Abstract
The TATA-binding protein (TBP) plays a crucial role in cellular transcription catalyzed by all three DNA-dependent RNA polymerases. Previous studies have shown that TBP is targeted by the poliovirus (PV)-encoded protease 3Cpro to bring about shutoff of cellular RNA polymerase II-mediated transcription in PV-infected cells. The processing of the majority of viral precursor proteins by 3Cpro involves cleavages at glutamine-glycine (Q-G) sites. We present evidence that suggests that the transcriptional inactivation of TBP by 3Cpro involves cleavage at the glutamine 104-serine 105 (Q104-S105) site of TBP and not at the Q18-G19 site as previously thought. The TBP Q104-S105 cleavage by 3Cpro is greatly influenced by the presence of an aliphatic amino acid at the P4 position, a hallmark of 3Cpro-mediated proteolysis. To examine the importance of host cell transcription shutoff in the PV life cycle, stable HeLa cell lines were created that express recombinant TBP resistant to cleavage by the viral proteases, called GG rTBP. Transcription shutoff was significantly impaired and delayed in GG rTBP cells upon infection with poliovirus compared with the cells that express wild-type recombinant TBP (wt rTBP). Infection of GG rTBP cells with poliovirus resulted in small plaques, significantly reduced viral RNA synthesis, and lower viral yields compared to the wt rTBP cell line. These results suggest that a defect in transcription shutoff can lead to inefficient replication of poliovirus in cultured cells.
Poliovirus (PV) is the prototype virus of a large group of medically important viruses (picornaviruses) that include those inducing poliomyelitis (polioviruses), infectious hepatitis (hepatitis A virus), the common cold (rhinoviruses), and encephalitis and myocarditis (coxsackie viruses) (31). The single-stranded, plus-polarity RNA genome (∼7,500 nucleotides) of PV (18, 30) is translated into one large polyprotein, which is cotranslationally processed by virally encoded proteases 2Apro, 3Cpro, and 3CDpro into mature viral structural and nonstructural proteins (22). The viral proteases have been studied extensively and found to be very specific in polyprotein cleavage; 3Cpro and 3CDpro cleave the polyprotein at glutamine-glycine (Q-G) bonds while the 2Apro cleaves only at tyrosine-glycine (Y-G) bonds (20). The proteases do not cleave every potential cleavage site within the polyprotein; other determinants such as accessibility and context of the cleavage site are also important.
Accurate initiation of transcription by RNA polymerase (Pol) II requires the assembly of a multiprotein complex on the core promoter around the mRNA start site (13). The multiprotein complex, consisting of at least seven to eight general transcription factors (GTFs) and RNA Pol II form a preinitiation complex at specific cis-acting elements (promoters) on the DNA template. The most common cis-acting element, the TATA box, is situated approximately 25 nucleotides upstream of the transcription start site. Among the GTFs in the complex, the TATA-binding protein (TBP) has been studied extensively (29). The TBP was first identified through its role in Pol II transcription, where it associates with 13 or 14 TBP-associated factors to form the TFIID complex (34). The unique aspect of TBP is its additional presence in complexes required for RNA Pol I (SL1) and RNA Pol III (TFIIIB) transcription. In both SL1 and TFIIIB complexes, TBP associates with a distinct set of TBP-associated factors (7, 33). Thus, TBP appears to be involved in all three RNA polymerase-mediated transcriptions. TBP has a bipartite structure with a highly conserved C-terminal core domain (amino acids 159 to 339), which folds as a molecular saddle, and is responsible for DNA binding via the concave underside. Additionally, the core also interacts with GTFs and other regulatory proteins via the solvent-exposed convex side. In contrast, the N-terminal region (amino acids 1 to 141) is variable in both length and sequence but is well conserved among vertebrates. It modulates DNA binding by interacting with the core domain and is characterized by a glutamine repeat region (Fig. 1). Recent studies have underscored the importance of the TBP N-terminal domain in transcriptional regulation both in yeast and mammalian systems (15, 21). Moreover, deletion of 55 and 96 amino acids from the N-terminal domain of TBP leads to inactivation of TATA-mediated transcription from the U6 small nuclear RNA promoter (24).
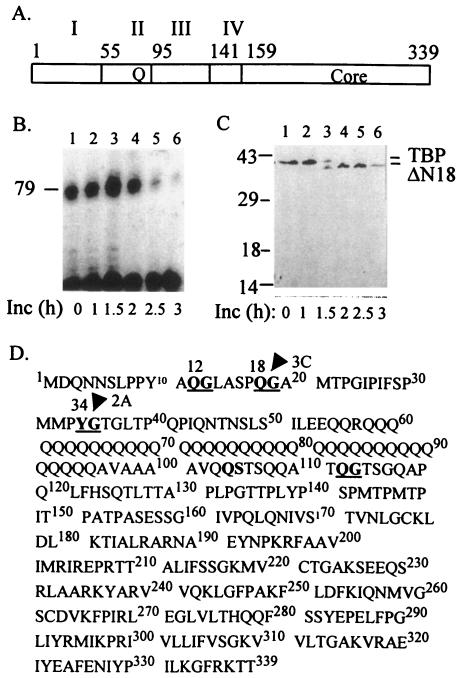
FIG. 1.
