to infect HPDA.
FIG. 6.
Viral load in HPDA 7 days after infection with HHV-6A strain U1102 and effect of blocking monoclonal antibodies against CD46. Preincubation of HPDA with the monoclonal antibody M177 (against SCR2), but not with M160 (against SCR3), J4.48 (against SCR1), or antibody anti-HLA-DR, significantly decreases the viral load in these cells compared to control (no antibody).
DISCUSSION
The aim of the present study was to compare and define the tropism of the HHV-6A and HHV-6B variants for human CNS material. This has become increasingly significant, as there is a greater appreciation for the role that these viruses play in neurological disease (11, 33). An extensive study on a large cohort of children and adults documented a relative predominance of HHV-6A in cerebral spinal fluid from children. Those authors claimed that this was suggestive of a greater neurotropic potential of HHV-6A (19) and is in agreement with the detection of HHV-6A in autopsy brain material three times more frequently than HHV-6B (9). The results in this study are strongly supportive for a variant-specific tropism and infection pattern for HHV-6 in HPDA. We have demonstrated substantial differences in infection efficacy and outcome between the two viral variants. HHV-6A gave rise to a productive infection with visible CPE, IFA positivity, and maturing viral particles. In contrast, variant B-infected HPDA had low intra- and extracellular viral DNA that decreased over time with no CPE or viral particles detectable by electron microscopy.
The results of HHV-6A infection in HPDA are in agreement with previous investigations on variant-specific infection of fetal astrocytes where HHV-6A (GS strain) was associated with a productive virus infection (20). However, HHV-6B was also stated (data not shown) to induce visible CPE and syncytia on fetal astrocytes, though at a lower level than HHV-6A-infected cultures (20). The data reported in the present study, demonstrating nonproductive infection of astrocytes with HHV-6B, are consistent with more recent reports on HHV-6 variant infection of astrocytic cells (10, 42). Using a chemiluminescent assay, viral DNA could be detected (10) in U373 MG astrocytic cell line infected with HHV-6A but not with HHV-6B. Additionally, decreasing titers of HHV-6B DNA were demonstrated in infected astrocytoma cell line U251 (42). As these latter studies used transformed astrocytic cell lines, we endeavored to utilize a possibly more relevant model system of human astrocytes derived from human fetal progenitor cells. HPDA are well characterized, and all cells were shown to be GFAP positive and lack markers for neurons and oligodendrocytes (23). Collectively, these results support the hypothesis that the two variants of HHV-6 have differential effects in human astrocytes.
The inability of HHV-6B to productively infect HPDA lends insight into possible mechanisms by which the virus fails to grow in these cells. One possibility could be a decreased ability or inability to bind and enter into infected HPDA, or alternatively, there could be a block in viral replication after entry. It has been shown that the HHV-6 receptor CD46 is necessary but not sufficient to enable HHV-6 fusion/entry, since some human T-cell lines were found to be nonpermissive for HHV-6-mediated fusion and replication despite the presence of significant surface levels of CD46 (31). Therefore, HHV-6B might require an additional factor(s) (coreceptor) in addition to CD46 to enter cells and induce cell-cell fusion. This possibility might explain the different cell-specific tropism and fusion capacity between HHV-6A and HHV-6B. Evidence has been presented in this report that underlines the crucial relevance of the SCR-2 domain of CD46 in HHV-6A binding to HPDA. In contrast, it has been reported that the SCR-2 and SCR-3 domains of CD46 were involved in HHV-6A binding to lymphocytes (18, 30). Ongoing experiments are exploring mechanisms of HHV-6B in HPDA.
The data presented in this study show that HHV-6B was able to bind to its cellular receptor and enter cells. Therefore, it is likely that a block in HHV-6B viral replication after entry is associated with nonproductive infection of this variant in HPDA, which is supported by the observation that HPDA expresses CD46 (data not shown) and that viral DNA and RNA transcripts could be detected early in infection. As quantitative real-time PCR for viral load determinations was used to measure HHV-6B growth in infected HPDA, we took great lengths to remove viral particles that may adhere to cell surfaces (the cells were washed several times and detached using trypsin prior to DNA extraction). Therefore, any viral DNA detected in HHV-6B-infected HPDA, though decreasing over time, corresponded to HHV-6 after entry (31). Moreover, HHV-6B-infected HPDA were positive for the immediate-early RNA transcript U94 and for the early transcripts U27 and U33 up to the second passage and demonstrated weak reactivity for the viral glycoprotein gp116/54/64 expression detected by IFA during the first week of infection.
