Abstract
Several recent reports indicate that cholesterol might play an important role in human immunodeficiency virus type 1 (HIV-1) replication. We investigated the effects of HIV-1 infection on cholesterol biosynthesis and uptake using microarrays. HIV-1 increased gene expression of cholesterol genes in both transformed T-cell lines and primary CD4+ T cells. Consistent with our microarray data, 14C-labeled mevalonate and acetate incorporation was increased in HIV-1-infected cells. Our data also demonstrate that changes in cholesterol biosynthesis and uptake are only observed in the presence of functional Nef, suggesting that increased cholesterol synthesis may contribute to Nef-mediated enhancement of virion infectivity and viral replication.
Several studies have highlighted the importance of cholesterol and lipid rafts in the human immunodeficiency virus type 1 (HIV-1) life cycle (1, 10, 13-15). Rafts are proposed to act as platforms for viral entry, facilitating interactions between CD4, coreceptors, and incoming virions (2, 12). Although this role has been questioned recently (16, 17), there is mounting evidence that rafts are important for HIV-1 assembly and budding. HIV-1 Gag and Env viral structural components are concentrated in rafts, facilitating assembly (4, 6, 8, 11, 14, 15, 19). It has also been suggested that Nef increases synthesis and transport of cholesterol to rafts and progeny virions (23). It is therefore reasonable to hypothesize that HIV-1 may have evolved the capacity to up-regulate intracellular cholesterol. We have addressed this hypothesis here, using gene expression profiling and metabolic labeling of HIV-1-infected cells.
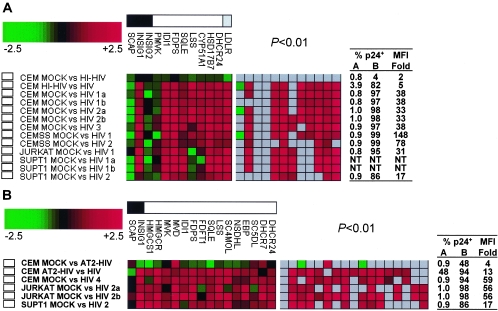
We infected CCRF-CEM, CEMss, Jurkat clone E6-1, and SupT1 cells with HIV-1LAI at a multiplicity of infection of 2 and confirmed infection levels by flow cytometry as described (22). Infection levels were >82% (mean, 96% ± 6%) with median fluorescence intensity increased 17- to 148-fold (mean, 55-fold ± 45-fold) in HIV-1-infected cells compared to mock-infected cells (Fig. 1A). Total RNA extraction, probe labeling, microarray processing, and data analysis were performed essentially as described (22). Complete microarray data sets are available at http://expression.microslu.washington.edu.
FIG. 1.
HIV-1 infection alters transcripts involved in cholesterol biosynthesis and uptake. Changes in expression of SREBF-2-regulated transcripts were determined using microarrays (A and B) or real-time reverse transcription-PCR (C and D). (A and B) Expression of SREBF-2-regulated transcripts was determined using in-house microarrays representing ≈4,500 unique human genes (A), or commercial microarrays representing ≈15,000 unique human genes (B). Gene groupings are indicated in the bar at the top of each panel for sterol biosynthesis regulators (black box), enzymes (white box) and the LDL receptor (grey box). Shown in color code below these bars are fold changes in mRNA levels in 24-h HIV-1LAI-infected CD4+ T-cell lines compared with mock-infected cells. Numbers 1 to 4 in the experiment name denote biological replicates, while letters a and b denote technical replicates. Ratios of change in mRNA levels in infected cells versus controls are depicted as green (down-regulated) or red (up-regulated) boxes. Left panels show all ratios, and the right panel shows box colors only for those ratios with P values of <0.01. The tables on the right of the panels depict the percentage of p24gag-expressing cells in controls (column A) and infected cells (column B) and the fold increase in median fluorescence intensity (MFI) for each of the conditions being compared in the corresponding microarray experiment. Heat-inactivated (HI-HIV) and chemically inactivated HIV-1 (AT2-HIV) were compared to both mock and infectious HIV-1 in separate experiments. (C and D) Expression of SREBF-2 and 3 SREBF-2-regulated transcripts in HIV-1-infected CD4+ T-cell lines (C) and primary CD4+ T cells (D) as determined by real-time reverse transcription-PCR (N = 2 to 5). Results were normalized by subtracting β-actin cycle threshold (Ct) values measured in the same samples from the experimental gene Ct values, resulting in normalized Δ Ct values for each mock or infected sample. Differences between corresponding mock and infected samples were expressed as ΔΔ Ct, subtracting Δ Ct (infected) from Δ Ct (mock). As each Ct difference corresponds to a twofold change in mRNA levels, this was translated to the fold changes depicted in the graph using 2ΔΔCt. Each sample was tested in triplicate (mean and standard deviation are depicted). P values: *, <0.05; **, <0.01; ***, <0.001.