Shutoff of RNA polymerase II-mediated transcription does not correlate with 3Cpro-induced cleavage at the 18th glutamine-glycine site in TBP. (A) The domain structure of TBP consisting of the core, N-terminal with the glutamine stretch (Q), and acidic domain (IV) is shown. (B) Transcription shutoff in vitro does not correlate with TBP cleavage at the 18th Q-G site. Kinetics of transcription shutoff was examined in HeLa transcription extracts (80 μg) using a template DNA containing the TATA element (37) in the presence of 1 μg of purified 3Cpro, and the reactions were stopped at indicated times. The transcribed RNA was analyzed by primer extension using a 32P-labeled Sp6 promoter primer (37). The arrowhead indicates the size (79 nucleotides) of the correctly initiated transcription product. (C) The same reactions as shown in panel B were examined by Western blotting using a polyclonal antibody to human TBP. The positions of migration of wt TBP and ΔN18 TBP are indicated. (D) The primary sequence of TBP is shown. The Q-G and Y-G sites in TBP are underlined. The arrow and the arrowhead indicate the known 2Apro and 3Cpro cleavage sites within TBP, respectively.
Infection of HeLa cells with PV causes a severe decrease in cellular transcription catalyzed by all three cellular RNA polymerases (10). Transcription mediated by RNA polymerase I (Pol I) is inhibited first, at 1 to 2 h postinfection, followed by inhibition of Pol II and Pol III transcription at approximately 3 and 4 h postinfection, respectively. Crawford et al. first showed that PV-induced inhibition of transcription observed in vivo could be recapitulated in vitro (8). Further studies showed that the viral protease 3Cpro alone was responsible for the shutoff of transcription by all three cellular RNA polymerases (36). Recent studies have shown that 3Cpro enters the nucleus in the form of its precursor 3CD (32), which presumably undergoes autocatalysis to generate 3Cpro in the nucleus of infected cells. A primary target of 3Cpro in PV-infected cells was previously identified to be the TATA-binding protein (6). Consistent with this observation, the TFIID complex isolated from PV-infected HeLa cells was transcriptionally inactive in an in vitro reconstituted transcription assay compared to the TFIID isolated from uninfected cells (19). Moreover, purified TBP could be directly cleaved by the purified 3Cpro in vitro, and the addition of purified TBP could completely restore both basal and activated transcription from the TATA and initiator promoters in HeLa cell extracts from PV-infected cells (37). Examination of the human TBP sequence revealed three Q-G sites at positions 12, 18, and 112 (Fig. 1). Subsequent in vitro studies using purified components showed that only the 18th Q-G site in TBP could be efficiently cleaved by 3Cpro both in vitro and in vivo (9).
To determine if cleavage of TBP at the 18th Q-G bond is responsible for the shutoff of transcription in PV-infected cells, we expressed and purified a recombinant TBP that lacks the N-terminal 18 amino acids (called ΔN18 TBP). If cleavage at the 18th Q-G bond is the primary cause of transcriptional inactivation of TBP, then ΔN18 TBP should not be able to restore transcription in PV-infected cell extracts. We found that ΔN18 TBP was as active as the wild-type (wt) TBP in fully restoring Pol II transcription in PV-infected cell extracts from the adenovirus major late promoter (Ad MLP). We also found that the transcriptional activity of ΔN18 TBP was comparable to that of wt TBP in an in vitro transcription reconstitution assay. Since both genetic and biochemical evidence suggested that 3Cpro was involved in the shutoff of transcription and that TBP was the primary target of 3Cpro, we set out to determine if TBP was being cleaved by 3Cpro at sites other than the 18th Q-G bond. We report here identification of a Q-S site at position 104 to 105 of TBP, which appears to be cleaved by the viral protease 3Cpro. We also demonstrate that an alanine residue at P4 is critical for 3C-mediated cleavage of the 104th Q-S bond. A truncated form of TBP lacking the first 100 amino acids is significantly less active transcriptionally than the wt TBP in vitro. Finally, we demonstrate that a stable HeLa cell line expressing a recombinant TBP (rTBP) resistant to cleavage by the viral proteases is significantly refractive to the shutoff of transcription by poliovirus compared to the control cell line expressing wt TBP. Infection with poliovirus of the HeLa cell line expressing the noncleavable form of TBP shows small plaques and significantly reduced viral RNA synthesis compared to the control TBP cell line, suggesting that a defect in the shutoff of transcription can lead to inefficient replication of the virus.