Of major interest in this study was that nonproductive HHV-6B infection in HPDA also resulted in long-term cultures (at least six passages) that contained low levels of HHV-6B viral DNA in the absence of detectable message. The ability to detect viral DNA without mRNA transcripts might suggest either a latent or abortive infection. Although the mechanisms of HHV-6 latency are poorly understood, it has been suggested that the U94 gene, coding for a human parvovirus, adeno-associated virus type 2 (36), is a latency-associated transcript (28). In HHV-6B-infected HPDA, the U94 mRNA transcript was undetectable after the first week of infection. Attempts to reactivate HHV-6 from these cultures with PMA did not result in detectable viral transcription and were consistent with a recent report where PMA also failed to reactivate HHV-6B in the U373 astrocytoma cell line (10). The inability to reactivate HHV-6B would suggest an abortive rather than latent infection of HPDA; however, there is the possibility that stimuli other than PMA or phorbol esters might be effective in HHV-6B reactivation.
The differential tropism of HHV-6A or HHV-6B in human astrocytes has implications for the role of these variants in human neurological disease. While human astrocytes are susceptible for HHV-6 B infection, HHV-6A appears to be more lytic and is associated with a productive infection in this cell type. In disease in which HHV-6A has been implicated (34), this viral variant could reach the CNS via infected inflammatory cells. The hallmark of the MS lesion is inflammation in which both oligodendrocytes (17) and astrocytes have been shown to contain HHV-6 (7, 27). Direct viral infection of these cells may be associated with abundant expression of viral antigens and might contribute to an immune-mediated response against viral antigens or shared sequence or conformational homology with self-proteins (molecular mimicry). It has long been suggested that a possible triggering mechanism in the pathogenesis of MS may be molecular mimicry between microbial and self (myelin)-antigens, leading to the activation of autoreactive T cells (38). Consistent with this hypothesis is an increased HHV-6A-specific T-cell responses in the peripheral blood of MS patients (34) and cross-reactive anti-HHV-6 CD4+ T cells with myelin basic proteins in MS patients (8, 35).
In contrast to HHV-6A-infected astrocytes, astrocytes infected with HHV-6B may preferentially establish a nonproductive infection. Persistent virus infection of the CNS with subsequent reactivations (a common feature of herpesviruses) may give rise to a productive infection and recurrence of periodic immune responses that may be immunopathogenic. Triggers that can reactivate HHV-6B in astrocytes are unknown and are currently being investigated, although profound immunosuppression has been associated with virus reactivation. This may account for HHV-6-associated encephalitis that has been demonstrated in some patients after allogeneic bone marrow transplantation (37, 39). The ability of viruses to establish latency in cells can also result in aberration of so called “luxury functions” without cytopathology (26). If, for example, HHV-6B-infected astrocytes have altered glutamate uptake (15), this could adversely affect the homeostatic balance of excitotoxic mediators possibly resulting in neuronal dysfunction (14).
In conclusion, the data presented in this study provide insight into the understanding of HHV-6 and CNS disease. Astrocytes may be an important reservoir for this virus in which differential tropism of HHV-6A and HHV-6B may be associated with different disease outcomes.
Acknowledgments
We thank Susan Cheng, Virginia Crocker, and Rite Azzam at the NINDS Electron Microscopy facility for technical help. We also thank Nahid Akhyani and Julie Fotheringham for their help and discussion.
J.A. was supported by the KI-NIH Graduate Partnership Program.