Expression of seven cholesterol enzymes (IDI1, FDPS, SQLE, LSS, CYP51, HSD17B7, and DHCR24), the low-density lipoprotein receptor (LDLR), and one cholesterol regulator (INSIG1) was increased in infected cells (Fig. 1A). No significant increase was observed in mRNA levels for three other genes involved in cholesterol biosynthesis (PMVK, SCAP, and INSIG2). Expression changes were observed 24 h postinfection, but not at the earlier time points tested (1, 4, 8, and 12 h postinfection; data not shown). Treatment of cells with heat-inactivated HIV-1 (2 h at 56°C) did not result in regulation of the cholesterol genes (Fig. 1A, first row), suggesting that intact virus particles are required. Regulations were observed in all cell lines, suggesting that induction of cholesterol biosynthesis and uptake might be a general consequence of HIV-1 infection.
The cholesterol biosynthesis pathway consists of more than 20 enzymes (Table 1), whose expression is regulated by the sterol-responsive element binding factor 2 (SREBF-2) (9). The rate-limiting step in this pathway is the conversion of 3-hydroxy-3-methylglutaryl coenzyme A into mevalonate by 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGCR). Activation of SREBF-2 also increases expression of LDLR, resulting in increased uptake of extracellular LDL.
TABLE 1.
SREBF-2 and SREBF-2-regulated genes
| Product | Gene name | HUGO | LocusLink identification no. | EC no. | RefSeqb | HIV induced |
|---|---|---|---|---|---|---|
| Sterol-responsive element binding factor 2 | SREBF2 | 6721 | NM_004599 | Yes | ||
| SREBP cleavage activating protein | SCAP | 22937 | NM_012235 | No | ||
| Insulin-induced gene 1 | INSIG1 | 3638 | NM_005542 | Yes | ||
| Insulin-induced gene 2 | INSIG2 | 51141 | NM_016133 | No | ||
| Acetyl-CoA | Acetyl-CoA acetyltransferase-2 | ACAT2 | 39 | 2.3.1.9 | NM_005891 | No |
| Acetoacetyl-CoA synthetase | AACS | 65985 | 6.2.1.16 | NM_023928 | NTa | |
| Acetoacetyl-CoA | HMG-CoA synthase 1 (soluble) | HMGCS1 | 3157 | 2.3.3.10 | NM_002130 | Yes |
| HMG-CoA synthase 2 (mitochondrial) | HMGCS2 | 3158 | 4.1.3.5 | NM_005518 | No | |
| HMG-CoA reductase | HMGCR | 3156 | 1.1.1.34 | NM_000859 | Yes | |
| Mevalonate | Mevalonate kinase | MVK | 4598 | 2.7.1.36 | NM_000431 | Yes |
| Phosphomevalonate kinase | PMVK | 10654 | 2.7.4.2 | NM_006556 | No | |
| Mevalonate pyrophosphate decarboxylase | MVD | 4597 | 4.1.1.33 | NM_002461 | Yes | |
| Isopentenyl diphosphate delta isomerase | IDI1 | 3422 | 5.3.3.2 | NM_004508 | Yes | |
| Farnesyl diphosphate synthetase | FDPS | 2224 | 2.5.1.1. | NM_002004 | Yes | |
| Squalene synthase | FDFT1 | 2222 | 2.5.1.21 | NM_004462 | Yes | |
| Squalene | Squalene epoxidase | SQLE | 6713 | 1.14.99.7 | NM_003129 | Yes |
| Lanosterol cyclase | LSS | 4047 | 5.4.99.7 | NM_002340 | Yes | |
| Lanosterol | Lanosterol 14 alpha demethylase | CYP51A1 | 1595 | 1.14.14.1 | NM_000786 | Yes |
| Sterol C4 methyl oxidase | SC4MOL | 6307 | NM_006745 | Yes | ||
| NAD(P)H steroid dehydrogenase-like | NSDHL | 50814 | NM_015922 | Yes | ||
| 17-Beta hydroxysteroid dehydrogenase 7 | HSD17B7 | 51478 | 1.1.1.62 | NM_016371 | Yes | |
| Emopamil binding protein (sterol isomerase) | EBP | 10682 | 5.3.3.5 | NM_006579 | Yes | |
| Sterol C5 desaturase (lathosterol oxidase) | SC5DL | 6309 | 1.3.3.2 | NM_006918 | Yes | |
| 7-Dehydrocholesterol reductase | DHCR7 | 1717 | 1.3.1.21 | NM_001360 | Yes | |
| Desmosterol reductase | DCE | 1631 | NT | |||
| Cholesterol | Low-density lipoprotein receptor | LDLR | 3949 | NM_000527 | Yes |
NT, not tested.
RefSeq, reference sequence identifier.
To determine whether other SREBF-2-regulated genes are also regulated by HIV-1 infection at 24 h, we used a commercial microarray platform representing ≈15,000 unique human genes (Agilent Technologies, Palo Alto, CA). HIV-1 infection levels were >85% (mean, 94% ± 5%) with median fluorescence intensity increased 17- to 59-fold (mean, 47-fold ± 20-fold) in HIV-1-infected cells compared to mock-infected cells (Fig. 1B). Results for SCAP, INSIG1, IDI1, FDPS, SQLE, and DHCR24 were confirmed. INSIG2, PMVK, CYP51, HSD17B7, and LDLR were not tested in these experiments. However, expression of 10 additional sterol enzymes (HMGCS1, HMGCR, MVK, MVD, FDFT1, SC4MOL, NSDHL, EBP, SC5DL, and DHCR7) was increased by HIV-1 infection, including the enzyme that is responsible for the rate-limiting step in sterol biosynthesis, HMGCR (Fig. 1B). Exposure of cells to aldrithiol-2-treated noninfectious HIV-1 (18) did not change the expression of this gene set (Fig. 1B, first row), suggesting that productive infection is required. No expression changes in SREBF-2-regulated genes were detected in response to influenza A virus, interferon, or heat shock, providing additional evidence that SREBF-2 induction is specific to HIV-1 infection (data not shown).
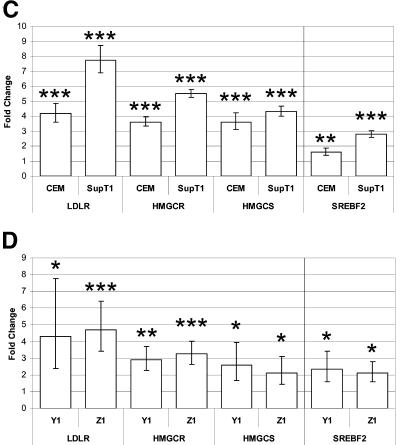
To confirm HIV-1-induced gene expression changes, we used Taqman primer-probe sets (Applied Biosystems, Foster City, CA). In each case the mRNA levels for LDLR, HMGCR, and HMGCS1 were higher in HIV-1-infected SupT1 and CCRF-CEM cells than in mock-infected cells, providing independent validation of our microarray results (Fig. 1C, left panel). Increased expression of LSS, FDPS, and IDI1 in HIV-1-infected CEMss cells had previously been confirmed using semiquantitative reverse transcription-PCR (22). In addition, increased LDLR expression in HIV-1-infected CEM and SupT1 cells were confirmed by fluorescence-activated cell sorting (data not shown). Moreover, expression of SREBF-2, the transcription factor regulation regulating the cholesterol biosynthesis pathway, was also up-regulated in HIV-1-infected cells (Fig. 1C, right panel).
Finally, we also verified induction of LDLR, HMGCR, HMGCS1, and SREBF-2 by HIV-1 in primary CD4+ T cells. CD4+ T cells were purified from peripheral blood mononuclear cells by negative selection (StemCell Technologies, Vancouver, Canada) to >96% purity. Purified cells were stimulated with phytohemagglutinin (Murex HA16, Remel Inc., Lenexa, KS) for 3 days before infection with HIV-1 at a multiplicity of infection of 2. Results for two representative donors (of four to five donors tested) are shown in Fig. 1D. In each case expression of LDLR, HMGCR, HMGCS1, and SREBF-2 was higher in HIV-1-infected cells than in mock-infected cells, indicating that the regulation is not merely observed in transformed T-cell lines, but also in primary cells.