MATERIALS AND METHODS
Cell culture.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% newborn calf serum and 5% fetal calf serum. Cells were split at a 1:10 ratio in fresh medium every 3 days. HeLa suspension cells were grown in minimum essential Eagle medium (Sigma Inc.) with 5% newborn calf serum at 37°C with constant stirring. Cells were reinoculated in fresh medium every 3 days at ratios of 1:5 to 1:10.
Mutant constructs.
All mutagenesis studies were done using the pGem3:wtTBP recombinant transcription vector carrying the wt TBP construct (39). For deletion of the C-terminal 68 (ΔC68TBP) and 172 (ΔC172TBP) amino acid (aa) residues, the BamHI-HincII and BamHI-SspI fragments were subcloned into the BamHI and HincII sites of the pGem3Zf− vector (Promega Inc.). To generate TBP with C-terminal deletions of 201 aa (ΔC201TBP), 139 aa (ΔC139TBP), and 105 aa (ΔC105TBP), fragments were PCR amplified using T7 promoter primer and a gene-specific primer carrying restriction sites. Fragments were digested with BamHI and HindIII or HincII and cloned into the same sites of the pGEM3Zf− vector. A gene-specific primer with a PstI site and SP6 promoter primer were used to amplify a fragment from 388 nucleotides (nt) to the end of the stop codon of TBP. It was digested with PstI and HindIII and ligated with PstI-HindIII-cut pGem3:wtTBP. Thus, an in-frame deletion of 101 to 129 aa of TBP was created. A PCR product 400 nt long with an SmaI site in the 3′ end was amplified using T7 promoter primer and a gene-specific primer. It was digested with BamHI and SmaI and ligated with an SspI-HindIII-cut fragment of TBP, and then the ligated product was religated into the BamHI-HindIII-cut pGem3Zf− vector to generate an in-frame deletion of 132 to 167 amino acid residues. All these deletion mutation clones, therefore, were under the control of the T7 promoter.
Amino acid substitution mutagenesis was performed using the double-stranded site-directed mutagenesis protocol. PCR primers carrying the mutated sequences were designed for both strands, and PCR was performed using Pfu Turbo DNA polymerase (Stratagene Inc.) in a 50-μl reaction volume. Product was digested with DpnI, and 2 μl of that was used to transform DH5α competent cells. All mutations were confirmed by subsequent sequencing. The mutated TBP in which all three protease cleavage sites were altered was generated by sequential mutagenesis of wt TBP and named rTBPGG (QG18,19AA, YG34,35AA, and AA100,101GG) or rTBPLL (QG18,19AA, YG34,35AA and AA100,101LL).
rTBPGG and rTBPLL along with the wt TBP were subcloned into the pET28a+ vector in BamHI and HindIII sites for bacterial expression and purification. From pET28a+ clones, BamHI and NotI fragments carrying TBP were subcloned into the same sites of mammalian expression vector pEF6/HisC (Invitrogen Inc.). Expression from the T7 promoter was confirmed in an in vitro assay using both plasmid clones.
To generate a ΔN100 TBP (TBP with a deletion of the N-terminal 100 amino acids) construct, the PCR fragment was amplified with a gene-specific forward primer (designed to amplify from 301 nt) and an SP6 promoter primer as reverse primer. The gene-specific primer had NdeI and BamHI sites at the 5′ end. For cloning of ΔC239TBP (a deletion of the C-terminal 239 aa) a fragment was amplified with T7 promoter primer as a forward primer and a gene-specific primer designed to amplify up to 300 nt of TBP. The latter primer had an HindIII site. The ΔN100 and ΔC239 PCR products were digested with NdeI-HindIII and BamHI-HindIII, respectively, and cloned into the pET28a+ vector cut with same restriction sites.
In vitro transcription translation and protease assays.
In vitro coupled transcription and translation in the presence of [35S]methionine was performed with TNT Quick coupled transcription/translation system (Promega) as described previously (38). In a 25-μl reaction mixture, 20 μl of TNT master mix, 10 μCi of [35S]methionine, and 1 μg plasmid DNA were added. The reaction was carried out at 30°C for 90 min. One microliter of this reaction mix was digested with 1 μg of purified 3C protease for 3 to 4 h at 30°C in a 10-μl reaction volume. The mix also contained 20 mM HEPES, 100 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT). A control reaction was incubated for same time but without 3C protease. Products were separated on either 10% or 12.5% polyacrylamide gels containing 0.1% sodium dodecyl sulfate (SDS), fixed, and detected after fluorography.
SDS-PAGE.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 16- by 18-cm gels (1.5 mm thick). Polyacrylamide gels (125 mM Tris-HCl, pH 8.8, 10 to 12.5% polyacrylamide with a 30:0.8 ratio of acrylamide to bisacrylamide) were prepared by standard procedures using ammonium persulfate (0.05%, wt/vol) and 0.2 ml of TEMED (N,N,N′,N′-tetramethylethylenediamine) for initiation. The stacking gel consisted of 4% acrylamide. The composition of buffers was as follows: 5× sample buffer consisted of 0.2 M Tris-HCl, pH 6.8, 10 mM DTT, 20% (vol/vol) glycerol, 10% (wt/vol) SDS, and 0.05% bromophenol blue; 1× running buffer contained 25 mM Tris-HCl, 200 mM glycine, and 0.1% (wt/vol) SDS. The gels were run at 400 to 500 V until the bromophenol blue dye reached the bottom of the gel.