REFERENCES
- 1.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545-552. [DOI] [PubMed] [Google Scholar]
- 2.Akhyani, N., R. Berti, M. B. Brennan, S. S. Soldan, J. M. Eaton, H. F. McFarland, and S. Jacobson. 2000. Tissue distribution and variant characterization of human herpesvirus (HHV)-6: increased prevalence of HHV-6A in patients with multiple sclerosis. J. Infect. Dis. 182:1321-1325. [DOI] [PubMed] [Google Scholar]
- 3.Albright, A. V., E. Lavi, J. B. Black, S. Goldberg, M. J. O'Connor, and F. Gonzalez-Scarano. 1998. The effect of human herpesvirus-6 (HHV-6) on cultured human neural cells: oligodendrocytes and microglia. J. Neurovirol. 4:486-494. [DOI] [PubMed] [Google Scholar]
- 4.Araque, A., G. Carmignoto, and P. G. Haydon. 2001. Dynamic signaling between astrocytes and neurons. Annu. Rev. Physiol. 63:795-813. [DOI] [PubMed] [Google Scholar]
- 5.Braun, D. K., G. Dominguez, and P. E. Pellett. 1997. Human herpesvirus 6. Clin. Microbiol. Rev. 10:521-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattaneo, R. 2004. Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens' magnet. J. Virol. 78:4385-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cermelli, C., R. Berti, S. S. Soldan, M. Mayne, J. M. D'Ambrosia, S. K. Ludwin, and S. Jacobson. 2003. High frequency of human herpesvirus 6 DNA in multiple sclerosis plaques isolated by laser microdissection. J. Infect. Dis. 187:1377-1387. [DOI] [PubMed] [Google Scholar]
- 8.Cirone, M., L. Cuomo, C. Zompetta, S. Ruggieri, L. Frati, A. Faggioni, and G. Ragona. 2002. Human herpesvirus 6 and multiple sclerosis: a study of T cell cross-reactivity to viral and myelin basic protein antigens. J. Med. Virol. 68:268-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuomo, L., P. Trivedi, M. R. Cardillo, F. M. Gagliardi, A. Vecchione, R. Caruso, A. Calogero, L. Frati, A. Faggioni, and G. Ragona. 2001. Human herpesvirus 6 infection in neoplastic and normal brain tissue. J. Med. Virol. 63:45-51. [PubMed] [Google Scholar]
- 10.De Bolle, L., J. Van Loon, E. De Clercq, and L. Naesens. 2005. Quantitative analysis of human herpesvirus 6 cell tropism. J. Med. Virol. 75:76-85. [DOI] [PubMed] [Google Scholar]
- 11.Dewhurst, S. 2004. Human herpesvirus type 6 and human herpesvirus type 7 infections of the central nervous system. Herpes 11:105A-111A. [PubMed] [Google Scholar]
- 12.Dietrich, J., B. M. Blumberg, M. Roshal, J. V. Baker, S. D. Hurley, M. Mayer-Proschel, and D. J. Mock. 2004. Infection with an endemic human herpesvirus disrupts critical glial precursor cell properties. J. Neurosci. 24:4875-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donati, D., N. Akhyani, A. Fogdell-Hahn, C. Cermelli, R. Cassiani-Ingoni, A. Vortmeyer, J. D. Heiss, P. Cogen, W. D. Gaillard, S. Sato, W. H. Theodore, and S. Jacobson. 2003. Detection of human herpesvirus-6 in mesial temporal lobe epilepsy surgical brain resections. Neurology 61:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotheringham, J., J. Ahlqvist, R. Bonwetsch, N. Akhyani, and S. Jacobson. 2004. Infection of glia with human herpesvirus-6 dysregulates glutamate uptake. J. Neurovirol. 10:I49. [Google Scholar]
- 16.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 17.Goodman, A. D., D. J. Mock, J. M. Powers, J. V. Baker, and B. M. Blumberg. 2003. Human herpesvirus 6 genome and antigen in acute multiple sclerosis lesions. J. Infect. Dis. 187:1365-1376. [DOI] [PubMed] [Google Scholar]
- 18.Greenstone, H. L., F. Santoro, P. Lusso, and E. A. Berger. 2002. Human herpesvirus 6 and measles virus employ distinct CD46 domains for receptor function. J. Biol. Chem. 277:39112-39118. [DOI] [PubMed] [Google Scholar]
- 19.Hall, C. B., M. T. Caserta, K. C. Schnabel, C. Long, L. G. Epstein, R. A. Insel, and S. Dewhurst. 1998. Persistence of human herpesvirus 6 according to site and variant: possible greater neurotropism of variant A. Clin. Infect. Dis. 26:132-137. [DOI] [PubMed] [Google Scholar]
- 20.He, J., M. McCarthy, Y. Zhou, B. Chandran, and C. Wood. 1996. Infection of primary human fetal astrocytes by human herpesvirus 6. J. Virol. 70:1296-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hourcade, D., M. K. Liszewski, M. Krych-Goldberg, and J. P. Atkinson. 2000. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology 49:103-116. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. S., K. S. Lee, J. H. Park, M. Y. Kim, and W. S. Shin. 2000. Detection of human herpesvirus 6 variant A in peripheral blood mononuclear cells from multiple sclerosis patients. Eur. Neurol. 43:170-173. [DOI] [PubMed] [Google Scholar]
- 23.Messam, C. A., J. Hou, R. M. Gronostajski, and E. O. Major. 2003. Lineage pathway of human brain progenitor cells identified by JC virus susceptibility. Ann. Neurol. 53:636-646. [DOI] [PubMed] [Google Scholar]
- 24.Mori, Y., T. Seya, H. L. Huang, P. Akkapaiboon, P. Dhepakson, and K. Yamanishi. 2002. Human herpesvirus 6 variant A but not variant B induces fusion from without in a variety of human cells through a human herpesvirus 6 entry receptor, CD46. J. Virol. 76:6750-6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitsche, A., C. W. Muller, A. Radonic, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2001. Human herpesvirus 6A DNA is detected frequently in plasma but rarely in peripheral blood leukocytes of patients after bone marrow transplantation. J. Infect. Dis. 183:130-133. [DOI] [PubMed] [Google Scholar]
- 26.Oldstone, M. B. 2002. Travels along the viral-immunobiology highway. Immunol. Rev. 185:54-68. [DOI] [PubMed] [Google Scholar]
- 27.Opsahl, M. L., and P. G. Kennedy. 2005. Early and late HHV-6 gene transcripts in multiple sclerosis lesions and normal appearing white matter. Brain 128:516-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rotola, A., T. Ravaioli, A. Gonelli, S. Dewhurst, E. Cassai, and D. Di Luca. 1998. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. USA 95:13911-13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salahuddin, S. Z., D. V. Ablashi, P. D. Markham, S. F. Josephs, S. Sturzenegger, M. Kaplan, G. Halligan, P. Biberfeld, F. Wong-Staal, B. Kramarsky, et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596-601. [DOI] [PubMed] [Google Scholar]
- 30.Santoro, F., H. L. Greenstone, A. Insinga, M. K. Liszewski, J. P. Atkinson, P. Lusso, and E. A. Berger. 2003. Interaction of glycoprotein H of human herpesvirus 6 with the cellular receptor CD46. J. Biol. Chem. 278:25964-25969. [DOI] [PubMed] [Google Scholar]
- 31.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 32.Seth, P., F. Diaz, J. H. Tao-Cheng, and E. O. Major. 2004. JC virus induces nonapoptotic cell death of human central nervous system progenitor cell-derived astrocytes. J. Virol. 78:4884-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soldan, S. S., and S. Jacobson. 2001. Role of viruses in etiology and pathogenesis of multiple sclerosis. Adv. Virus Res. 56:517-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soldan, S. S., T. P. Leist, K. N. Juhng, H. F. McFarland, and S. Jacobson. 2000. Increased lymphoproliferative response to human herpesvirus type 6A variant in multiple sclerosis patients. Ann. Neurol. 47:306-313. [PubMed] [Google Scholar]
- 35.Tejada-Simon, M. V., Y. C. Zang, J. Hong, V. M. Rivera, and J. Z. Zhang. 2003. Cross-reactivity with myelin basic protein and human herpesvirus-6 in multiple sclerosis. Ann. Neurol. 53:189-197. [DOI] [PubMed] [Google Scholar]
- 36.Thomson, B. J., F. W. Weindler, D. Gray, V. Schwaab, and R. Heilbronn. 1994. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology 204:304-311. [DOI] [PubMed] [Google Scholar]
- 37.Wainwright, M. S., P. L. Martin, R. P. Morse, M. Lacaze, J. M. Provenzale, R. E. Coleman, M. A. Morgan, C. Hulette, J. Kurtzberg, C. Bushnell, L. Epstein, and D. V. Lewis. 2001. Human herpesvirus 6 limbic encephalitis after stem cell transplantation. Ann. Neurol. 50:612-619. [DOI] [PubMed] [Google Scholar]
- 38.Wucherpfennig, K. W., and J. L. Strominger. 1995. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell 80:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa, T. 2003. Human herpesvirus-6 and -7 infections in transplantation. Pediatr. Transplant. 7:11-17. [DOI] [PubMed] [Google Scholar]
- 40.Yoshikawa, T. 2003. Significance of human herpesviruses to transplant recipients. Curr. Opin. Infect. Dis. 16:601-606. [DOI] [PubMed] [Google Scholar]
- 41.Yoshikawa, T., and Y. Asano. 2000. Central nervous system complications in human herpesvirus-6 infection. Brain Dev. 22:307-314. [DOI] [PubMed] [Google Scholar]
- 42.Yoshikawa, T., Y. Asano, S. Akimoto, T. Ozaki, T. Iwasaki, T. Kurata, F. Goshima, and Y. Nishiyama. 2002. Latent infection of human herpesvirus 6 in astrocytoma cell line and alteration of cytokine synthesis. J. Med. Virol. 66:497-505. [PubMed] [Google Scholar]
- 43.Zerr, D. M., L. C. Yeung, R. M. Obrigewitch, M. L. Huang, L. M. Frenkel, and L. Corey. 2002. Case report: primary human herpesvirus-6 associated with an afebrile seizure in a 3-week-old infant. J. Med. Virol. 66:384-387. [DOI] [PubMed] [Google Scholar]