To assess changes in cholesterol production occurring as a result of HIV-1 infection, we labeled cells infected at a multiplicity of infection of 2 as above for 6 h with 14C-labeled mevalonate (Perkin-Elmer Life Sciences) (5, 7). Cells were washed with phosphate-buffered saline, resuspended in H2O, and extracted with chloroform/methanol (2:1 vol/vol) and petroleum ether. Samples were then spotted onto silica gel 60 thin-layer chromatography plates and chromatographed in toluene/diethylether (9:1 vol/vol). Labeled products were visualized using a STORM phosphorimager.
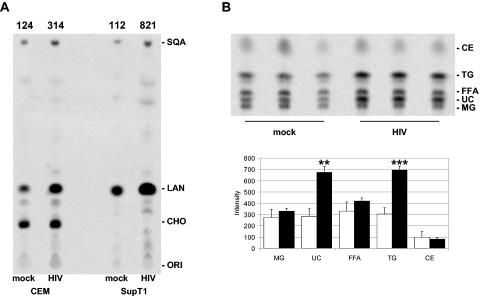
As expected, key biosynthetic intermediates downstream from mevalonate (squalene, lanosterol, and cholesterol) and several minor intermediate products were detected. Consistent with the microarray results, the rate of 14C-labeled mevalonate incorporation in downstream intermediates was increased 2.5-fold in infected CCRF-CEM cells and 7.4-fold in infected SupT1 cells (Fig. 2A). Because the rate-limiting step of cholesterol biosynthesis is not assessed using 14C-labeled mevalonate, we also labeled HIV-1-infected cells for 2 h with 14C-labeled acetate. Cells were washed with phosphate-buffered saline and extracted with hexane:isopropanol (3:1 vol/vol) and chloroform/methanol (2:1 vol/vol). Samples were spotted on thin-layer chromatography plates and run in hexane/glacial acetic acid/diethylether (90:1:25 vol/vol). Similar to 14C-labeled mevalonate labeling, 2.4-fold increased cholesterol production was measured in infected CCRF-CEM cells using 14C-labeled acetate (Fig. 2B). Therefore, increased expression levels of cholesterol enzymes observed in HIV-1-infected cells indeed resulted in increased cholesterol production.
FIG. 2.
HIV-1 infection increases 14C-labeled mevalonate and acetate incorporation. Cells were mock or HIV-1LAI infected for 24 h before addition of 14C-labeled mevalonate (A) or 14C-labeled acetate (B). After 6 or 2 h, respectively, cells were harvested, and lipids were extracted and separated using thin-layer chromatography. (A) Mevalonate results are representative of three separate experiments using CCRF-CEM, CEMss, Jurkat, or SupT1 cells. Numbers above the image denote signal intensity. Note that CYP51 mRNA expression was not detected in SupT1 cells and as a result no 14C-labeled cholesterol was produced. (B) Signal intensities for acetate labeling of mock-infected (open bars) or HIV-1-infected (solid bars) CEM cells are plotted in the graph below the image. The mean and standard deviation for the triplicate experiments are shown. Individual products were identified by comparison of the observed Rf factor with those of known sterols. ORI, origin; CHO, cholesterol; LAN, lanosterol; SQA, squalene; MG, monoglycerides; UC, free cholesterol; FFA, free fatty acids; TG, triglycerides; CE, cholesterol esters. P values: **, <0.01; ***, <0.001.
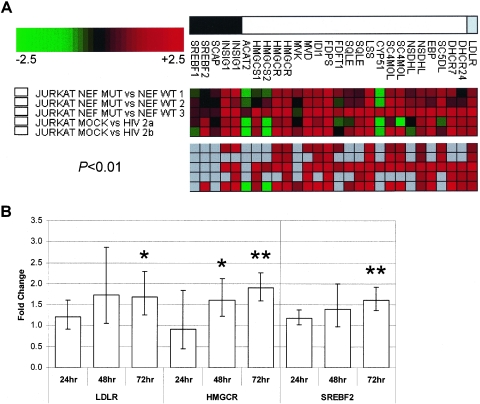
It has been suggested that Nef might increase synthesis and transport of cholesterol to rafts and progeny virions to increase viral infectivity (23), although this role of Nef was not confirmed in acutely infected T cells (21). To further investigate the effect of Nef on cholesterol biosynthesis, we determined gene expression profiles of Jurkat cells expressing HIV-1 nef-intact or nef-defective reading frames, using HIV-1 internal ribosome entry site (IRES)-enhanced green fluorescent protein (eGFP) replication-competent reporter viruses as described (20). Transduction efficiency was confirmed by fluorescence-activated cell sorting and was similar for both Nef constructs (% GFP-expressing cells: nef-intact, 78.1 ± 1.4; nef-defective, 81.7 ± 4.2).
Most genes affected by HIV-1LAI infection of Jurkat cells are also regulated by HIV-1NL4-3 infection of these cells, but only in the presence of a functional Nef (Fig. 3A). This was confirmed by real-time reverse transcription-PCR for LDLR, HMGCR and SREBF-2 in two additional time course experiments (Fig. 3B). In addition, increased expression of LDLR in the presence of functional Nef was confirmed by fluorescence-activated cell sorting (data not shown). This indicates that Nef indeed up-regulates SREBF-2 and SREBF-2-dependent cholesterol biosynthesis and uptake in transformed cell lines. These findings extend the observations by Zheng and colleagues (23), who reported induction of the CYP51 promoter by Nef. In addition, we find that Nef causes up-regulation of SREBF-2 and almost all SREBF-2-regulated genes, including the enzyme responsible for the rate-limiting step, HMGCR, and LDLR. This suggests that the increased infectivity of virus carrying wild-type Nef may be the result of this induction. It will be important to evaluate this function of Nef in primary cells and to further understand the mechanism through which regulation occurs.
FIG. 3.
HIV-1 Nef induces multiple genes involved in cholesterol biosynthesis and uptake. (A) Gene expression profiles of Jurkat T cells transduced with vesicular stomatitis virus G-pseudotyped HIV-1NL4-3 IRES-eGFP replication-competent reporter viruses containing nef-intact (WT) or nef-defective (MUT) reading frames or infected with HIV-1LAI or mock infected. Shown are fold changes in mRNA levels for sterol biosynthesis regulators (black box), enzymes (white box), and the LDL receptor (grey box). Ratios of change in mRNA levels in infected versus controls or wild-type versus mutant are depicted as green (down-regulated), red (up-regulated), or grey (not significantly changed at P < 0.01). The top panel shows all ratios irrespective of significance, whereas the bottom panel shows only those ratios with P values of < 0.01. Results for three independent transductions are shown. (B) Real-time reverse transcription-PCR analysis of SREBF-2 and SREBF-2-regulated transcripts in wild-type nef versus mutant nef at 24, 48, and 72 h postinfection (N = 2). Fold changes were determined as in Fig. 1. Each sample was tested in triplicate (mean and standard deviation are depicted). P values: *, <0.05; **, <0.01.
In summary, our results demonstrate that nef-intact HIV-1 infection induces a complex set of genes to increase cellular cholesterol levels. Consistent with an important role of cholesterol in the HIV-1 life cycle in vivo, it was recently reported that treatment of six HIV-1-infected patients with lovastatin induced a clear reduction in serum viral RNA loads that rebounded after discontinuation (3). Thus, cholesterol-lowering drugs might complement existing therapies to inhibit HIV-1 transmission and replication.
Acknowledgments
We thank Ginger K. Lehrman, Gemma C. O'Keeffe, and Dawn M. Cunningham for excellent technical assistance, Fred S. Buckner, John F. Oram, and Maria M. Culala for metabolic labeling protocols, and Michael G. Katze and colleagues for help with microarray data analysis. We are also indebted to Gary K. Geiss and Roger E. Bumgarner for valuable discussions, encouragement, and grant support.
This work was supported by the National Institutes of Health (R21 AI52028, P50 HG002360, and R01 AI058894), the University of Washington Center for AIDS Research and STDs, and grants from the Deutsche Forschungsgemeinschaft and the Wilhelm-Sander Foundation.
REFERENCES
- 1.Aloia, R. C., H. Tian, and F. C. Jensen. 1993. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc. Natl. Acad. Sci. USA 90:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Real, G., S. Jimenez-Baranda, E. Mira, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, M. Alegret, J. M. Pena, M. Rodriguez-Zapata, M. Alvarez-Mon, A. C. Martinez, and S. Manes. 2004. Statins inhibit HIV-1 infection by down-regulating Rho activity. J. Exp. Med. 200:541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding, L., A. Derdowski, J. J. Wang, and P. Spearman. 2003. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. J. Virol. 77:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folch, J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497-509. [PubMed] [Google Scholar]
- 6.Guyader, M., E. Kiyokawa, L. Abrami, P. Turelli, and D. Trono. 2002. Role for human immunodeficiency virus type 1 membrane cholesterol in viral internalization. J. Virol. 76:10356-10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haughan, P. A., M. L. Chance, and L. J. Goad. 1995. Effects of an azasterol inhibitor of sterol 24-transmethylation on sterol biosynthesis and growth of Leishmania donovani promastigotes. Biochem. J. 308:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holm, K., K. Weclewicz, R. Hewson, and M. Suomalainen. 2003. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55gag associates with membrane domains that are largely resistant to Brij98 but sensitive to Triton X-100. J. Virol. 77:4805-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton, J. D., J. L. Goldstein, and M. S. Brown. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Investig. 109:1125-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao, Z., L. M. Cimakasky, R. Hampton, D. H. Nguyen, and J. E. Hildreth. 2001. Lipid rafts and HIV pathogenesis: host membrane cholesterol is required for infection by HIV type 1. AIDS Res. Hum. Retroviruses 17:1009-1019. [DOI] [PubMed] [Google Scholar]
- 11.Lindwasser, O. W., and M. D. Resh. 2001. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. J. Virol. 75:7913-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manes, S., G. del Real, R. A. Lacalle, P. Lucas, C. Gomez-Mouton, S. Sanchez-Palomino, R. Delgado, J. Alcami, E. Mira, and A. C. Martinez. 2000. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep. 1:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maziere, J. C., J. C. Landureau, P. Giral, M. Auclair, L. Fall, A. Lachgar, A. Achour, and D. Zagury. 1994. Lovastatin inhibits HIV-1 expression in H9 human T lymphocytes cultured in cholesterol-poor medium. Biomed. Pharmacother. 48:63-67. [DOI] [PubMed] [Google Scholar]
- 14.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Percherancier, Y., B. Lagane, T. Planchenault, I. Staropoli, R. Altmeyer, J. L. Virelizier, F. Arenzana-Seisdedos, D. C. Hoessli, and F. Bachelerie. 2003. HIV-1 entry into T-cells is not dependent on CD4 and CCR5 localization to sphingolipid-enriched, detergent-resistant, raft membrane domains. J. Biol. Chem. 278:3153-3161. [DOI] [PubMed] [Google Scholar]
- 17.Popik, W., and T. M. Alce. 2004. CD4 receptor localized to non-raft membrane microdomains supports HIV-1 entry. Identification of a novel raft localization marker in CD4. J. Biol. Chem. 279:704-712. [DOI] [PubMed] [Google Scholar]
- 18.Rossio, J. L., M. T. Esser, K. Suryanarayana, D. K. Schneider, J. W. Bess, Jr., G. M. Vasquez, T. A. Wiltrout, E. Chertova, M. K. Grimes, Q. Sattentau, L. O. Arthur, L. E. Henderson, and J. D. Lifson. 1998. Inactivation of human immunodeficiency virus type 1 infectivity with preservation of conformational and functional integrity of virion surface proteins. J. Virol. 72:7992-8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rousso, I., M. B. Mixon, B. K. Chen, and P. S. Kim. 2000. Palmitoylation of the HIV-1 envelope glycoprotein is critical for viral infectivity. Proc. Natl. Acad. Sci. USA 97:13523-13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindler, M., S. Wurfl, P. Benaroch, T. C. Greenough, R. Daniels, P. Easterbrook, M. Brenner, J. Munch, and F. Kirchhoff. 2003. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sol-Foulon, N., C. Esnault, Y. Percherancier, F. Porrot, P. Metais-Cunha, F. Bachelerie, and O. Schwartz. 2004. The effects of HIV-1 Nef on CD4 surface expression and viral infectivity in lymphoid cells are independent of rafts. J. Biol. Chem. 279:31398-31408. [DOI] [PubMed] [Google Scholar]
- 22.van 't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng, Y. H., A. Plemenitas, C. J. Fielding, and B. M. Peterlin. 2003. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proc. Natl. Acad. Sci. USA 100:8460-8465. [DOI] [PMC free article] [PubMed] [Google Scholar]