Abstract
The role of the herpes simplex virus tegument protein VP22 is not yet known. Here we describe the characterization of a virus in which the entire VP22 open reading frame has been deleted. We show that VP22 is not essential for virus growth but that virus lacking VP22 (Δ22) displays a cell-specific replication defect in epithelial MDBK cells. Virus particles assembled in the absence of VP22 show few obvious differences to wild-type (WT) particles, except for a moderate reduction in glycoproteins gD and gB. In addition, the Δ22 virus exhibits a general delay in the initiation of virus protein synthesis, but this is not due to a glycoprotein-related defect in virus entry. Intriguingly, however, the absence of VP22 has an obvious effect on the intracellular level of the immediate-early (IE) protein ICP0. Moreover, following translocation from the nucleus to the cytoplasm, ICP0 is unable to localize to the characteristic cytoplasmic sites observed in a WT infection. We demonstrate that, in WT-infected cells, VP22 and ICP0 are concentrated in the same cytoplasmic sites. Furthermore, we show that, while ICP0 and ICP4 are components of WT extracellular virions, the altered localization of ICP0 in the cytoplasm of Δ22-infected cells correlates with an absence of both ICP0 and ICP4 from Δ22 virions. Hence, while a role has not yet been defined for virion IE proteins in virus infection, our results suggest that their incorporation is a specific event requiring the tegument protein VP22. This report provides the first direct evidence that VP22 influences virus assembly.
The tegument of the herpes simplex virus particle is the proteinaceous layer located between the capsid and envelope and is made up of at least 15 virus-encoded polypeptides (3). Although it comprises a major part of the virion, there are many aspects of tegument assembly and organization that are not understood. For instance, the cellular site(s) of tegument acquisition by the virus particle has not yet been conclusively identified, while the many molecular interactions involved in assembling this multicomponent compartment are not yet clear (reviewed in reference 27). The role of the tegument at virus entry has also received little attention despite the fact that the tegument is delivered to the cell immediately upon infection, implying its components may function at this very early stage of the infectious process. With a few notable exceptions, such as the transactivator of immediate-early gene expression VP16 (16, 17, 32) or the host shut-off protein encoded by UL41 (21, 29), there is little known about the role of individual tegument components at virus entry. Interestingly, there have been a number of reports that the immediate-early proteins ICP4 and ICP0 are also packaged into the tegument (34, 35), suggesting that the tegument could function as a direct source of these important transactivators prior to their de novo synthesis within the cell.
The herpes simplex virus type 1 (HSV-1) structural protein VP22 is one of the three major components of the HSV-1 tegument together with VP16 and VP13/14. In spite of its abundance within the virion, previously estimated to be around 2,400 molecules per particle (18), its role in virus infection, be it at virus entry or later in virus replication, has not yet been defined. Previous studies on VP22 have shown that it displays a complex range of localization patterns within the cell, some of which may provide insight into its role in virus infection. For example, we have previously shown that VP22 binds and stabilizes microtubules when expressed in isolation in the cell (8). We have also used a virus expressing green fluorescent protein (GFP)-tagged VP22 to show that it is predominantly cytoplasmic throughout infection and that it localizes to rapidly moving trafficking complexes that we believe to represent a stage of the tegument assembly pathway (10). Interestingly, we have also shown that VP22 binds to another major tegument protein, VP16 (6), an interaction which may provide some understanding of the overall process of tegument assembly.
The VP22-encoding gene UL49 is conserved throughout the alphaherpesvirus family. While a VP22-null virus for HSV-1 has not yet been described, the homologous UL49 genes have been successfully deleted from a number of other alphaherpesviruses, including bovine herpesvirus type 1 (BHV-1), pseudorabies virus (PRV), and Marek's disease virus (MDV), resulting in viruses with contrasting phenotypes. In the case of PRV, the deletion of the UL49 gene had no apparent effect on virus growth in either culture or animals (4). By contrast, BHV-1 grown in the absence of its UL49 gene replicated slowly in culture and exhibited greatly reduced virulence in cows (23), while the MDV UL49 gene is considered absolutely essential for growth in culture (5). It is unclear why the consequence of deleting the UL49 gene should be so different for these individual alphaherpesviruses, but the range of outcomes may provide useful insights into the replication strategies used by these viruses.
In this paper we describe the construction and analysis of an HSV-1 recombinant virus in which the GFP gene has replaced the entire UL49 gene. We show that the absence of VP22 has little effect on the virion content of major structural proteins, with the exception of a decrease in glycoproteins gD and gB. We also show that while this virus replicates in Vero cells to the level of wild-type virus, it exhibits a delay in the onset of viral protein synthesis and a replication defect in MDBK cells. Strikingly, the absence of VP22 is shown to alter the expression and subcellular localization of the immediate-early protein ICP0. Furthermore, we show that while our wild-type (WT) virions package both ICP0 and ICP4, as demonstrated by others (34, 35), neither of these immediate-early proteins is assembled into our Δ22 virions. Hence, we suggest that VP22 is required for the incorporation of these proteins into the virus structure and that the phenotype of the Δ22 virus may in part be due to its indirect effect on ICP0 and/or ICP4.
MATERIALS AND METHODS
Cells and viruses.
Vero, BHK, and MDBK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum. Viruses were routinely grown in BHK cells or Vero cells and titrated on Vero cells. Extracellular virions were purified on Ficoll gradients from the infected cell medium of 5 × 108 BHK cells, as described previously (10). Infectious virus DNA was also produced from extracellular virus, as described previously (10).
The VP22-expressing complementing cell line was constructed in EcR-293 cells using the inducible Ecdysone system (Invitrogen) and was grown in DMEM supplemented with 10% fetal calf serum, 400 μg/ml zeocin (Invitrogen), and 300 μg/ml Geneticin (Invitrogen). Cells were induced 24 h before virus infection using 1 μg/ml muristerone A (Invitrogen).
The parental virus strain used in this study was strain 17 (s17) of HSV-1. The recombinant virus expressing GFP-tagged VP22, based on s17, has been described previously (10). The Δ22 virus was constructed using the plasmid pGE169, which was based on plasmid pGE120 described previously (10), and consisted of the GFP open reading frame surrounded by the UL49 flanking sequences. Cotransfection was carried out between pGE169 and infectious s17 DNA in preinduced VP22-expressing 293 cells, and resulting fluorescent recombinant virus was plaque purified three times on the same cells before analysis. To construct a Δ22 virus rescue mutant, an intermediate virus expressing monomeric red fluorescent protein instead of GFP was first constructed. The monomeric red fluorescent protein gene was amplified by PCR from a plasmid kindly provided by Roger Tsien (1) and inserted into plasmid pGE120 (10) to make plasmid pWH3. This was cotransfected into Vero cells with infectious 169v DNA, and red fluorescent plaques were purified as virus WH3v. Infectious WH3v DNA was then cotransfected into Vero cells with plasmid pCP6 encoding the full-length UL49 gene within its flanking sequences, and resulting progeny virus was screened for the production of nonfluorescing plaques. The resulting rescued virus (Δ22R) was sequenced across the UL49 gene to ensure that recombination had taken place correctly.
Virus genomic DNA screening.
Virus DNA for restriction digestion was purified from 5 × 107 infected cells as described previously (10) and digested for 8 h with the appropriate enzymes in the presence of RNase A. Electrophoresis was carried out overnight in a 0.8% agarose gel, and the gel was transferred to a nylon membrane by standard procedures. The Southern blots were then hybridized with a 32P-labeled DNA probe synthesized by the random priming of fragments specific for UL49, UL47, or GFP.
Virus growth curves.
Vero or MDBK cells grown in a six-well plate (1 × 106 cells per well) were infected at a multiplicity of either 10 or 0.1 in 1 ml medium per well. After 1 h (taken as 1 h postinfection), the inoculum was removed, the cells were washed with phosphate-buffered saline (PBS), and 2 ml fresh medium was added to each well. At a range of times after infection, one well of infected cells was harvested for either total virus yield or both extracellular virus from the cell medium and intracellular virus from the cells. Each virus sample was then titrated on Vero cells. All plaque assays were carried out in DMEM supplemented with 2% newborn calf serum and 1% human serum (Harlan Sera-Lab).
Measurement of adsorption and penetration rates.
To measure virus adsorption, monolayers of Vero cells were chilled to 4°C for 1 h. Approximately 300 PFU of virus was added to each dish in 0.5 ml of medium, and the dishes were left at 4°C. At various time points ranging from 5 to 180 min, the inoculum was removed and the cells were washed twice with cold medium and overlaid with medium containing 1% human serum. To measure penetration rate, monolayers of Vero cells were chilled to 4°C for 1 h prior to infection with 300 PFU of virus. The dishes were incubated at 4°C for 2 h to allow virus adsorption and then transferred to 37°C, and at various times ranging from 0 to 60 min, the monolayers were washed with a pH 3 citrate buffer before being overlaid with medium containing 1% human serum.
Antibodies.
Monoclonal antibodies to α-tubulin, GFP, and HSV-1 major capsid protein VP5 were obtained from Sigma, Clontech, and Autogen Bioclear, respectively. The monoclonal antibodies against ICP4 and ICP0 used in Western blotting were obtained from Virusys. The anti-VP22 polyclonal antibody AGV30 has been described previously (9). The monoclonal antibodies against VP16 (LP1) and gD (LP14) were kindly provided by Tony Minson and Helena Browne. Monoclonal antibody 11060, used to detect ICP0 by immunofluorescence, was kindly provided by Roger Everett.
In vivo cell radiolabeling.
Medium from infected Vero cells grown in six-well plates was removed and replaced with 1 ml methionine-free DMEM supplemented with 2% dialyzed calf serum. After 30 min at 37°C, 25 μCi of [35S]methionine was added to the medium and the cells were labeled for an additional hour. Cells were washed three times in PBS to remove unincorporated radiolabel, resuspended in 400 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and analyzed by SDS-PAGE.
SDS-PAGE.
Protein samples were analyzed on 10% polyacrylamide gels. Following electrophoresis, gels were either stained with Coomassie blue or transferred to nitrocellulose for analysis by Western blotting. Western blots were processed using horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Bio-Rad) and developed using an enhanced chemiluminescence kit (Pierce).
Immunofluorescence.
Cells for immunofluorescence were grown on 16-mm coverslips in individual wells of a six-well plate. Cells were fixed for 10 min in 100% methanol or 20 min in 4% paraformaldehyde, followed by permeabilization for 10 min with 0.5% Triton X-100. The fixed cells were blocked by incubation for 30 min in PBS containing 10% newborn calf serum, and primary antibody was added for 30 min in the same solution. Following extensive washing in PBS, fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G or Texas Red-conjugated anti-mouse immunoglobulin G (Vector Labs) was added in block solution and the mixture was incubated for a further 30 min. The coverslips were then washed extensively in PBS and mounted in Vectashield (Vector Labs). Images were acquired using a Zeiss LSM410 confocal microscope and processed using Adobe Photoshop software.
RESULTS
Deletion of the VP22-encoding gene (UL49) from the HSV-1 genome.
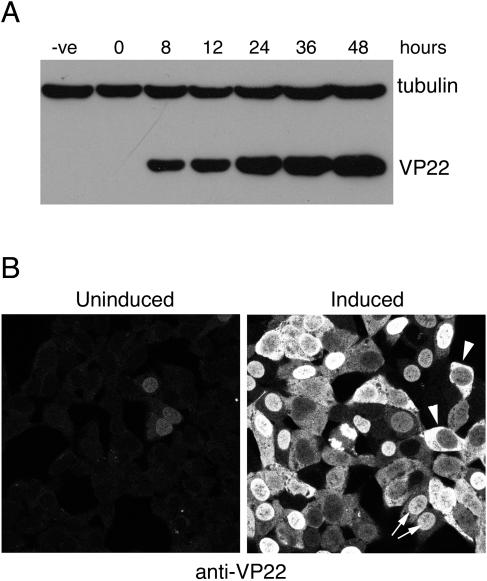
While it was not yet known whether VP22 was essential for the replication of HSV-1, our previous attempts to construct a UL49 deletion mutant had consistently failed. Therefore, to facilitate the propagation of a UL49 deletion mutant, we first decided to produce a complementing cell line that expressed high levels of VP22. Our initial attempts to develop cell lines constitutively expressing VP22 also failed, suggesting that high levels of VP22 may not be favorable for cell growth. Thus, we decided to utilize the Ecdysone inducible expression system (Invitrogen) to produce a 293 cell line expressing VP22 under the control of an inducible promoter. Characterization of these cells showed that a high level of VP22 was expressed as early as 8 h after induction with the Ecdysone analogue muristerone A (Fig. 1A). Furthermore, when we examined the subcellular localization of VP22 in these cells by immunofluorescence, it was clear that in some cells the protein was in the nucleus (Fig. 1B), while in others it was located in the cytoplasm (Fig. 1B). Interestingly, these contrasting localization patterns can probably be explained by our previous studies on the subcellular localization of VP22, where we demonstrated a cytoplasmic to nuclear translocation of the protein through the cell cycle (7). As it was not yet clear whether VP22 functioned in the cytoplasm or nucleus during the HSV-1 replication cycle, we reasoned that at least a subpopulation of the complementing cell line would contain VP22 in the correct subcellular compartment to rescue any defect produced by the absence of virus-encoded VP22.
FIG. 1.
Characterization of an inducible VP22-expressing 293 cell line. (A) The VP22-293-EcR cell line was induced to express VP22 with 1 μg/ml muristerone A, and samples were harvested at various times after induction and analyzed by SDS-PAGE and Western blotting with antibodies against VP22 and α-tubulin. (B) VP22-293-EcR cells grown on coverslips were left uninduced or induced with muristerone A, fixed 24 h later, and stained with polyclonal anti-VP22. Arrows indicate cells with VP22 in the nucleus; arrowheads indicate cells with VP22 in the cytoplasm. -ve, parental 293-EcR cell line.
To delete the UL49 gene from the HSV-1 genome, we constructed a plasmid containing the open reading frame for GFP surrounded by the flanking sequences of the UL49 open reading frame, such that GFP was under the control of the UL49 promoter. This plasmid was cotransfected with infectious HSV-1 DNA (strain 17) into VP22-expressing 293 cells that had been induced to express 24 h earlier. Potential recombinant plaques were screened by GFP fluorescence and subjected to three rounds of plaque purification on the same cells.
Characterization of the HSV-1 UL49 deletion mutant (Δ22).
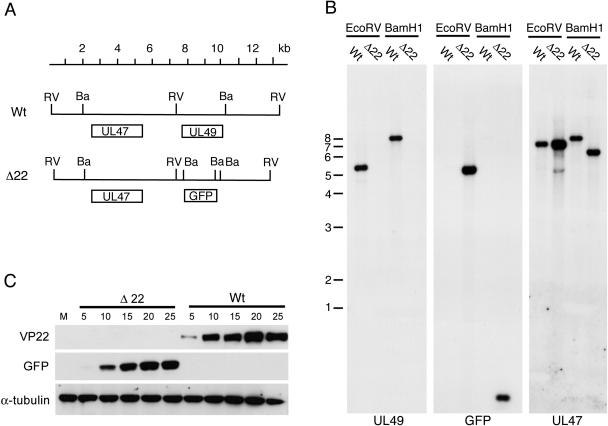
Recombinant virus was first checked by Southern blotting of restriction enzyme-digested genomic DNA, indicating that the GFP open reading frame had recombined into the HSV-1 genome in place of the UL49 gene (Fig. 2A and B). We next tested the ability of this Δ22 virus to grow on noncomplementing Vero cells. Interestingly, titrations of the Δ22 virus on monolayers of Vero cells resulted in the formation of plaques in this noncomplementing cell line, indicating that HSV-1 VP22 is dispensable for virus growth in Vero cells (data not shown). The lack of VP22 expression from the Δ22 virus was confirmed by Western blotting of Vero cell extracts taken at various times following infection at a high multiplicity (Fig. 2C). These samples were also blotted for GFP to show that this protein was expressed in the Δ22 virus-infected cells in place of VP22 (Fig. 2C). Taken together, these results demonstrate that VP22 is not essential for the growth of HSV-1 in Vero cells, and hence, all further experiments in this study were carried out with virus stocks grown on noncomplementing cells.
FIG. 2.
Characterization of the Δ22 virus. (A) DNA map of the HSV-1 genome across the UL49 gene, showing the relevant BamHI (Ba) and EcoRV (RV) sites. (B) Southern blotting of EcoRV- and BamHI-digested genomic DNA from WT and Δ22 viruses. The blots have been hybridized with probes for the UL49, GFP, and UL47 genes. (C) Monolayers of Vero cells were infected with either WT or Δ22 virus at a multiplicity of 10 and harvested at various time points ranging from 5 to 25 h. Equal amounts of total cell lysates were analyzed by SDS-PAGE followed by Western blotting with antibodies against VP22, GFP, and α-tubulin. Numbers to the left indicate DNA size markers in kilobases.
Virus assembled in the absence of VP22 exhibits reduced levels of glycoproteins gD and gB.
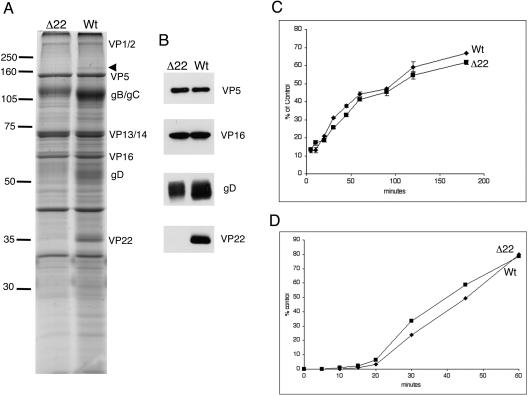
To investigate the effect of deleting the VP22-encoding gene on the virus assembly process, extracellular virions were purified from BHK cells infected with either WT or Δ22 virus and analyzed by SDS-PAGE. Notably, the absence of VP22 in these infected cells had little effect on the overall profile of the virus particles (Fig. 3A), in spite of VP22 being considered a major tegument protein. Importantly, there was no apparent difference in the incorporation of the other major tegument proteins, VP1/2, VP13/14, and VP16, suggesting that VP22 is not required to maintain the correct ratios of any of these proteins in the structure of the virus (Fig. 3A). However, the major differences that we observed in the virion profile of particles lacking VP22 was a reduction in the intensity of the bands present around the size of the envelope proteins gB/gC and gD, as indicated by the Coomassie blue-stained profile of the virions (Fig. 3A). Western blotting of virions was used to show a reduction in the level of gD in the Δ22 virions compared to the relative levels of the major capsid protein VP5 and the tegument protein VP16 (Fig. 3B). Further studies showed that the level of gD incorporated into the Δ22 virions was generally two- to threefold lower than in WT virions as judged by Western blotting, although this value varied between different virion preparations (data not shown). While the Δ22 virions also showed an alteration in their profile around the gB/gC region of the stained gel (Fig. 3A), Western blotting with anti-gB or anti-gC antibodies (data not shown) did not show a convincing reduction in the levels of either of these glycoproteins in the Δ22 virions. This result may be due to a lack of linearity in the transfer and detection of these glycoproteins during the Western blotting process. Nonetheless, Coomassie staining of a number of different virion preparations consistently indicated a reduction in the levels of the gD and gB/gC bands present in the Δ22 virions.
FIG. 3.
Characterization of HSV-1 virions assembled in the absence of VP22. (A) Equivalent amounts of purified extracellular WT and Δ22 virions were analyzed by SDS-PAGE and stained with Coomassie blue. Major virion proteins are labeled. The arrowhead denotes a species around 175 kDa that is absent from Δ22 virions. Numbers on the left indicate molecular mass in kilodaltons. (B) The same samples shown in panel A were analyzed by Western blotting with antibodies against the indicated virion proteins. (C) Monolayers of Vero cells were infected at 4°C with approximately 300 PFU of virus stock. At the indicated times, the cells were washed and transferred to 37°C, and plaques were allowed to develop. (D) Monolayers of Vero cells were infected with approximately 300 PFU of virus stock and incubated at 4°C for 2 h. Cells were transferred to 37°C and washed with citrate buffer (pH 3) at the indicated times, and plaques were allowed to develop.
As the Δ22 virus particles exhibited reduced levels of glycoproteins, we next wanted to investigate whether this was reflected in the ability of the virus to enter cells. Thus, we carried out both attachment and penetration assays on the WT and Δ22 viruses in Vero cells, the results of which are shown in Fig. 3C and D, respectively. These data clearly show that the Δ22 virus attaches to cells and penetrates the cell membrane as efficiently as WT virus, in spite of its reduced glycoprotein content.
HSV-1 lacking VP22 exhibits a delay in the onset of virus protein synthesis.
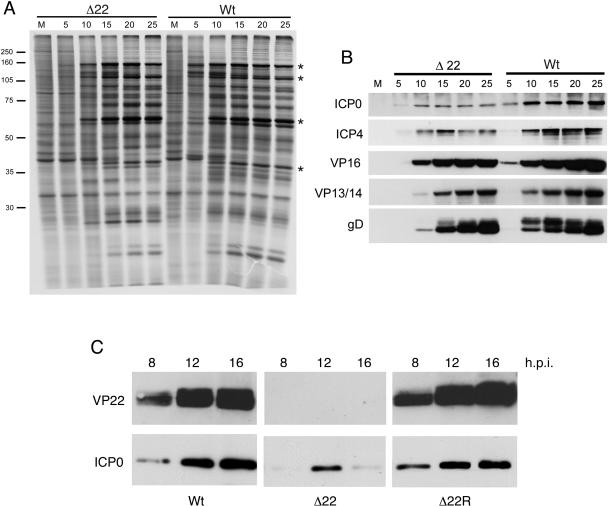
We next wished to investigate the characteristics of the Δ22 virus during the replication process. We first examined the Δ22 virus by investigating the overall levels of virus protein synthesis within the infected cell. Monolayers of Vero cells were infected with either WT or Δ22 virus at a multiplicity of 10, and then at a range of times after infection, the monolayers were pulse-labeled with [35S]methionine for a period of 2 h. The monolayers were then lysed and the samples subjected to SDS-PAGE. Under these conditions, it can be seen that in a WT infection, virus-specific proteins were detectable as early as 5 h after infection (Fig. 4A, compare M and 5-h profiles of WT samples). By contrast, in the Δ22 virus-infected cells, virus-specific proteins were not detectable until 10 h after infection (Fig. 4A). Nonetheless, by 20 h, the protein expression profiles for the two viruses were similar, suggesting that at later times the Δ22 virus had recovered from any early defects in its ability to express virus genes (Fig. 4A, compare 20-h profiles in Δ22 and WT).
FIG. 4.
The Δ22 virus exhibits an early delay in the infectious process. (A) Monolayers of Vero cells were infected with either WT or Δ22 virus stocks at a multiplicity of 10 followed by pulse-labeling with [35S]methionine at various times after infection. The cells were harvested and analyzed by SDS-PAGE and autoradiography. Asterisks indicate virus-induced proteins. Numbers on the left indicate molecular masses in kilodaltons. (B) Monolayers of Vero cells were infected with the same WT or Δ22 virus stocks at a multiplicity of 10, and samples were harvested at the indicated times after infection and analyzed by SDS-PAGE and Western blotting with antibodies against virus proteins as indicated. (C) Monolayers of Vero cells were infected with WT, Δ22, or Δ22R virus at a multiplicity of 10, and samples were harvested at the indicated times after infection and analyzed by SDS-PAGE and Western blotting with antibodies against VP22 and ICP0. h.p.i., hours postinfection.
To determine whether the delay in virus protein synthesis was a general effect or a specific down-regulation of individual genes, we next examined the expression rates of several individual virus proteins in a high multiplicity time course. Western blots for a number of representative proteins indicated a slight delay in the synthesis of the tegument protein VP16, which was detectable in WT virus-infected cells at 5 h after infection but not in Δ22 virus-infected cells (Fig. 4B). This was also the case for the tegument protein VP13/14 and the glycoprotein gD (Fig. 4B). However, there were clear differences in the expression profiles of the immediate-early protein ICP4 and, in particular, the immediate-early protein ICP0 (Fig. 4B). Hence, not only was the expression of ICP0 delayed but its intracellular level was also greatly reduced in Δ22 virus-infected cells compared to WT-infected cells (Fig. 4B). To ensure that this defect in ICP0 expression was specific to the deletion of the VP22-encoding gene, we went on to isolate a revertant of the Δ22 virus by rescuing the virus with a plasmid encoding the UL49 region of the HSV-1 genome (Δ22R). Western blotting of an infected cell time course confirmed that VP22 expression had been repaired in the Δ22R virus (Fig. 4C). We then investigated the level of ICP0 expression in cells infected with this revertant virus compared to the level of ICP0 in Δ22 virus-infected cells (Fig. 4C). As can be seen, ICP0 expression in Δ22R-infected cells is similar to the levels detected in WT-infected cells, confirming that this defect is specific to the absence of VP22.
The Δ22 virus replicates poorly in MDBK cells.
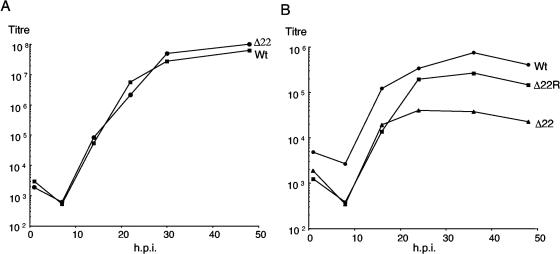
To investigate the overall replication efficiency of the Δ22 virus compared to WT virus, we carried out a number of growth curves with Vero cells. Monolayers of Vero cells were infected at either high or low multiplicity, and virus was harvested at a range of times after infection. In all cases, there was little difference between the replication efficiency of the two viruses, even when a low-multiplicity growth curve was carried out (Fig. 5A). Furthermore, we detected no difference in the ability of the Δ22 virus to be released from cells when we measured intracellular versus extracellular released virus levels (data not shown). Thus, in Vero cells at least, the absence of VP22 has little detectable effect on the HSV-1 replication cycle. By contrast, a low-multiplicity time course carried out with MDBK cells indicated a clear difference between the replication efficiency of the WT and Δ22 viruses (Fig. 5B), with a reduction in virus yield of up to 50-fold in the Δ22 virus-infected cells. In addition, the defect in virus replication exhibited by the Δ22 virus was rescued in cells infected with the revertant virus described above (Fig. 5B). Hence, the replication defect of the Δ22 virus appeared to be specific to the deletion of the VP22-encoding gene.
FIG. 5.
The Δ22 virus exhibits a replication defect in MDBK cells compared to Vero cells. (A) Monolayers of Vero cells were infected with WT or Δ22 virus at a multiplicity of 0.1. At various times after infection, total virus was harvested by combining the cells and medium, and the samples were titrated on Vero cells. (B) Monolayers of MDBK cells were infected with WT, Δ22, or Δ22R virus at a multiplicity of 0.02. At various times after infection, total virus was harvested by combining the cells and medium, and the samples were titrated on Vero cells. h.p.i., hours postinfection.
Localization of ICP0 in Δ22 virus-infected cells.
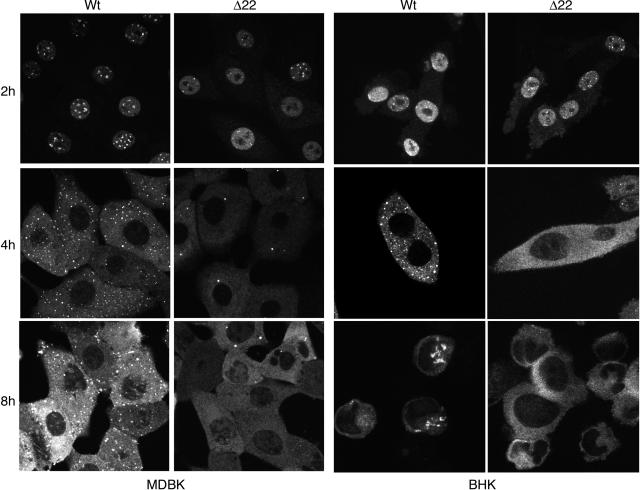
As we have shown above that there is a difference in ICP0 levels in cells infected with the Δ22 virus compared to WT virus, we next wished to investigate whether this was reflected in the subcellular localization of ICP0. MDBK and BHK cells grown on coverslips were infected with WT or Δ22 virus at a multiplicity of 10, cells were fixed at various times after infection, and immunofluorescence was carried out with an antibody against ICP0. At early times in infection (2 h), there was little difference in the characteristic nuclear localization patterns of ICP0 in either cell type infected with WT or Δ22 virus (Fig. 6, 2 h). Two hours later, ICP0 had undergone a dramatic translocation from the nucleus to the cytoplasm in both infections (Fig. 6, 4 h). However, there was a distinct difference in the ICP0 pattern exhibited in the two infections. In WT-infected cells, ICP0 was present in numerous specific punctate cytoplasmic domains, as have been described previously for the protein (19), while in Δ22-infected cells, the protein was mainly diffuse in the cytoplasm, with one or two large punctate domains also present per cell (Fig. 6, 4 h, compare WT with Δ22). Because BHK cells had already begun to round up at this stage, the different patterns of localization were clearer in the MDBK infections. Nonetheless the same features can be seen in infected BHK cells (Fig. 6, 4 h, compare MDBK and BHK). This difference in ICP0 localization was not due simply to a delay in ICP0 reaching these sites, as by 8 h, the differences between the two viruses were even more apparent (Fig. 6). Once again, even though the BHK cells were rounded up, the difference in ICP0 localization was clearly detectable in this cell line as well as in the flatter MDBK cells (Fig. 6, compare MDBK and BHK). These results suggest that the absence of VP22 from infected cells prevents the localization of ICP0 to its characteristic cytoplasmic domains.
FIG. 6.
Localization of ICP0 in Δ22 virus-infected cells. MDBK and BHK cells grown on coverslips were infected with either WT virus or Δ22 virus at a multiplicity of 10. At 2, 4, and 8 h after infection, the cells were fixed and processed for immunofluorescence with the anti-ICP0 antibody 11060.
ICP0 localizes to the same cytoplasmic sites as VP22.
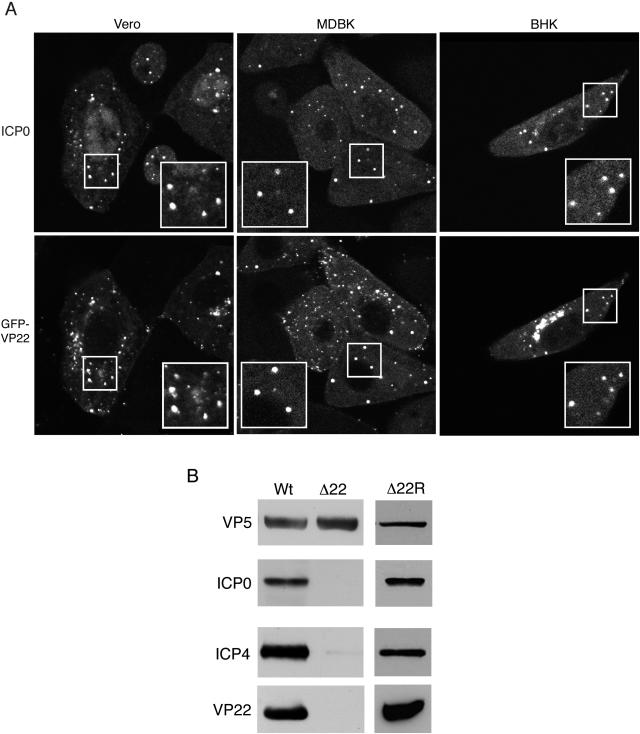
We have previously used a virus expressing GFP-VP22 to investigate the localization and trafficking of this tegument protein in live infected cells. Our data have shown that VP22 localizes to numerous cytoplasmic punctate sites that move rapidly around the cytoplasm of the cell and are reminiscent of vesicles trafficking in the secretory pathway (10). Upon examining the localization of ICP0, we realized there was a similarity between the ICP0 pattern and that observed with GFP-VP22. Thus, we carried out immunofluorescence studies on Vero, MDBK, and BHK cells that had been infected with our GFP-VP22-expressing virus at high multiplicity and fixed cells at 6 h after infection. The results from these experiments clearly show that VP22 and ICP0 localize to the same cytoplasmic sites in all cell types examined (Fig. 7A).
FIG. 7.
VP22 colocalizes with ICP0 and is required for ICP0 incorporation into virions. (A) Vero, MDBK, and BHK cells grown on coverslips were infected with the GFP-VP22-expressing virus 166v at a multiplicity of 10. At 6 h, the cells were fixed and processed for immunofluorescence with the anti-ICP0 antibody 11060. The insets show magnified images of the indicated regions. (B) Virion samples of WT, Δ22, and Δ22R viruses grown in BHK21 cells were analyzed by SDS-PAGE followed by Western blotting with antibodies against VP5, ICP0, ICP4, and VP22.
ICP0 is assembled into WT but not Δ22 virions.
As it has previously been shown that ICP0 may be present within the tegument of the virus particle (34), we postulated that it may be recruited into the virus in the cytoplasmic sites where it colocalized with VP22. Accordingly, because ICP0 does not localize to these sites in the Δ22 virus-infected cells, ICP0 would not be present in virions produced by the Δ22 virus. Thus, we next determined the ICP0 content of virions purified from cells infected with either WT or Δ22 viruses by Western blotting equivalent amounts of each with antibodies against the major capsid protein, VP5, and ICP0. As can be seen, ICP0 is clearly present in the WT virions (Fig. 7B), confirming the previous results of Yao and Courtney (34). However, although the VP5 blot showed that the virion samples were equally loaded on the gel, no ICP0 was detectable in the Δ22 virion preparation, even upon overexposure of the blot (Fig. 7B). Because ICP4 has also been shown to be a component of virions (35) and is known to interact with ICP0, we also tested both virion preparations for the presence of ICP4. Interestingly, we found that while ICP4 was clearly present in the WT virion preparation, its levels were also greatly reduced in the Δ22 virions (Fig. 7B). Moreover, a second analysis of the virion profiles of WT and Δ22 virions revealed the presence of a 175-kDa species, the correct size for ICP4, in WT but not Δ22 virions (Fig. 3A). Western blotting of virions purified from Δ22R-infected cells demonstrated that assembly of both ICP0 and ICP4 had been recovered in the revertant virus, confirming that the effect is specific to VP22. It is noteworthy that it has previously been suggested that ICP0 and ICP4 may be assembled into light particles produced during virus infection instead of, or in addition to, intact virions (26, 33). However, our technique for virion purification using a Ficoll rather than a sucrose gradient is the accepted method of separating these two types of particle, and while we cannot rule out slight L particle contamination of our virion preparations, we think it unlikely that it would be sufficient to account for the level of these immediate-early proteins detected in our particles. Taken together then, these data suggest that VP22 is involved in recruiting ICP0 (and potentially ICP4) to specific cytoplasmic domains and is required for the assembly of both of the immediate-early proteins, ICP0 and ICP4, into the tegument compartment of HSV particles. Hence, this provides further evidence that the cytoplasmic sites containing VP22 and ICP0 are involved in tegument assembly.
DISCUSSION
VP22 is a major structural component of all alphaherpesviruses. However, like so many other tegument proteins, the role of VP22 in virus infection has remained elusive. While the homologous UL49 genes have been deleted from other alphaherpesviruses such as BHV-1, PRV, and MDV (4, 5, 23), with varying consequences for the viruses in question, there have been no reports of a corresponding UL49 knockout for HSV. Here we provide the first description of HSV-1 in which the UL49 gene has been entirely deleted and show that, as for BHV-1 and PRV, VP22 is not essential for the replication of HSV-1 in tissue culture. This being the case, it is not clear why the HSV-1 Δ22 virus was so difficult to generate and ultimately had to be isolated on a high-level complementing cell line. It is possible that although when grown separately the WT and Δ22 viruses appear to behave similarly, when present in a mixed population, the WT virus possesses a definite growth advantage. This may be linked to the defect that the Δ22 virus exhibits at early times in infection. Indeed, we have shown that the absence of VP22 does affect virus growth in at least one cell type, MDBK cells, suggesting that there may be a differential requirement for VP22 in HSV-1 replication. The basis for the defect in MDBK cells is not yet known, but we are currently investigating the possibility that the Δ22 virus defect in replication is specific to polarized epithelial cells. Furthermore, we are also investigating the growth of the Δ22 virus in a mouse model to determine whether, as for BHV-1 (22), VP22 plays a crucial role in HSV-1 replication in the host.
Our studies on the growth characteristics of the Δ22 virus have, rather unexpectedly, demonstrated a relationship between VP22 and the immediate-early protein ICP0. We have shown that VP22 influences both the intracellular levels and localization of ICP0. The reduced level of ICP0 in Δ22 virus-infected cells may be due to a reduction in efficiency of expression from the ICP0 promoter, and as we have previously shown that VP22 interacts directly with VP16 (6), it is tempting to speculate that VP22 from the tegument of incoming virus may function as a cofactor of VP16 in its role as a transactivator of immediate-early genes. Alternatively, the reduced levels of ICP0 in Δ22 virus-infected cells may be due to an increased turnover rate of the protein, with VP22 somehow protecting ICP0 from degradation in WT infection.
ICP0 is a well-studied general activator of virus gene expression that early in infection localizes to ND10 nuclear structures, dispersing promyelocytic leukemia protein and inducing its proteasome-dependent degradation (13, 14). However, the cytoplasmic sites to which ICP0 is targeted after it has left the nucleus have received little attention. It is clear from a number of studies that the rate and efficiency of ICP0 translocation from the nucleus to the cytoplasm vary between cell types. In our own studies, the translocation was particularly fast and efficient in MDBK cells, with complete translocation occurring by 4 h after infection, but even in BHK and Vero cells, the ICP0 translocation to the cytoplasm was obvious if not complete by 4 h. An important role for ICP0 in the cytoplasm was first suggested by Kawaguchi and coworkers (19) when they demonstrated an interaction between ICP0 and elongation factor 1δ of the translational machinery. These authors proposed that ICP0 may play a role in altering the efficiency of translation of viral mRNAs, a theory that may be supported by our data suggesting an overall decrease in virus protein expression levels when the level of ICP0 is lowered and its cytoplasmic localization is altered in Δ22 virus-infected cells.
It has been previously suggested that the retention of ICP0 in the cytoplasm may be caused by the cellular proteasomal machinery (24), and indeed, ICP0 has been shown to induce the accumulation of colocalizing conjugated ubiquitin in both the nucleus and the cytoplasm of the infected cell (11). It was intriguing then to discover that the ICP0 cytoplasmic domains are the same sites to which VP22 localizes in infection. The role of such sites in the replication and assembly of the virus remains to be determined, but our data presented here indicate a correlation in ICP0, at least, between localization to these sites and assembly into the virion. It is interesting that we have now shown that a number of the major tegument proteins, including VP13/14 and VP16, also localize to the same sites, albeit at lower levels than VP22 (unpublished data). In our studies on tegument protein assembly, we have generally assumed that these localized concentrations of tegument proteins represent some aspect of the virus assembly pathway. Indeed, live cell studies of cells infected with viruses expressing fluorescent tegument protein, where these complexes can be observed to move rapidly around the cytoplasm, serve to emphasize the dynamic nature of them and support the theory that they may be involved in virus maturation. The colocalization of ICP0 and VP22 in the infected cell cytoplasm thus led us to determine the virion content of ICP0 in both our WT and Δ22 viruses. As a consequence, we have now shown not only that ICP0 is assembled into the virion but that its assembly correlates with localization to these specific cytoplasmic locations. Furthermore, we have also shown that the immediate-early protein ICP4, already known to interact directly with ICP0 (36), is also absent from the Δ22 virions. Although we have not determined the exact relationship between these three proteins, in that it is not clear which protein VP22 might interact with to bring into the particle, there is no doubt that VP22 must be present for ICP0 and ICP4 to be packaged into the virus. Intriguingly, studies on varicella-zoster virus (VZV) have shown that its UL49 homologue interacts with its ICP4 homologue, IE62, which, in the case of VZV, is a major component of the virion (20, 30). It has been suggested that VZV packages such high levels of ICP4 to compensate for the reduced ability of its VP16 homologue to transactivate immediate-early promoters (25). Thus, it may follow that the VZV VP22 homologue would be important for virus replication and, in particular, the incorporation of ICP4 into the tegument. In addition, the requirement for respective alphaherpesvirus VP22 homologues in virus replication may also be indicative of their need to package immediate-early proteins into the virus structure. This in turn may reflect a necessity to bypass immediate-early gene transactivation by virion-associated VP16, especially if the particular VP16 homologue is not functional.
Another interesting consequence of deleting the VP22-encoding gene from HSV-1 was that, although the amounts of the major tegument proteins VP1/2, VP13/14, and VP16 in the virion were unaffected by the absence of VP22, the levels of glycoproteins gD and gB incorporated usually showed a two- to threefold reduction. Very recently, a study was published suggesting that VP22 interacts directly with the cytoplasmic tail of gD and that this interaction may be involved in binding the capsid to the envelope (2). Furthermore, a similar situation has been suggested previously for PRV, where it has been shown that UL49 interacts with the cytoplasmic tails of gE and gM (15). Nonetheless, a level of redundancy must exist in this tegument-envelope interaction in HSV-1, as has been already shown for PRV (16), because the growth of virus in the absence of VP22 does not have a dramatic effect on the incorporation of the respective glycoproteins proposed to interact with VP22.
In summary, we have shown that although VP22 is not essential for the replication of HSV-1, its presence enhances virus growth in certain cell types. This situation is not unusual among HSV-1 genes, with proteins such as ICP22 and, indeed, ICP0 itself, exhibiting similar variations for their requirement (12, 28, 31). It is possible that the Δ22 virus phenotype can be explained solely by the alterations observed in ICP0 during infection, but this has yet to be conclusively proven, and there are likely to be other as yet undefined consequences of deleting the VP22-encoding gene. Furthermore, the realization that VP22 is involved in packaging ICP0 (and ICP4) into the virus particle increases the complexity of the defect observed in the knockout virus at early times in infection. Importantly, the fact that we can now assemble virus without ICP0 and ICP4 affords us the opportunity to investigate the role for these virion-packaged immediate-early proteins in virus infection.
Acknowledgments
We thank Tony Minson, Helena Browne, Roger Everett, and Roger Tsien for providing reagents used in this study. We also thank Janneke Verhagen and Corinne Potel for assistance with experiments.
A.W. was funded by the NLCB (now Community Fund), and W. H. was funded by Innovative Medizinische Forschung (IMF) (identification no. HA620202). G.E. and E.B. were funded by Marie Curie Cancer Care.
REFERENCES
- 1.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 3.Dargin, D. 1986. The structure and assembly of herpes viruses, p. 359-437. In J. R. Harris and R. W. Horne (ed.), Electron microscopy of proteins, vol. 5. Virus structure. Academic Press, London, United Kingdom. [Google Scholar]
- 4.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., C. Boutell, and A. Orr. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 78:1763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerster, T., and R. G. Roeder. 1988. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc. Natl. Acad. Sci. USA 85:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves, R. F., and P. O'Hare. 1990. Structural requirements in the herpes simplex virus type 1 transactivator Vmw65 for interaction with the cellular octamer-binding protein and target TAATGARAT sequences. J. Virol. 64:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwong, A. D., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang, X., B. Chow, and L. A. Babiuk. 1997. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine 15:1057-1064. [DOI] [PubMed] [Google Scholar]
- 23.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, P., C. Van Sant, and B. Roizman. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKee, T. A., G. H. Disney, R. D. Everett, and C. M. Preston. 1990. Control of expression of the varicella-zoster virus major immediate early gene. J. Gen. Virol. 71(Pt 4):897-906. [DOI] [PubMed] [Google Scholar]
- 26.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73(Pt 2):269-276. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poffenberger, K. L., P. E. Raichlen, and R. C. Herman. 1993. In vitro characterization of a herpes simplex virus type 1 ICP22 deletion mutant. Virus Genes 7:171-186. [DOI] [PubMed] [Google Scholar]
- 29.Smibert, C. A., D. C. Johnson, and J. R. Smiley. 1992. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J. Gen. Virol. 73(Pt 2):467-470. [DOI] [PubMed] [Google Scholar]
- 30.Spengler, M., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella zoster virus. Arch. Virol. Suppl. 17:71-79. [DOI] [PubMed] [Google Scholar]
- 31.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 32.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, T. Y., and R. J. Courtney. 1995. Influence of the host cell on the association of ICP4 and ICP0 with herpes simplex virus type 1. Virology 211:209-217. [DOI] [PubMed] [Google Scholar]
- 34.Yao, F., and R. J. Courtney. 1992. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J. Virol. 66:2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, F., and R. J. Courtney. 1989. A major transcriptional regulatory protein (ICP4) of herpes simplex virus type 1 is associated with purified virions. J. Virol. 63:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]