Bacterial expression and purification.
Wild-type and mutated TBPs were purified from BL21(DE3) codon-plus Escherichia coli host carrying the recombinant pET28a+:wt TBP or rTBPGG or rTBPLL. Expressed protein was purified from soluble fractions by nickel affinity chromatography using a standard protocol. Purified protein was finally dialyzed against suspension buffer containing 20 mM HEPES, pH 7.6, 0.5 mM EDTA, 2 mM MgCl2, 50 mM KCl, 1 mM DTT, 20% glycerol, and 0.01% NP-40.
Transfection and cell line.
Two micrograms of plasmid DNA was transfected to HeLa cells using Lipofectamine-Plus reagent (Invitrogen Inc.) following the manufacturer's protocol. Cells were transferred to a medium containing 3 μg/ml of blasticidin 24 h after transfection, and medium was replaced every 48 h until the colonies appeared. Distinct bigger colonies were picked up with clone-disk and multiplied.
Pol II transcription assay.
Two to four liters of HeLa suspension culture at a concentration of 4 × 105 cells/ml was used to make nuclear extract. Nuclear extract was prepared using a standard protocol (19). Template DNAs were p1634, p2038, or p126 as described earlier (37) carrying the Ad MLP TATA box and terminal deoxy transferase initiator sequence, only the TATA box, or the Sp1 activator sequence upstream to TATA box, respectively. Transcription reaction mixtures contained 700 ng of DNA template, 0.4 mM nucleoside triphosphate mix, 5 mM MgCl2, 1 mM DTT, 8.8% glycerol, 8.8 mM HEPES, pH 7.6, 40 U of RNAsin (Promega), and 100 μg of HeLa extract in a 50-μl reaction volume. It was incubated for 90 min at 30°C and purified by phenol-chloroform-isoamylalcohol extraction and alcohol precipitation. Primer extension with labeled SP6 promoter primer was performed using avian myeloblastosis virus reverse transcriptase (Promega Inc.) following the manufacturer's protocol. The product was subjected to 8% Tris-borate-EDTA-Urea-PAGE and detected by autoradiography.
Plaque assay.
About 1 × 106 cells were plated in a 60-mm plate the day before the assay. Blasticidin at a concentration of 5 μg/ml was added to cell lines. Before infection, cells were washed once with phosphate-buffered saline (PBS) and once with serum-free DMEM. Virus was diluted in DMEM without serum, and 200 μl of virus suspension was added per plate. Virus was allowed to adsorb for 30 min with agitation at 37°C and then was removed from the plate. A solution of 0.9% noble agar prepared in DMEM was poured on top of the plate and allowed to set at room temperature for 15 min, followed by incubation at 37°C for 48 h. The agar overlay was removed, and plaques were stained with a solution of 0.1% crystal violet in 20% ethanol.
[3H]uridine incorporation assay.
The day before assay, wt and GG cell lines were plated in six-well plates at a concentration of approximately 0.3 × 106 cells per well in the presence of 5 μg/ml blasticidin. After 24 h, cells were washed once with PBS and once with serum-free DMEM and infected with poliovirus at a multiplicity of infection of 30 cells per well in a 100-μl volume at 37°C with constant agitation. After adsorption 1 ml of DMEM was added and incubated at 37°C. At 3 h postinfection, actinomycin D was added at a concentration of 5 μg/ml and incubated for 15 min at 37°C, followed by the addition of 5 μCi of [3H]uridine as previously described (11). Infection was stopped by the addition of ice-cold PBS. Cells were washed twice with ice-cold PBS, and detached cells were collected by centrifugation. Cell lysis was performed by the addition of 0.5 ml of 0.5% SDS followed by scraping, and nucleic acids were precipitated with 0.5 ml of 10% cold trichloroacetic acid. The precipitate was collected on Whatman GF/C glass fiber filters, air dried, and counted in a biodegradable counting cocktail.
Northern blot analysis.
Twenty micrograms of total RNA isolated from PV-infected cells using Trizol reagent (Life Technologies) was separated by denaturing gel electrophoresis, transferred to nylon membranes, and hybridized with [α-32P]dCTP-labeled DNA probes. The hybridization probe radiolabeled with a High Prime labeling kit (Roche) consisted of PV cDNA encompassing the 3Dpol coding region.
RESULTS
Shutoff of RNA polymerase II transcription does not correlate with TBP cleavage at the 18th Q-G bond.
To determine whether 3Cpro-mediated cleavage at the 18th Q-G site of TBP was the primary cause for shutoff of Pol II transcription, a time course of transcription from a plasmid containing the TATA element (37) was performed in HeLa nuclear extracts following treatment with the purified recombinant 3Cpro. Transcription from the template DNA remained almost unaffected up to 2 h following treatment with 3Cpro (Fig. 1B, lanes 1 to 4), with the exception of the 1.5-h time point, at which time slight stimulation of transcription was observed (lane 3). Significant shutoff of transcription in vitro was first detected at 2.5 h following treatment with 3C and was almost complete by 3 h (lanes 5 and 6). Western blot analysis of endogenous TBP in the transcription extract, however, revealed a significant amount of TBP cleavage at 1.5 h of 3Cpro treatment without any concomitant decrease in transcription (Fig. 1B and C, lanes 3). Although almost all of the full-length TBP had been cleaved to a truncated form by 2 h of 3Cpro treatment, there was hardly any inhibition of transcription at this time point compared to the control (Fig. 1B and C, lanes 4). Our previous results have identified the faster-migrating TBP species as one that lacks the N-terminal 18 amino acids as a result of 3Cpro cleavage at the 18th Q-G bond (Fig. 1C) (9). Thus, there was no correlation between the shutoff of transcription and the cleavage of TBP at the 18th Q-G bond in the in vitro transcription assay.
To further investigate whether 3Cpro is capable of cleaving TBP at an alternative site, in vitro translated, [35S]methionine-labeled TBP was incubated with purified 3Cpro for 4 h, and the products were analyzed by SDS-PAGE. As can be seen in Fig. 2A, two proteolysis products of TBP having approximate molecular masses of 27 and 24 kDa were clearly evident in addition to ΔN18 TBP in this reaction. We were unable to detect the 27- and 24-kDa cleaved products by Western analysis (Fig. 1C). This could be due to the inability of the TBP antibody to react with these truncated TBP molecules on the Western blot. Alternatively, these polypeptides might have been degraded quickly in the translation lysates.
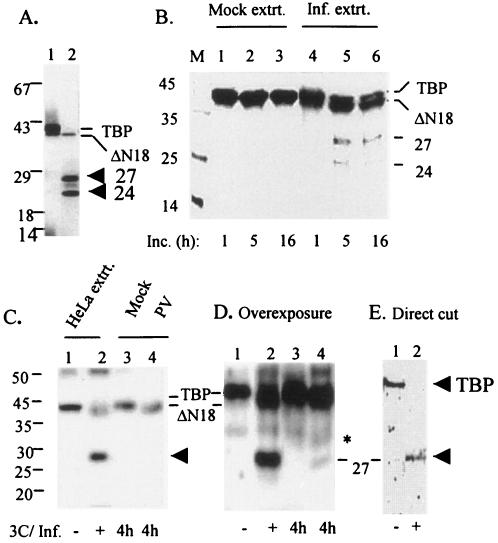
FIG. 2.
TBP cleavage by 3Cpro generates other proteolytic products in addition to ΔN18 TBP. (A) In vitro 3Cpro induced cleavage of TBP at sites other than the 18th Q-G site. One microliter of in vitro translated [35S]methionine-labeled TBP was incubated with buffer (lane 1) or 1 μg of purified 3Cpro (lane 2) for 4 h, and products were analyzed by SDS-PAGE. The positions of migration of TBP, Δ18 TBP, and two additional bands at 27 and 24 kDa are indicated. (B) Incubation of TBP with PV-infected extract generates the 27- and 24-kDa products. [35S]methionine-labeled TBP (∼150,000 cpm) was incubated with 40 μg of 4-h mock- or PV-infected (lanes 1 to 3 and lanes 4 to 6, respectively) extracts for 1, 5, and 16 h as indicated, and the reactions were analyzed by SDS-PAGE. (C and D) Detection of the 27-kDa polypeptide in PV-infected cells. In a Western blot using anti-TBP, the cell extracts from 4-h mock- or PV-infected (lanes 3 and 4, respectively) cells were compared with HeLa cell extract treated with buffer (lane 1) or purified 3Cpro (lane 2) in vitro. An overexposure of the blot shown in panel C is shown in panel D. The migrations of TBP, ΔN18TBP, and the 27-kDa polypeptide are indicated. (E) Generation of 27-kDa polypeptide by direct cleavage of TBP. Approximately 60 ng of purified TBP was incubated with buffer alone (lane 1) or with 250 ng of purified 3Cpro (lane 2) for 4 h, and the TBP-related products were visualized by Western blot analysis using a polyclonal anti-TBP antibody.
To determine if the 27- and 24-kDa TBP-related polypeptides could be detected in PV-infected cells, cell extracts from 4-h mock- or PV-infected cells were compared with purified TBP treated with buffer or 3Cpro in vitro in Western blotting using anti-TBP polyclonal antibody (Fig. 2C and D). A diffused 27-kDa band, which comigrated with the in vitro 3Cpro-generated 27-kDa product, was detected in PV-infected extract only when the blot was overexposed (Fig. 2D, lane 4). This band was not present in mock-infected extract (lane 3). The 27-kDa band was not detected in a short exposure of the same blot (Fig. 2C, lane 4). We were unable to detect the 24-kDa band in PV-infected extract even after overexposure of the immunoblot (Fig. 2D). Since TBP is limiting in HeLa cells, a similar experiment was conducted where [35S]methionine-labeled in vitro translated TBP was incubated with 4-h PV- or mock-infected extracts, and the products were analyzed by SDS-PAGE. As can be seen in Fig. 2B, incubation of the labeled TBP with PV-infected cell extract, but not mock-infected extract, for 5 h resulted in the generation of 27 and 24-kDa products (compare lanes 2 and 5). At 16 h of incubation, only the 27-kDa band was visible (lane 6).
To determine whether TBP is directly cleaved by 3Cpro to generate the 27- and 24-kDa polypeptides, purified TBP and 3Cpro were incubated, and the products were examined by Western blot analysis using anti-TBP. Only the 27-kDa product was detected in the reaction containing 3Cpro (Fig. 2E). Again, the 24-kDa product could not be detected in the direct cleavage assay. Taken together, these results suggest that TBP is likely cleaved by 3Cpro at a site(s) other than the 18th Q-G bond.
Identification of the alternative 3Cpro cleavage site within TBP.
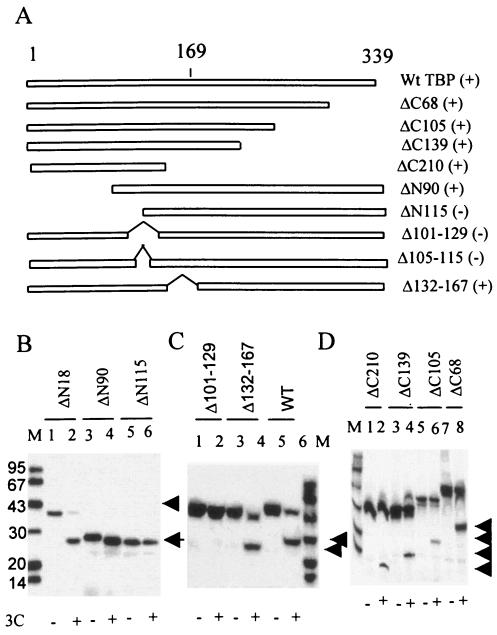
To localize the alternative 3Cpro cleavage site(s) within TBP, we initially prepared both NH2 and COOH-terminal deletion mutants of TBP. Various truncated TBP molecules (Fig. 3A) were translated in vitro in the presence of [35S]methionine and then incubated with purified 3Cpro, and the products were analyzed by SDS-PAGE. Incubation of the ΔN18 TBP with 3Cpro resulted in the generation of the corresponding 27-kDa product (Fig. 3B, lanes 1 and 2). A similar result was obtained with the ΔN90 TBP (lanes 3 and 4). In both reactions the faster-migrating (24 kDa with the wt TBP) product was not detected. This is most probably due to relatively quick degradation of this product in translation extracts. Deletion of the N-terminal 115 amino acids prevented further cleavage of TBP by 3Cpro (lanes 5 and 6). These results suggested that the alternative 3Cpro cleavage site was most probably present between amino acids 90 to 115 of TBP. Carboxy terminal deletions of up to 210 amino acids had no significant effect on TBP cleavage (Fig. 3A and D), suggesting that the alternative 3Cpro cleavage site was situated within the NH2-terminal 129 amino acids. The deletion of the C-terminal 68 amino acids of TBP did not have any significant effect on the generation of the corresponding 27-kDa product (Fig. 3D, lanes 7 and 8). TBP molecules with C-terminal deletions of 105, 129, and 210 amino acids still generated the corresponding faster-migrating product upon incubation with 3Cpro but with lower efficiency compared with the ΔC68 mutant (Fig. 3D). This is most probably due to changes in the protein structure induced by relatively large deletions in the TBP molecule.
FIG. 3.
Effects of various TBP terminal and internal deletions on 3Cpro-induced cleavage of TBP. (A) Schematic representation of amino- and carboxy-terminal and internal deletion mutants of TBP. The pluses and minuses in parentheses indicate qualitatively whether these polypeptides are cleaved (+) or not cleaved (−) to generate the 27- and 24-kDa products by 3Cpro. (B) Various N-terminal deletion mutants of TBP were translated in vitro in the presence of [35S]methionine and subjected to digestion with buffer only (lanes 1, 3, and 5) or 1 μg of purified 3Cpro (lanes 2, 4, and 6). (C) The wild-type (lanes 5 and 6) and internal deletion mutants of TBP, Δ101-129 (lanes 1 and 2), and Δ132-167 (lanes 3 and 4) were digested with buffer alone (lanes 1, 3, and 5) or purified 3Cpro (lanes 2, 4, and 6). The arrows indicate the proteolyzed products. (D) The in vitro translated, [35S]methionine-labeled C-terminal deletion mutants ΔC210 (lanes 1 and 2), ΔC139 (lanes 3 and 4), ΔC105 (lanes 5 and 6), and ΔC68 (lanes 7 and 8) of TBP were digested with buffer alone (odd-numbered lanes) or purified 3Cpro (even-numbered lanes).
To confirm the results of amino- and carboxy-terminal deletions, internal deletions of amino acids spanning residues 101 to 129 and 132 to 167 of TBP were made. While Δ132-167 TBP was cleaved by 3Cpro to the same extent as wt TBP, the cleavage of the Δ101-129 TBP mutant that generates the 27-kDa product was not apparent (Fig. 3C, lanes 1 to 6). These results suggest that the alternative 3Cpro cleavage site within TBP was possibly present between amino acid residues 105 and 129.
Double and single mutations of amino acid residues 101 to 129 identify the 3Cpro alternative cleavage site.
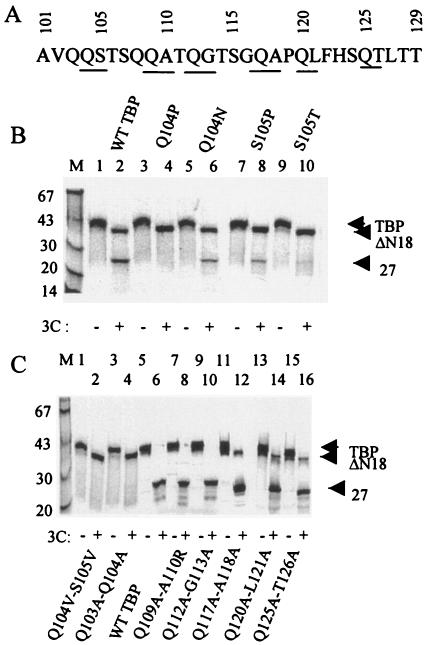
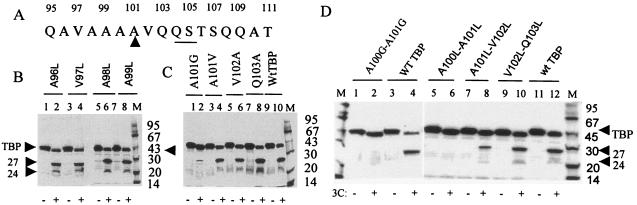
Examination of the amino acid sequence between residues 101 and 129 of TBP identified a number of potential 3Cpro (alternative) cleavage sites including Q104-S105, Q109-A110, Q112-G113, Q117-A118, Q120-L121, and Q125-T126. All potential primary and alternative 3Cpro cleavage sites within the amino acid sequence 101 to 129 of TBP were altered by double amino acid substitutions. Double amino acid substitutions of the Q109-A110, Q112-G113, Q117-A118, Q120-L121, and Q125-T126 pairs did not prevent formation of the 27-kDa polypeptide when incubated with purified 3Cpro (Fig. 4C, lanes 5 to 16, and Table 1). However, mutating the Q104-S105 and Q103-Q104 pairs almost completely blocked formation of the 27-kDa product, although the 3Cpro-induced 18th Q-G cleavage was apparent with both double mutants (Fig. 4C, lanes 1 to 4, and Table 1). These results suggested that amino acid residues 103 to 105 were important for 3Cpro-mediated generation of the 27-kDa product.
FIG. 4.
Effects of double and single amino acid substitution mutations of potential 3Cpro cleavage sites between amino acid residues 101 and 126 of TBP. (A) The primary sequence of amino acid residues 101 through 129 of TBP is shown. The potential 3Cpro alternative cleavage sites are underlined. (B) In vitro translated, [35S]methionine-labeled wild type or various point mutants of TBP were digested with buffer alone (odd-numbered lanes) or 1 μg of 3Cpro (even-numbered lanes) prior to SDS-PAGE. (C) wt TBP or various double mutants were digested with buffer (odd-numbered lanes) or 3Cpro (even-numbered lanes). The proteolysis products are indicated by arrowheads to the right.
TABLE 1.
Effect of mutations on 3C-mediated cleavage of TBP
| TBP mutation | 27-kDa band (% of wt control)a |
|---|---|
| Q104V-S105V | 2 |
| Q103A-Q104A | 4 |
| Q109A-A110R | 93 |
| Q112A-G113A | 101 |
| Q117A-A118A | 107 |
| Q120A-L121A | 97 |
| Q125A-T126A | 99 |
| A100G-A101G | 0 |
| A100L-A101L | 0 |
| A101L-V102L | 35 |
| V102L-Q103L | 93 |
| Q104P | 1 |
| Q104N | 30 |
| S105P | 31 |
| S105T | 12 |
| A96L | 98 |
| V97L | 95 |
| A98L | 102 |
| A99L | 100 |
| A101G | 10 |
| A101V | 99 |
| V102A | 95 |
| Q103A | 115 |
The values shown were obtained by densitometric scanning of the 27-kDa band generated by incubation of mutant TBP with 3Cpro relative to the value of the same band generated from wt TBP by 3Cpro digestion.
Because the picornaviral 3C protease is known to cleave Q-S bonds in the viral polyprotein precursors (17), we carried out point mutagenesis of the Q104-S105 site in TBP. The Q104 or the S105 were mutated individually, and the effects of these mutations were determined in the in vitro 3Cpro cleavage assay. As can be seen in Fig. 4B, substitution of Q104 with proline almost completely blocked the generation of the 27-kDa band compared to the wt TBP (Fig. 4B, lanes 1 to 4, and Table 1). However, replacement of Q104 with a very similar amino acid (Q104N) only partially affected TBP cleavage (lanes 5 and 6). Substitution of the S105 with proline also partially blocked TBP cleavage; however, substituting the S105 with threonine almost totally blocked generation of the 27-kDa product (Fig. 4C, lanes 7 to 10, and Table 1). Taken together, these results suggest that the Q104-S105 bond is the most likely alternative 3Cpro cleavage site in TBP.
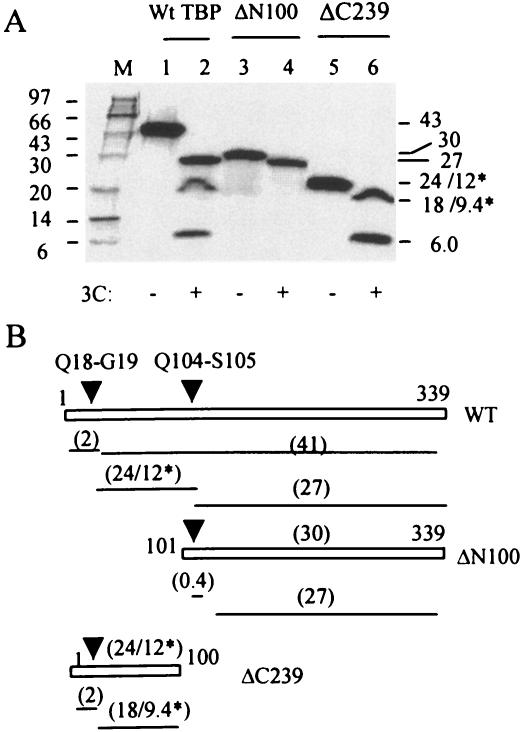
To further confirm our results, we compared the 3Cpro cleavage pattern of wt TBP with the patterns of ΔN100 and ΔC239 TBP mutants consisting of the C-terminal 239 and N-terminal 100 amino acid residues, respectively. The wt and ΔC239 TBP (both with intact N termini) had an extra 3.5-kDa N-terminal peptide, while the ΔN100 TBP (deletion of N-terminal 100 amino acids) had 12 N-terminal extra amino acids that resulted from cloning manipulations. The amino acid sequence of the extra N-terminal amino acids in these TBP molecules did not reveal any potential sequence with respect to either cleavage site or context. In this experiment, the labeled TBP or mutants synthesized by in vitro translation were digested to completion by purified 3Cpro. The products were then examined by SDS-PAGE followed by autoradiography. Complete digestion of wt TBP generated three polypeptides migrating at approximately 27, 24, and 6 kDa (Fig. 5A, lane 2). Amino acid composition and N-terminal sequencing of the 24-kDa band revealed that it had an actual molecular mass of ∼12-kDa and contained G19 as the N-terminal residue (data not shown). The 24-kDa polypeptide presumably consisted of amino acids 19 through 104 of TBP. Amino acid sequencing of both the C terminus of the 24-kDa as well as the N terminus of the 27-kDa polypeptides was inconclusive (data not shown). Relatively slower migration of the 24-kDa fragment was most probably due to the presence of the 38 consecutive glutamine residues in this molecule (Fig. 1A). The 6-kDa band most probably represented the N-terminal 18 amino acids (∼2.1 kDa) linked to an additional 3.5-kDa peptide generated during the cloning procedure. The 27-kDa band was presumed to represent the C-terminal TBP fragment (amino acids 105 to 339). Consistent with the origin of the 27-kDa band (235 amino acids), the ΔN100 TBP (C-terminal 239 amino acids plus 12 extra N-terminal amino acids generated by cloning manipulations) migrated slightly more slowly than the 27-kDa polypeptide at approximately 30 kDa (Fig. 5A, lane 3, and B). The 3Cpro cleavage of ΔN100 generated a band that comigrated with the 27-kDa product generated by incubation of full-length TBP with 3Cpro (compare lanes 2 and 4). The difference in the molecular weights between the uncleaved and cleaved ΔN100 TBP could be explained by the loss of the TBP N-terminal four amino acids linked to an additional 12 amino acids incorporated at the N terminus due to cloning manipulations. The 6-kDa band was not generated from the ