Abstract
Epstein-Barr virus latent protein EBNA3C has been shown to bind Nm23-H1, a known suppresser of cell migration and metastasis and a regulator of the guanine exchange factor Tiam-1. This interaction results in cellular translocation of Nm23-H1 to the nucleus and suppression of the antimigratory effect in vitro. Furthermore, these proteins can synergistically increase transcription of a basal promoter when targeted to DNA by fusion to a Gal4 DNA binding domain. In this report, we show that EBNA3C and Nm23-H1 can cooperate to upregulate expression of MMP-9, known to be expressed in aggressive forms of lymphomas. This upregulation resulted in increased levels of MMP-9 mRNA, as well as a detectable increase in MMP-9 gelatinolytic activity. Specific mutations in the MMP-9 promoter showed that the Ap1 and NFκB binding sites are important for upregulation by the proteins. Additionally, it was shown for the first time that EBNA3C and Nm23-H1 can bind subunits of these transcription factors. This suggests that the ability of EBNA3C to reverse the antimigratory effects of Nm23-H1 is likely to be in part through the synergistic upregulation of MMP-9, mediated through interactions with the AP1 and NFκB transcription factors.
Epstein-Barr virus (EBV) is a human gammaherpesvirus which predominantly targets B cells and epithelial cells and is associated with a number of cancers, including Burkitt's lymphoma, nasopharyngeal carcinoma, Hodgkin's disease, AIDS-associated and transplant-associated immunoblastic lymphoma, and somewhat controversially, invasive breast carcinoma (3, 13, 29). EBV is also known to be the causative agent of infectious mononucleosis (3, 13, 29). In vitro infection of B cells with EBV gives rise to lymphoblastoid cell lines (LCLs) which express a subset of 12 latent viral transcripts (13, 29). These 12 transcripts encode six nuclear antigens (EBNAs), three latent membrane proteins (LMPs), two early RNAs (EBERs), and the BARF transcripts (13, 29). Of these genes, six, EBNA1, EBNA-LP, EBNA-2, EBNA-3A, EBNA-3C, and LMP1, have been shown to be critical for growth transformation and immortalization of human primary B cells in vitro (5, 11, 43).
The EBNA3 family contains a set of three genes tandemly arranged on the EBV genome. The three antigens primarily function as transcriptional regulators through interactions with other cellular and viral factors (13). The three proteins contain similar structural motifs and have a region of limited homology in the amino terminus (13, 49). This domain contains the binding site for the cellular repressor RBP-Jκ, also referred to as CSL (31, 49). Previous studies demonstrated that EBNA3 binding to RBP-Jκ disrupts its ability to bind to its cognate DNA sequence, suggesting that the EBNA3 proteins can regulate genes with responsive RBP-Jκ binding sites (31). Additional studies have also shown that EBNA3C can prevent binding of RBP-Jκ to EBNA2 and can downregulate RBP-Jκ-mediated EBNA2 transactivation of the LMP1 promoter (20, 22). Furthermore, EBNA3C can regulate Cp, the major latent promoter controlling EBNA expression, through RBP-Jκ and other corepressors (27). EBNA3C has also been shown to play a role in regulating the acetylation and coactivation activity of the p300/ProTα complex (37). When tethered to DNA as a Gal4 fusion protein, EBNA3C has been shown to function as a transcriptional repressor and in vitro has been shown to interact with the Spi-1 and Spi-B transcription factors (50).
The addition of a stop codon after amino acid (aa) 365 in the EBNA3C open reading frame has been shown to nullify the growth transformation properties of EBV in terms of B-cell immortalization in vitro (43). The region downstream of aa 365 to 992 was therefore important in mediating B-cell immortalization (43). The metastasis suppressor Nm23-H1 was found to bind to a short stretch of amino acids located between the glutamine- and proline-rich domains of EBNA3C (36, 39). This interaction between Nm23-H1 and EBNA3C has been shown to result in an increase in transcriptional activity on a responsive promoter (39). Nm23-H1 tethered to DNA by a Gal4 DNA binding domain (DBD) can activate transcription from a basal promoter at relatively low levels. However, when EBNA3C was introduced, the transactivation activity was shown to be substantially increased (39). These results suggest that Nm23-H1 may possess transcriptional regulatory activities independent of a possible role in directly binding to DNA or through its interaction with EBNA3C. Interestingly, the presence of EBNA3C mediates the cellular translocation of Nm23-H1 from a mostly cytoplasmic to a predominantly nuclear signal (36). Moreover, EBNA3C can reverse the antimigratory effects of Nm23-H1 in Burkitt's lymphoma and breast carcinoma cells (36).
The Nm23 gene family is a closely related group of nucleoside dinucleotide phosphate kinases for which eight distinct genes are known in humans (Nm23-H1 to -H8) (15). The proteins are characterized by a wide variety of functions, including transcriptional regulation, differentiation, proliferation, and suppression of tumor metastasis (15). Importantly, changes in cellular levels of Nm23-H1 have been correlated with decreased metastasis in a number of cancers, including breast, gastric, and cervical cancers (25). In some cancers, such as colorectal carcinoma, mutations in Nm23-H1 are associated with increased metastasis (18). Transfection of Nm23-H1 genes into breast, melanoma, colon, and oral squamous cells has been shown to suppress metastasis in the nude mouse model in vivo (17, 18, 23, 40). While there is a significant amount of data implicating Nm23-H1 in the regulation of metastasis, the biochemical mechanism for this activity is still poorly understood.
An important step in invasion and metastasis is degradation of the basement membrane to enable tumor cells to escape from the primary growth site (35). The matrix metalloproteinase (MMP) family of proteins is involved in the degradation of all components of the basement membrane, and members of every family of metalloproteinases have been implicated in malignancy and metastasis (35, 45). The activity of MMPs is tightly controlled, with regulation occurring mainly at the transcriptional level (35). The proteins are further regulated through activation of the proenzyme and the tissue inhibitors of metalloproteinase (35). A number of MMPs have been found to be associated with cell migration and invasion, including MMP-2 and MMP-9, both of which degrade type IV collagen (8, 28). The ability to degrade type IV collagen, a major component of the basement membrane, indicates that these proteins may play an important role in metastasis by degrading the basement membrane at sites of malignant tumor growth. Specifically, MMP-9 has been implicated as an essential molecule required for tumor cell intravasation and extravasation during metastasis (24).
The role of Nm23-H1 in regulating MMP-9 has not been investigated in depth. One report indicated that in oral squamous cell carcinoma cells, Nm23-H1 can suppress cell migration without affecting MMP-2 or MMP-9 levels (12). Importantly, EBV-immortalized B cells have been shown to synthesize MMP-9 and the addition of TIMP-1 to EBV-infected B cells has been shown to inhibit their migration in vitro (7, 44). Moreover, LMP1 expression increases levels of MMP-9 in cervical carcinoma cells (24, 41, 44, 46). LMP1-mediated MMP-9 expression occurs primarily through the NFκB and Ap1 signaling pathways (46). Additionally, LMP1 was shown to increase invasiveness of cells in vitro, as well as to enhance MMP-9 levels in tumor cells grown in mice (24). In this report, we show that the interaction of the essential EBV latent antigen EBNA3C with Nm23-H1 also leads to increased expression of MMP-9. This provides, in part, an important clue toward understanding the mechanism by which EBNA3C reverses the antimigratory effect of Nm23-H1 in the infected cell.
MATERIALS AND METHODS
Cell lines and antibodies.
BJAB cells are EBV-negative B cells isolated from Burkitt's lymphoma and were provided by Elliott Kieff (Brigham and Women's Hospital, Boston, MA). The EBNA3C-expressing cell lines have been previously described (6, 30). LCL1 and LCL2 are transformed B-cell lines previously described (6). All B-cell lines were grown in RPMI 1640 medium (HyClone, Logan, UT) supplemented with 10% fetal bovine serum, 2 mM glutamine, and 25 U/ml penicillin-streptomycin. The EBNA3C-expressing cells were grown in medium with an additional 200 ng/ml G418. Human embryonic kidney fibroblast 293T cells transformed with E1A and E1B were obtained from Jon Aster (Brigham and Women's Hospital, Boston, MA). C33a cells are epithelial cells isolated from cervical carcinoma and were provided by Joseph Pagano (University of North Carolina School of Medicine, Chapel Hill). 293T and C33a cells were grown in Dulbecco modified Eagle medium (HyClone, Logan, UT) supplemented with 10% bovine growth serum, 2 mM glutamine, and 25 U/ml penicillin-streptomycin.
A mouse monoclonal antibody against Sp1 (E-3), a rabbit polyclonal antibody against c-Fos (sc-52), and a rabbit polyclonal antibody against the NFκB p50 subunit (H-119) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). A rabbit monoclonal antibody against c-Jun was obtained from Cell Signaling Technology (Beverly, MA).
Transfection.
BJAB, 293T, and C33a cells were transfected by electroporation using a Bio-Rad Gene Pulser II electroporator. Ten million cells were collected and washed once in phosphate-buffered saline (PBS). The cells were then resuspended in 400 μl of either Dulbecco modified Eagle medium or RPMI 1640 containing DNA normalized to balance total DNA, and efficiency determined by green fluorescent protein (GFP) expression as an internal control. Once resuspended, the cells were transferred to 0.4-cm electroporation cuvettes and electroporated at 975 μF and 210 V for 293T and C33a cells and 220 V for BJAB cells. Following electroporation, the cells were plated in 10 ml of supplemented medium and grown at 37°C and 5% CO2 for 20 h before being harvested.
Luciferase assay.
Ten million BJAB cells were collected at a concentration of 5 × 105/ml and transfected as described above. The MMP-9 reporter plasmid and specific mutants were provided by Joseph Pagano (University of North Carolina School of Medicine, Chapel Hill) and originally constructed by Hiroshi Sato (Cancer Research Institute, Kanazawa University, Kanazawa, Japan) (34). The −670 to +54 fragment of the MMP-9 promoter, GenBank accession no. D10051, was inserted into the pGL3-basic vector with point mutations made in the Ap1, Sp1, and NFκB binding sites. Twenty hours posttransfection, the cells were harvested and then washed in PBS and lysed in 400 μl of reporter lysis buffer (Promega, Inc., Madison, WI). A 40-μl aliquot of the lysate was then mixed with 100 μl of luciferase assay reagent in an Opticomp Luminometer (MGM Instruments, Inc., Hamden, CT), and luminescence was read for 10 s. Diluted lysates were also measured to ensure that the values were within the linear range of the assay. The presented results represent experiments performed in triplicate.
IP and Western blotting.
Immunoprecipitation (IP) assays were performed as previously reported (16). Thirty million 293T cells were transfected with pA3M-EBNA3C and/or pA3M-Nm23-H1 and incubated for 20 h. The cells were subsequently lysed in radioimmunoprecipitation assay (RIPA) buffer for 1 h on ice and then precleared by a 1-h incubation with protein A-Sepharose beads. Anti-p50, anti-c-Jun, anti-c-Fos, or anti-Sp1 antibodies were incubated with the lysates overnight at 4°C. The immunoprecipitates were collected by a 1-h rotation with protein A-Sepharose beads, followed by four washes with RIPA buffer. Sodium dodecyl sulfate (SDS)-β-mercaptoethanol lysis buffer was added, and the protein was heated and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Western blot assays were performed using antibodies specific to EBNA3C and Nm23-H1 and fluorescence-labeled secondary antibodies (Rockland, Inc., Gilbertsville, PA), followed by detection with an Odyssey imager (LiCor, Inc., Lincoln, NE). For BJAB, EBNA3C stable cells, LCL1, and LCL2, 20 million cells were used and IP was performed as described above.
GST fusion protein preparation, cellular lysate binding, and in vitro binding assays.
DH5α cells were transformed by heat shock with the plasmid construct for glutathione S-transferase (GST)-Nm23-H1 and selected with ampicillin. A 2-ml overnight culture, inoculated with a single colony and grown in LB medium at 37°C with shaking, was then used to inoculate 500 ml of LB medium, and the culture was grown at 37°C with shaking until the mid-exponential growth phase. The cells were then induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) overnight at 30°C with shaking. The cells were subsequently harvested and sonicated, and the protein was solubilized. The protein was then incubated with glutathione-Sepharose beads overnight at 4°C with rotation. The beads were collected by centrifugation and then washed four times with NETN (50% NP-40, 20 mM Tris, 1 mM EDTA, 100 mM NaCl) containing protease inhibitors. The protein-bound beads were stored at 4°C in NETN containing protease inhibitors.
The cellular lysates used in the binding experiments were prepared by collecting 30 million cells, washing them once in PBS, and then lysing them in RIPA buffer for 1 h with vortexing every 15 min. The lysates were then precleared with glutathione-Sepharose beads for 30 min at 4°C with rotation. The lysates were additionally precleared with GST-bound glutathione-Sepharose beads for 1 h at 4°C with rotation. The lysates were then incubated with an amount of GST-Nm23-bound beads equivalent to the GST-bound beads used to preclear and rotated overnight at 4°C. SDS lysis buffer with heating was used to elute the bound protein from the beads, followed by 10% SDS-PAGE. Western blot assays using anti-c-Jun, -c-Fos, -p50, and -Sp1 antibodies were then performed to detect the transcription factors associated with Nm23-H1.
The full-length and truncated pA3M clones of EBNA3C were transcribed and translated in vitro with [35S]methionine-cysteine in the T7 TNT system (Promega, Inc., Madison, WI). The in vitro-translated proteins were first precleared with glutathione-Sepharose beads in binding buffer (1× PBS, 0.1% NP-40, 0.5 mM dithiothreitol, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 2 μg aprotinin per ml, 1 μg pepstatin A per ml, 2 μg leupeptin per ml) for 30 min at 4°C with rotation, and the beads were removed by centrifugation. A second preclearing with GST-bound glutathione-Sepharose beads followed for 1 h at 4°C with rotation with the beads removed by centrifugation. The protein lastly was incubated with GST-Sp1 or GST-Sp1 truncations for 16 h at 4°C with rotation. The beads were then pelleted by centrifugation and washed four times with binding buffer. The beads and bound protein were then denatured with SDS-β-mercaptoethanol lysis buffer with boiling, followed by SDS-PAGE. The gel was then dried and exposed to a Storage Phosphor Screen (Amersham Biosciences, Piscataway, NJ).
Gelatin zymography.
Gelatin zymography was performed to assay for MMP-9 activity as previously described (46). Conditioned medium from C33a cells transfected with pA3M vector and/or pA3M-EBNA3C and/or pA3M-Nm23-H1 was collected after 24 h and concentrated 10 times with a Centricon 10 filter (Millipore, Corp., Billerica, MA). The concentrated medium was then mixed with SDS lysis buffer, followed by SDS-PAGE with gelatin present at a final concentration of 0.1%. The gel was subsequently washed twice for 30 min in 50 mM Tris (pH 7.6)-10 mM CaCl2-0.02% NaN3-0.001 mM ZnCl2-2.5% Triton X, followed by a 30-min wash with H2O. The gel was then incubated for 24 h at 37°C in a solution containing 50 mM Tris (pH 7.6), 10 mM CaCl2, 0.02% NaN3, and 150 mM NaCl. The gel was stained with Coomassie blue R250 in 30% methanol and 10% acetic acid and destained in 30% methanol and 10% acetic acid. The gelatinolytic activity shows up as a clear band against the dark background of stained gelatin.
Real-time quantitative PCR.
Total RNA from C33a cells stably expressing EBNA3C, Nm23-H1, or both proteins was collected using Trizol reagent (Invitrogen, Inc., Carlsbad, CA) following the manufacturer's instructions. cDNA was then made using a Superscript II RT kit (Invitrogen, Inc., Carlsbad, CA) following the manufacturer's instructions. The specific primers for MMP-9 used were as follows: sense, 5′-TGCGCTGCTGCTTCTCCAGA-3′; antisense, 5′-GGTCGCCCTCAAAGGTTTGG-3′. They yielded a 125-bp product. For β-actin, the primers were as follows: sense, 5′-GCTCGTCGTCGACAACGGCTC-3′; antisense, 5′-CAAACATGATCTGGGTCATCTTCTC-3′. They yielded a product 352 bp in length. The cDNA was amplified using SYBR green real-time mastermix (MJ Research Inc., Waltham, MA), 1 mM each primer, and 1 μl of the cDNA product in a total volume of 20 μl. Thirty-five cycles of 1 min at 94°C, 1 min at 56°C, and 30 s at 72°C, followed by 7 min at 72°C, were performed in an MJ Research Opticon II thermocycler. Each cycle was followed by two plate reads, with the first at 72°C and the second at 85°C. A melting curve analysis was performed to verify the specificity of the products, and the values for the relative quantitation were calculated by the ΔΔCt method. The experiment was performed in triplicate.
Electrophoretic mobility shift assays (EMSA).
The probes for the Ap-1 and NFκB binding sites within the MMP-9 promoter have been previously described (46). The probes were end labeled by Klenow fill-in reaction with [α-32P]dCTP and purified with NucTrap probe purification columns (Stratagene, Inc., La Jolla, CA). Radioactive probes were diluted in water to a final concentration of 100,000 cpm/μl. DNA binding reactions and preparation of nuclear extracts (NEs) were performed in a manner similar to that previously described (14). BJAB cells used to make NEs were treated with phorbol 12-myristate 13-acetate at 10 ng/ml for 24 h. Fifteen micrograms of protein from NEs was mixed with 1 μg poly(dI-dC) (Sigma) in DNA binding buffer (20 mM HEPES [pH 7.5], 0.01% NP-40, 5.0% glycerol, 10 mM MgCl2, 100 μg of bovine serum albumin, 100 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 40 mM KCl) to a total volume of 50 μl and incubated at room temperature for 5 min. One microliter of labeled probe was added to each reaction mixture, followed by an additional 15 min at room temperature. Unlabeled competitors (200×) were added prior to the initial incubation at room temperature. Rabbit polyclonal antibodies against c-Fos, p50, and Nm23-H1, as well as rabbit serum against EBNA3C and a rabbit monoclonal antibody against c-Jun, were used to supershift the probe. DNA-protein complexes were resolved by nondenaturing 6% PAGE run in 0.5× Tris-borate-EDTA buffer at a constant voltage of 150 V. Following electrophoresis, the gels were dried and exposed to a Storage Phosphor Screen for 48 to 72 h.
RESULTS
Nm23-H1 and EBNA3C can activate transcription of the MMP-9 promoter.
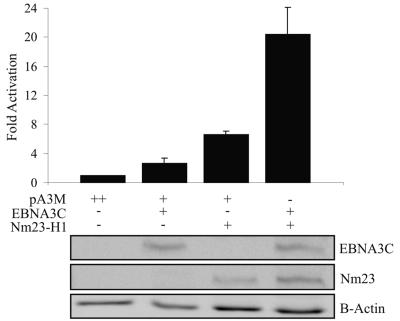
Previous studies by our group showed that EBNA3C can reverse Nm23-H1-mediated suppression of cell migration (36). To further elucidate the mechanism by which this occurs, we wanted to determine whether either or both of these proteins could modulate the expression levels of MMP-9, a protein found to play a key role in cell migration and metastasis (9, 19, 24, 26, 48). The effects of these proteins on the full-length MMP-9 promoter inserted into the pGL2 basic vector were assessed by luciferase assay. pA3M-EBNA3C, pA3M-Nm23-H1, or both were transiently transfected into BJAB cells along with pLpluc, the reporter plasmid containing the MMP-9 promoter. Total transfected DNA was balanced with empty vector, and transfection efficiency was monitored by the use of pEGFP and counting of transfected cells for GFP fluorescence. The results of the assay demonstrated that, individually, EBNA3C and Nm23-H1 activated the MMP-9 promoter at levels approximately twofold over the vector alone, with Nm23-H1 having a greater effect than EBNA3C (Fig. 1). However, when coexpressed, the level of activation of the promoter was significantly higher, with activity greater than the combined activation of each protein independently expressed to 20-fold above the vector alone (Fig. 1). This result indicates that EBNA3C and Nm23-H1 may function synergistically to transactivate the MMP-9 promoter in an in vitro reporter assay. The expression of EBNA3C and Nm23-H1 was detected using a Myc-specific antibody, while the protein levels were determined by β-actin Western blot assay.
FIG. 1.
EBNA3C and Nm23-H1 synergistically upregulate the transcriptional activity of the MMP-9 promoter. The MMP-9 reporter vector contains the 670 bases upstream of the site of translation initiation of the MMP-9 protein inserted into the pGL3 vector (34). A 2.5-μg sample of the MMP-9 reporter plasmid was transfected into BJAB cells along with 5 μg of pA3M-EBNA3C, pA3M-Nm23-H1, or both, with total DNA being normalized with empty pA3M vector. Promoter activity is expressed as n-fold activation relative to the reporter vector and pA3M alone. Means and standard deviations are derived from three independent experiments. Protein lysates were analyzed by Western blotting, with both Nm23-H1 and EBNA3C being detected by anti-Myc antibody and the total protein level being shown by β-actin blotting.
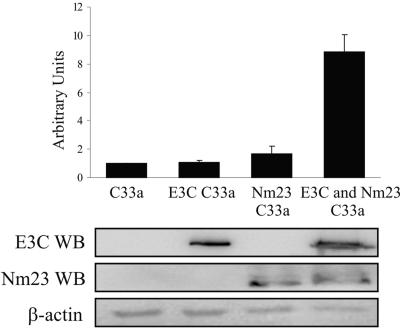
To further verify the results of the luciferase assay, real-time reverse transcriptase PCR (RT-PCR) was performed. C33a cells stably overexpressing Nm23-H1, as well as expressing EBNA3C or both proteins together, were harvested and total RNA isolated. Using a two-step process, cDNA was made, followed by real-time PCR for MMP-9. The results of the PCR indicated that EBNA3C and Nm23-H1 independently had little or no effect on MMP-9 transcript levels (Fig. 2). However, when they were coexpressed, the detected levels of MMP-9 mRNA were as much as 10-fold greater than the base levels (Fig. 2). These results correlated with the data from the luciferase assay described above. Thus, these data suggest that Nm23-H1 and EBNA3C, acting synergistically, can result in upregulation of MMP-9.
FIG. 2.
Increased MMP-9 transcripts in EBNA3C and Nm23-H1 C33a stable cell lines as detected by real-time RT-PCR analysis. Total RNA was isolated from 10 million C33a cells by Trizol reagent. Five micrograms of RNA was used in the Superscript First Strand synthesis system to construct the cDNA. Real-time PCR was performed using the DyNAmo SYBR green quantitative PCR kit with β-actin as the standard. The PCR data are expressed as n-fold activation relative to normal C33a cells as calculated by the ΔΔCt method. A series of three independent experiments was used to calculate the mean and standard deviation. WB, Western blotting.
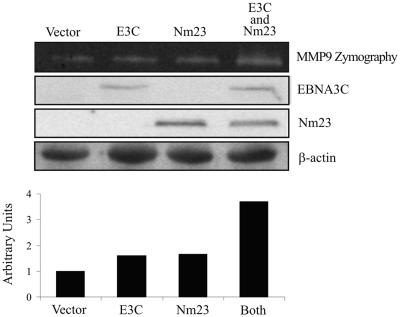
Nm23-H1 and EBNA3C can enhance MMP-9 activity in a transient in vitro assay.
MMP-9 gelatinolytic activity was assayed to determine if the observed increase in gene transcription resulted in a similar increase in protein activity. EBNA3C and Nm23-H1 expression plasmids were transiently transfected into C33a cells and compared to a pA3M-transfected control. The results suggest that there was a modest increase to 1.5-fold in expression observed in cells where only one protein was expressed (Fig. 3). However, in cells cotransfected with both proteins, a greater than threefold increase in MMP-9 activity was observed as determined by zymography (Fig. 3). Therefore, the overall increase in MMP-9 enzyme activity requires the cooperation of Nm23-H1 and EBNA3C expressed together in this in vitro assay. The exogenous expression of the transfected constructs was confirmed by Western blot assay for Myc, with the internal protein levels determined by β-actin Western blot analysis (Fig. 3).
FIG. 3.
Increased gelatinolytic activity of MMP-9 in C33a cells containing EBNA3C and Nm23-H1 as shown by zymography. Serum-free conditioned medium was collected from C33a-transfected cells after 48 h and subsequently concentrated by a Centricon 10 filter. The concentrated samples were then electrophoresed on an 8% SDS-PAGE gel containing 0.1% gelatin, followed by a 1-h wash in a 2.5% Triton X solution and then incubation at 37°C for 24 h. Gelatinolytic activity was resolved as white bands on a Coomassie blue R250-stained background at the predicted molecular mass of MMP-9 of 92 kDa. Protein levels of Nm23-H1 and EBNA3C were measured by Western blot assay.
The cellular transcription factors Ap1 and NFκB are important for Nm23-H1 and EBNA3C upregulation of the MMP-9 promoter.
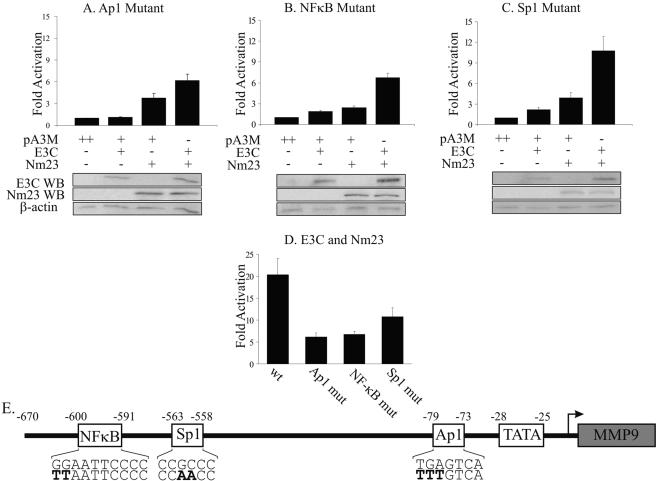
Since neither EBNA3C nor Nm23-H1 has been shown to regulate transcription by directly binding DNA, we looked for possible transcription factors that might be targeted, resulting in functional regulation of the MMP-9 promoter. Ap1, Sp1, and NFκB have been previously shown to be transactivators of MMP-9 expression, as well as being important in LMP1-enhanced expression (33, 46). EBNA3C has also been shown to contain a region with homology to the glutamine-rich activation domain of Sp1, which can function as a transcriptional activator when fused to the Gal4 DBD (1). Reporter plasmids with targeted mutations in the Sp1, Ap1, or NFκB binding sites of the MMP-9 promoter were used in luciferase assays as described above. The most dramatic effect was observed in the Ap1 and NFκB mutants, where activation was decreased by approximately 70% and 67%, respectively, in the coexpressed samples (Fig. 4A, B, and D), while a decrease of 47% was observed from the Sp1 mutant compared to wild-type data (Fig. 4C and D). The effects of the mutations were specific for the transfected molecule. The low level of activation by EBNA3C was not affected by mutations in the Sp1 or NFκB binding site but was almost completely ablated to background levels by mutation of the Ap1 binding site (compare Fig. 4A with B and C). Activation of the reporter by Nm23-H1 was decreased by mutations in both the Ap1 and NFκB binding sites, with the greatest effect observed in the case of the NFκB site, while mutation of the Sp1 site had little effect on Nm23-H1 activity (Fig. 4). These data seem to indicate that the Ap1 and NFκB binding sites in the MMP-9 promoter play an important role in Nm23-H1/EBNA3C-mediated upregulation of MMP-9, with each protein showing specificity for a particular transcription factor.
FIG. 4.
Ap1, Sp1, and NFκB transcription site-specific mutants reduce the ability of Nm23-H1 and EBNA3C to activate MMP-9 transcription. The wild-type MMP-9 reporter vector had site-specific mutations made in the Ap1 (A), NFκB (B), and Sp1 (C) transcription factor binding sites (34). Samples (2.5 μg) of the MMP-9 mutant reporter plasmids were transfected into BJAB cells along with 5 μg of pA3M-EBNA3C, pA3M-Nm23-H1, or both, with total DNA being normalized with empty pA3M vector. Promoter activity is expressed as n-fold activation relative to the reporter vector and pA3M alone. Means and standard deviations were derived from three independent experiments. Protein lysates were analyzed by Western blotting (WB), with both Nm23-H1 and EBNA3C being detected by anti-Myc antibody and total protein level being shown by β-actin blotting. (D) Comparison of wild-type (wt) versus mutant (mut) promoter activities from the MMP-9 reporter vector in EBNA3C- and Nm23-H1-cotransfected BJAB cells. (E) Schematic of the MMP-9 promoter indicating the transcription factor binding sites used for the mutant studies with the point mutations indicated in bold.
EBNA3C and Nm23-H1 associate with cellular transcription factors Sp1, c-Jun, c-Fos, and NFκB.
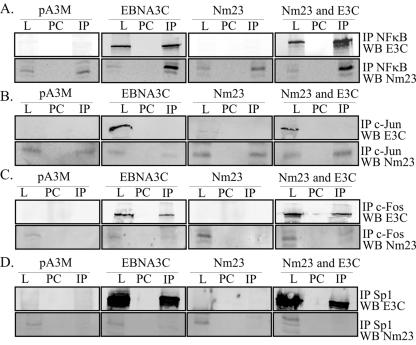
The data above strongly suggest that EBNA3C and Nm23-H1 can upregulate the expression of MMP-9 through interactions with known cellular transcription factors. To show that this occurs through association with these specific cellular factors, IP and GST binding experiments were performed using antibodies specific for the known transcription activators shown to have an effect in the reporter assays above. Nm23-H1 and EBNA3C expression vectors were transfected into 293T cells. Twenty-four hours posttransfection, the cells were harvested and lysed by RIPA buffer. Sp1, c-Jun, c-Fos, and the NFκB p50 subunit were immunoprecipitated using specific antibodies, and the complexes were resolved by SDS-PAGE. Fractionated proteins were transferred to nitrocellulose membrane. The blots were probed with mouse anti-Myc antibody to detect EBNA3C and a mouse monoclonal antibody for detection of Nm23-H1. Our studies showed that EBNA3C coimmunoprecipitated with p50, c-Fos, and Sp1, as detected from the EBNA3C-transfected cells (Fig. 5A, C, and D). Nm23-H1 coimmunoprecipitated with c-Jun and p50 in all cells, with no obvious difference in association between cells where Nm23-H1 was overexpressed and cells with only endogenous levels (Fig. 5). Strikingly, the levels of Nm23-H1 seen in cells where EBNA3C was transfected showed a dramatic increase in levels of Nm23-H1 associated with p50 (Fig. 5A).
FIG. 5.
(A to D) Both EBNA3C and Nm23-H1 coimmunoprecipitate with transcription factors shown to be important for the regulation of MMP-9. Thirty million 293T cells were transfected with EBNA3C, Nm23-H1, or both expression vectors. At 20 h posttransfection, the cells were harvested and lysed with RIPA buffer; the lysates were then used for the IP experiments. Mouse monoclonal antibodies against Sp1, rabbit monoclonal antibody against c-Jun, and rabbit polyclonal antibodies against c-Fos and NFκB p50 were used to show that either EBNA3C or Nm23-H1 immunoprecipitated with the transcription factor. L, lysate; PC, preclear; WB, Western blot.
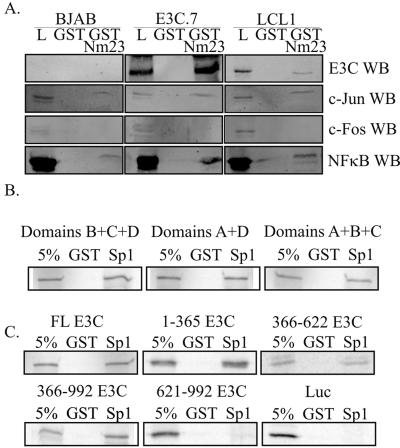
A GST-Nm23 pulldown assay was also performed from an EBV-transformed LCL, as well as an EBNA3C stable cell line created by stably expressing the EBNA3C protein in an EBV-negative B-cell line, BJAB. As expected, Nm23-H1 associated with EBNA3C in both the EBNA3C stable cell line and the EBV-transformed LCL (Fig. 6A). These results were also obtained with two additional cell lines stably expressing EBNA3C and another LCL (data not shown). Again, Nm23-H1 was also associated with c-Jun and the NFκB p50 subunit, although it was clear that endogenous levels were much lower for c-Jun and c-Fos. These studies strongly suggest that both EBNA3C and Nm23-H1 can associate with transcription activators Ap1, Sp1, and NFκB, mediating upregulation of MMP-9 transcription.
FIG. 6.
GST-Nm23-H1 interacts with transcription factor subunits important for the regulation of MMP-9 from EBV-infected and EBNA3C stably expressing cells. (A) Thirty million cells were collected and lysed in RIPA buffer. The lysates were incubated with GST, followed by GST-Nm23-H1, overnight. The pulldown products were boiled and then electrophoresed on 10% SDS-PAGE gels. Western blot analysis was used to detect the presence of EBNA3C, as well as the transcription factor subunits. Ten percent lysate controls were run for all samples. (B) Sp1-GST truncation mutants were tested for binding with EBNA3C. Transactivation domains A and B are identical glutamine-rich domains required for transcriptional activation, while domains C and D contain the DBD. (C) EBNA3C and all truncations except for the extreme carboxy end interact with SP1. Full-length (FL) or truncated forms of EBNA3C were in vitro translated with [35S]methionine labeling, followed by incubation with Sp1-GST. Bound proteins were run on 8% SDS-PAGE and detected by Storage Phosphor Screen (Amersham Biosciences). All samples have a 5% input lane and a GST preclear control lane. Luciferase (Luc) was used as a negative control. L, lysate; WB, Western blot.
Based on the strong interaction between Sp1 and EBNA3C observed by IP, we wanted to further map the interaction between the two proteins. Sp1 contains four distinct domains, two of which are identical glutamine-rich domains (A and B) and two of which contain the DBD (C and D). A series of GST deletion mutant forms of Sp1 have previously been described (10), with one mutant with domain A deleted, a second with domains B and C deleted, and the third with domain D deleted. In vitro binding assays with all three of these GST mutants, as well as full-length Sp1-GST, were performed with in vitro-translated EBNA3C full-length and truncated molecules. Binding was observed with full-length Sp1, as well as with the three truncated molecules (Fig. 6B and C). The inability of any of the mutants to knock out binding suggests that EBNA3C may be able to bind to either of the glutamine-rich domains.
In vitro binding reactions were also used to map the region of EBNA3C that binds to Sp1. Truncated EBNA3C clones representing aa 1 to 365, 366 to 622, 366 to 992, and 621 to 992, as well as full-length EBNA3C, were in vitro translated as described above and tested for the ability to bind full-length Sp1-GST. The amino-terminal 365 aa and the aa 366 to 622 region bound to Sp1. The truncation mutant with the first 365 aa deleted also bound to GST-Sp1. However, the 621 to 992 clone containing the 371 extreme carboxy-terminal amino acids was unable to bind Sp1 in this assay (Fig. 6C). These results indicate that Sp1 binds to EBNA3C at a region 5′ to aa 365 and also to another domain 3′ to aa 365, possibly the two acidic domains.
A complex of EBNA3C and Nm23-H1 interacts with NFκB and Ap1 bound to the respective cognate sequences within the MMP-9 promoter.
The results obtained with the luciferase reporter promoter mutants indicated that both the Ap1 and NFκB binding sites within the MMP-9 promoter are critical for EBNA3C- and Nm23-H1-mediated upregulation of MMP-9. Additionally through IP experiments we were able to show that EBNA3C and Nm23-H1 can bind various subunits of NFκB and the Ap1 complex. Therefore, we wanted to test whether these proteins were regulating MMP-9 through transcription factor-mediated DNA binding. The previously described double-stranded DNA probes for the NFκB and Ap1 binding sites (46) were labeled and tested for binding to NEs from BJAB cells transiently transfected with EBNA3C, Nm23-H1, or both proteins.
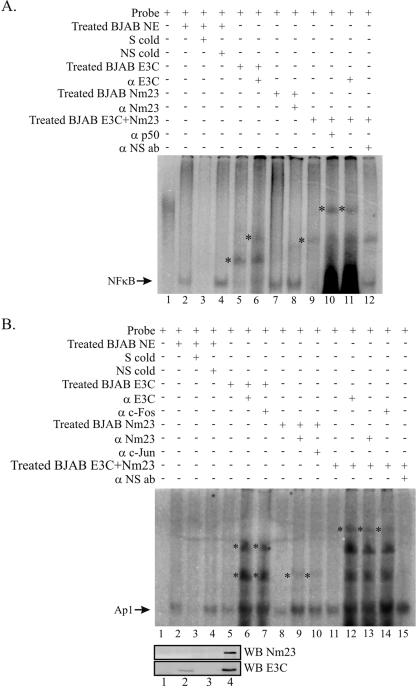
A specific NFκB shift was observed in NE from BJAB cells, and the specificity of the shift was verified through its disappearance in the presence of specific competitor (Fig. 7A, compare lanes 2 and 3). No effect was seen in the presence of nonspecific competitor (Fig. 7A, lane 4). The mobility of the NFκB probe was reduced by the NE from BJAB cells overexpressing EBNA3C (Fig. 7A, lane 5), with the presence of EBNA3C in the complex verified by additional supershifting in the presence of anti-EBNA3C antibody (Fig. 7A, lane 6). BJAB NE where Nm23-H1 was overexpressed had no obvious effect on the mobility of the NFκB probe at this level of detection (Fig. 7A, lanes 7 and 8). The lack of effect is likely due to the very low levels of Nm23-H1 that are normally found in the nucleus, as is indicated by the representative Western blot assays of the NEs (Fig. 7B). The BJAB NE from cells expressing both EBNA3C and Nm23-H1 reduced the mobility of the probe to a greater extent than that seen with the EBNA3C NE above, suggesting that EBNA3C and Nm23-H1 are likely to be part of a multiprotein complex bound to the probe (Fig. 7A, lane 9). The probe was additionally supershifted by anti-p50 and anti-EBNA3C antibodies, indicating their presence in the complex (Fig. 7A, lanes 10 and 11), while nonspecific antibody had no effect (Fig. 7A, lane 12).
FIG. 7.
EBNA3C and Nm23-H1 form complexes with Ap1 and NFκB bound to their cognate sequences. (A) The probe for the EMSA consisted of the NFκB binding site from the MMP-9 promoter along with the flanking bases. (B) The probe for the EMSA consisted of the Ap1 binding site from the MMP-9 promoter along with the flanking bases. The arrows indicate the position of either the Ap1/DNA or the NFκB/DNA complex, with the asterisks indicating the positions of shifted complexes with BJAB NEs transfected with either EBNA3C or EBNA3C and Nm23-H1 and supershifted complexes in the presence of antibody (ab) against EBNA3C, Nm23-H1, c-Fos, c-Jun, or p50. S, specific probe; NS nonspecific probe; cold, unlabeled; WB, Western blot.
To determine if EBNA3C and Nm23-H1 formed a complex with Ap1 bound to DNA, an EMSA was also performed as described above. The results showed a specific Ap1 shift in BJAB NE, with the specificity of the shift verified through its disappearance in the presence of specific competitor (Fig. 7B, compare lanes 2 and 3), with no effect seen in the presence of nonspecific competitor (Fig. 7B, lane 4). NEs from BJAB cells expressing EBNA3C had no obvious effect on the mobility of the probe (Fig. 7B, lane 5). However, the probe was clearly supershifted in the presence of anti-EBNA3C and anti-c-Fos antibodies, with the multiple bands probably due to the formation of higher-order complexes (Fig. 7B, lanes 6 and 7). BJAB NE where Nm23-H1 was expressed also had no effect on the mobility of the NFκB probe (Fig. 7B, lanes 8), but a faint supershifted band was observed in the presence of anti-Nm23-H1 and anti-c-Jun antibodies (Fig. 7B, lanes 9 and 10). The weak supershift is likely due to the very low levels of Nm23-H1 that are normally found in the nucleus, as indicated by the representative Western blot assays of the NEs (Fig. 7B). The BJAB NE from cells expressing both proteins also had no effect on the mobility of the probe, in line with the observed effect of the other NEs (Fig. 7B, lane 11). In the presence of anti-EBNA3C, anti-Nm23-H1, and anti-c-Fos antibodies, the same supershifted bands as with EBNA3C by itself were observed along with an additional higher band suggesting the presence of Nm23-H1 in the complex (Fig. 7B, lanes 12, 13, and 14). Addition of nonspecific antibody did not result in any supershifted complexes detected in our assay (Fig. 7B, lane 15). These results suggest that a multiprotein complex of EBNA3C and Nm23-H1 may regulate cellular promoters through binding to the Ap1 and NFκB transcription factors bound to their sequence-specific binding sites.
DISCUSSION
EBV transformation of B lymphocytes requires a subset of six latent proteins, including EBNA3C. EBNA3C has been shown to function as a transcriptional activator or repressor, depending on its interaction with specific cellular factors (38). When transfected with EBNA2, EBNA3C has been shown to significantly increase LMP1 expression in some assays while independently it has only a minimal effect on LMP1 expression (20). Additionally, previous studies have shown that EBNA3C can also repress the major LMP1 latent promoter in large part through its interaction with the RBP-Jκ transcription repressor, through disruption of the EBNA2-RBP-Jκ interaction, as well as the interaction of RBP-Jκ with its cognate sequence within the LMP1 promoter (22, 30, 31). In this report, we show additional interactions between EBNA3C and the cellular transcription factors Sp1, the NFκB p50 subunit, and the Ap-1 component c-Fos.
The interaction between EBNA3C and Nm23-H1 is of interest as it presents an effect where the interaction of this essential viral oncoprotein and the known cellular metastasis suppressor and regulator of cell proliferation can lead to reversion of cell migration in vitro and affect cell proliferation and metastasis in EBV-positive tumors (36). The region of EBNA3C which binds Nm23-H1 has been mapped to a 39-aa carboxy-terminal sequence located between the glutamine- and proline-rich domains of the protein (39). The location of this binding site is interesting because the glutamine-rich region is known to function as a transactivation domain, suggesting that its interaction with Nm23-H1 may affect the ability of EBNA3C to regulate transcription (32). Moreover, it has previously been shown that EBNA3C can increase Nm23-H1-mediated activation of a basal promoter and that this effect is dependent on the glutamine-rich region since the Nm23-H1 binding domain alone had little effect on activation (39). Thus, the interaction domain may serve as a recruitment domain for the glutamine-rich functional activator of EBNA3C.
The mechanism by which EBNA3C is able to reverse the cell migratory effects of Nm23-H1 is not known, though it has been suggested that it may in part be due to upregulation of adhesion molecules E-cadherin and α-integrins (36). Additionally, it is also possible that other functional effects of Nm23-H1 are affected by EBNA3C. In this report, we focus on regulation of MMP-9. The type IV collagenase MMP-9 is often expressed by malignant tumor cells, and its release has previously been correlated with metastasis (9, 19, 24, 26, 48). Furthermore, EBV LMP1 has been shown to cause increased expression of MMP-9 through the NFκB signaling pathway, with a lesser effect through the Ap1 pathway, but with Ap1 remaining important (41).
It should be noted that no clear link has been previously established between MMP-9 expression and Nm23-H1 levels. Here we report an observed increase in MMP-9 expression in the presence of Nm23-H1 and EBNA3C. In line with the observed synergistic nature of the regulatory effect of these two proteins, neither protein had relatively large effects when expressed independently; however, when they were expressed together a relatively high level of activation was observed (39). Therefore, a possible mechanism by which the association of EBNA3C with Nm23-H1 may lead to increased metastatic potential of tumor cells is through increasing expression of MMP-9, which results in increased degradation of collagen IV, a major component of the basement membrane. This is likely to result in increased metastasis from primary tumor sites.
A previous report of an Nm23 protein regulating an MMP was that of the rat homolog of Nm23-H1, Nm23-β, which was shown to downregulate MMP-2, the other member of the gelatinase MMP family (4). However, in oral squamous cell carcinoma cells, Nm23-H1 suppression of migration was found to be independent of MMP-9 expression (12). The significance of the later finding in the context of our observed results has yet to be resolved. Our model indicates that an Nm23/EBNA3C complex can regulate MMP-9. It is, however, possible that the formation of a complex of EBNA3C and Nm23-H1 may be independent of other mechanisms through which Nm23-H1 can suppress metastasis. Therefore, even if Nm23-H1 does not independently regulate MMP-9, the association with EBNA3C is likely to be sufficient to overcome its antimetastatic activities.
Mutational studies of the MMP-9 promoter revealed that NFκB and Ap1 may be critical for both Nm23-H1- and EBNA3C-mediated upregulation, while the effect with Sp1 was more subdued. The results of these transient reporter assays were further supported by gelatin zymography and RT-PCR results from EBV-negative C33a epithelial cells, which demonstrated the same general trend as the reporter assay. These studies corroborate the finding that Nm23-H1 and EBNA3C cooperate with each other by direct association or indirectly through association with other known transcription activators to upregulate MMP-9 activities.
Nm23-H1 has been shown to localize to the cytoplasm; however, in the presence of EBNA3C, its localization has been shown to shift to an almost exclusively nuclear signal (36). This change in localization strengthens the possibility that Nm23-H1 may function as a transcriptional regulator. However, in uninfected cells low levels have been detected in the nucleus, suggesting that a cellular partner may play a role in this translocation or that Nm23-H1 itself may be posttranslationally modified to mediate this event. Based on our IP data and GST results, we have determined that both Nm23-H1 and EBNA3C bind to the cellular transcription activators NFκB, Ap1, and Sp1. Additionally, promoter analyses suggest that these associations are important for the regulation of the MMP-9 promoter. This finding is the first report of the association of Nm23-H1 with cellular transcription activators. The reporter analyses provide strong evidence that Nm23-H1 is capable of regulating cellular proteins through its interaction with transcription activators bound to their responsive elements within the promoter of the target molecules. It has also been suggested that Nm23-H1 may directly bind DNA as reported, showing repression of transcriptional activity of platelet-derived growth factor A (21). However, this study has not been further confirmed and we have not been able to show that Nm23-H1 directly bound to DNA in our studies. Additional studies should help determine this potential dual effect on transcriptional regulation mediated by Nm23-H1.
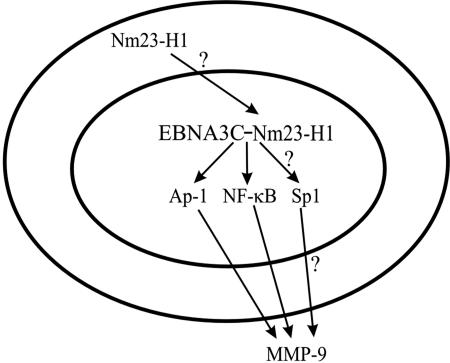
The EMSA data provide convincing evidence that EBNA3C and Nm23-H1 likely form a multiprotein complex capable of regulating MMP-9 primarily through the Ap1 and NFκB transcription factors bound to the MMP-9 promoter. These results are supported by previous data showing that Nm23-H1 and EBNA3C could enhance the transcriptional activation of a responsive Gal4 promoter (39). While these complexes also demonstrated that the proteins could independently form complexes with the binding sites within the promoter, the additional data from the reporter assays, enzymatic assay, and RT-PCR strongly support the possibility that the regulation occurs predominantly when these two proteins are in a complex and they can substantially upregulate MMP-9 transcription. Additionally, the upregulation of MMP-9 may also be due in part to the change in cellular localization of Nm23-H1 to the nucleus. This change in localization may result in a change in the level of signal transduction from an Nm23-H1-regulated pathway, thereby affecting the regulation of MMP-9 (Fig. 8). Previous studies involving specific transcription factors Ap-1 and NFκB within the urokinase plasmigen receptor (uPAR) promoter hint at a role for Nm23-H1 effects mediated through the Rho signaling pathway (2). However, a more detailed analysis should verify this involvement of Nm23-H1 in regulating MMP-9 in a Rho-dependent manner.
FIG. 8.
Hypothetical model for the regulation of MMP-9 by EBNA3C and Nm23-H1. Regulation of MMP-9 in EBNA3C-expressing cells first involves the recruitment of Nm23-H1 to the nucleus by EBNA3C through an undetermined mechanism. Once in the nucleus, these two proteins form a complex with one another, which enables them to transactivate the MMP-9 promoter through the binding of DNA-responsive elements in the promoter. The binding of these elements is mediated through the complex binding c-Jun, c-Fos, NFκB (p50), and possibly Sp1.
Our results suggest that the interaction between Nm23-H1 and EBNA3C can lead to upregulation of the metastasis-associated protein MMP-9. The role of EBV in metastasis is clearly very complex, given the difference in viral protein expression across the latency programs established by EBV, while the potential for metastasis exists with the associated cancers (29). The overall role of EBNA3C in this process may be only one aspect of this complicated regulatory pathway in that it is only expressed during type III latency (13, 47). Only a few known EBV-associated cancers are associated with the latency III program, and they are mostly seen in immunocompromised patients (29, 42). The prevalence of human immunodeficiency virus and transplants continues to rise; therefore, the importance of EBNA3C in EBV-associated malignancies is also expected to increase as the incidence of these cancers increases.
Acknowledgments
We thank Joseph Pagano (University of North Carolina School of Medicine, Chapel Hill) for providing the C33a cells, as well as all of the reporter plasmids, which were initially received from Hiroshi Sato (Cancer Research Institute, Kanazawa University, Kanazawa, Japan), as well as Satoru Kondo (University of North Carolina School of Medicine, Chapel Hill) for assistance with the zymography.
J.S.K. is supported by the Lady Tata Memorial Trust. This work was supported by NIH grants NCI CA72150-07, NIDCR DE14136-01, and NCI CA108461 to E.S.R. E.S.R. is a scholar of the Leukemia and Lymphoma Society of America.
REFERENCES
- 1.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benitah, S. A., P. F. Valeron, L. van Aelst, C. J. Marshall, and J. C. Lacal. 2004. Rho GTPases in human cancer: an unresolved link to upstream and downstream transcriptional regulation. Biochim. Biophys. Acta 1705:121-132. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet, M., J. M. Guinebretiere, E. Kremmer, V. Grunewald, E. Benhamou, G. Contesso, and I. Joab. 1999. Detection of Epstein-Barr virus in invasive breast cancers. J. Natl. Cancer Inst. 91:1376-1381. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, S., M. A. Alfonso-Jaume, P. R. Mertens, and D. H. Lovett. 2002. Tumour metastasis suppressor, nm23-β, inhibits gelatinase A transcription by interference with transactivator Y-box protein-1 (YB-1). Biochem. J. 366:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotter, M. A., II, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudin, P., C. Trocme, S. Berthier, S. Kieffer, J. Boutonnat, C. Lamy, A. Surla, J. Garin, and F. Morel. 2000. TIMP-1/MMP-9 imbalance in an EBV-immortalized B lymphocyte cellular model: evidence for TIMP-1 multifunctional properties. Biochim. Biophys. Acta 1499:19-33. [DOI] [PubMed] [Google Scholar]
- 8.Giannelli, G., J. Falk-Marzillier, O. Schiraldi, W. G. Stetler-Stevenson, and V. Quaranta. 1997. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science 277:225-228. [DOI] [PubMed] [Google Scholar]
- 9.Hiratsuka, S., K. Nakamura, S. Iwai, M. Murakami, T. Itoh, H. Kijima, J. M. Shipley, R. M. Senior, and M. Shibuya. 2002. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell 2:289-300. [DOI] [PubMed] [Google Scholar]
- 10.Kardassis, D., P. Papakosta, K. Pardali, and A. Moustakas. 1999. c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J. Biol. Chem. 274:29572-29581. [DOI] [PubMed] [Google Scholar]
- 11.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan, M. H., M. Yasuda, F. Higashino, S. Haque, T. Kohgo, M. Nakamura, and M. Shindoh. 2001. nm23-H1 suppresses invasion of oral squamous cell carcinoma-derived cell lines without modifying matrix metalloproteinase-2 and matrix metalloproteinase-9 expression. Am. J. Pathol. 158:1785-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kieff, E. 1996. Epstein-Barr virus and its replication, 3rd ed., vol. 2., p. 2343-2397. Lippincott-Raven, Philadelphia, Pa.
- 14.Knight, J. S., M. A. Cotter II, and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 15.Lacombe, M. L., L. Milon, A. Munier, J. G. Mehus, and D. O. Lambeth. 2000. The human Nm23/nucleoside diphosphate kinases. J. Bioenerg. Biomembr. 32:247-258. [DOI] [PubMed] [Google Scholar]
- 16.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leone, A., U. Flatow, C. R. King, M. A. Sandeen, I. M. Margulies, L. A. Liotta, and P. S. Steeg. 1991. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell 65:25-35. [DOI] [PubMed] [Google Scholar]
- 18.Leone, A., U. Flatow, K. VanHoutte, and P. S. Steeg. 1993. Transfection of human nm23-H1 into the human MDA-MB-435 breast carcinoma cell line: effects on tumor metastatic potential, colonization and enzymatic activity. Oncogene 8:2325-2333. [PubMed] [Google Scholar]
- 19.Liabakk, N. B., I. Talbot, R. A. Smith, K. Wilkinson, and F. Balkwill. 1996. Matrix metalloprotease 2 (MMP-2) and matrix metalloprotease 9 (MMP-9) type IV collagenases in colorectal cancer. Cancer Res. 56:190-196. [PubMed] [Google Scholar]
- 20.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma, D., Z. Xing, B. Liu, N. G. Pedigo, S. G. Zimmer, Z. Bai, E. H. Postel, and D. M. Kaetzel. 2002. NM23-H1 and NM23-H2 repress transcriptional activities of nuclease-hypersensitive elements in the platelet-derived growth factor-A promoter. J. Biol. Chem. 277:1560-1567. [DOI] [PubMed] [Google Scholar]
- 22.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazaki, H., M. Fukuda, Y. Ishijima, Y. Takagi, T. Iimura, A. Negishi, R. Hirayama, N. Ishikawa, T. Amagasa, and N. Kimura. 1999. Overexpression of nm23-H2/NDP kinase B in a human oral squamous cell carcinoma cell line results in reduced metastasis, differentiated phenotype in the metastatic site, and growth factor-independent proliferative activity in culture. Clin. Cancer Res. 5:4301-4307. [PubMed] [Google Scholar]
- 24.Murono, S., T. Yoshizaki, H. Sato, H. Takeshita, M. Furukawa, and J. S. Pagano. 2000. Aspirin inhibits tumor cell invasiveness induced by Epstein-Barr virus latent membrane protein 1 through suppression of matrix metalloproteinase-9 expression. Cancer Res. 60:2555-2561. [PubMed] [Google Scholar]
- 25.Ouatas, T., M. Salerno, D. Palmieri, and P. S. Steeg. 2003. Basic and translational advances in cancer metastasis: Nm23. J. Bioenerg. Biomembr. 35:73-79. [DOI] [PubMed] [Google Scholar]
- 26.Parsons, S. L., S. A. Watson, H. M. Collins, N. R. Griffin, P. A. Clarke, and R. J. Steele. 1998. Gelatinase (MMP-2 and -9) expression in gastrointestinal malignancy. Br. J. Cancer 78:1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy, K. B., J. S. Krueger, S. B. Kondapaka, and C. A. Diglio. 1999. Mitogen-activated protein kinase (MAPK) regulates the expression of progelatinase B (MMP-9) in breast epithelial cells. Int. J. Cancer 82:268-273. [DOI] [PubMed] [Google Scholar]
- 29.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, 3rd ed., vol. 2., p. 2397-2447. Lippincott-Raven, Philadelphia, Pa.
- 30.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sample, C., and B. Parker. 1994. Biochemical characterization of Epstein-Barr virus nuclear antigen 3A and 3C proteins. Virology 205:534-539. [DOI] [PubMed] [Google Scholar]
- 33.Sato, H., Y. Kida, M. Mai, Y. Endo, T. Sasaki, J. Tanaka, and M. Seiki. 1992. Expression of genes encoding type IV collagen-degrading metalloproteinases and tissue inhibitors of metalloproteinases in various human tumor cells. Oncogene 7:77-83. [PubMed] [Google Scholar]
- 34.Sato, H., and M. Seiki. 1993. Regulatory mechanism of 92 kDa type IV collagenase gene expression which is associated with invasiveness of tumor cells. Oncogene 8:395-405. [PubMed] [Google Scholar]
- 35.Stamenkovic, I. 2000. Matrix metalloproteinases in tumor invasion and metastasis. Semin. Cancer Biol. 10:415-433. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian, C., M. A. Cotter II, and E. S. Robertson. 2001. Epstein-Barr virus nuclear protein EBNA-3C interacts with the human metastatic suppressor Nm23-H1: a molecular link to cancer metastasis. Nat. Med 7:350-355. [DOI] [PubMed] [Google Scholar]
- 37.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian, C., J. S. Knight, and E. S. Robertson. 2002. The Epstein Barr nuclear antigen EBNA3C regulates transcription, cell transformation and cell migration. Front. Biosci. 7:d704-d716. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 76:8702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagashira, H., K. Hamazaki, N. Tanaka, C. Gao, and M. Namba. 1998. Reduced metastatic potential and c-myc overexpression of colon adenocarcinoma cells (Colon 26 line) transfected with nm23-R2/rat nucleoside diphosphate kinase alpha isoform. Int. J. Mol. Med. 2:65-68. [DOI] [PubMed] [Google Scholar]
- 41.Takeshita, H., T. Yoshizaki, W. E. Miller, H. Sato, M. Furukawa, J. S. Pagano, and N. Raab-Traub. 1999. Matrix metalloproteinase 9 expression is induced by Epstein-Barr virus latent membrane protein 1 C-terminal activation regions 1 and 2. J. Virol. 73:5548-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorley-Lawson, D. A., and A. Gross. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328-1337. [DOI] [PubMed] [Google Scholar]
- 43.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trocme, C., P. Gaudin, S. Berthier, C. Barro, P. Zaoui, and F. Morel. 1998. Human B lymphocytes synthesize the 92-kDa gelatinase, matrix metalloproteinase-9. J. Biol. Chem. 273:20677-20684. [DOI] [PubMed] [Google Scholar]
- 45.Westermarck, J., and V. M. Kahari. 1999. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 13:781-792. [PubMed] [Google Scholar]
- 46.Yoshizaki, T., H. Sato, M. Furukawa, and J. S. Pagano. 1998. The expression of matrix metalloproteinase 9 is enhanced by Epstein-Barr virus latent membrane protein 1. Proc. Natl. Acad. Sci. USA 95:3621-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, L. S., and P. G. Murray. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108-5121. [DOI] [PubMed] [Google Scholar]
- 48.Zeng, Z. S., A. M. Cohen, and J. G. Guillem. 1999. Loss of basement membrane type IV collagen is associated with increased expression of metalloproteinases 2 and 9 (MMP-2 and MMP-9) during human colorectal tumorigenesis. Carcinogenesis 20:749-755. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]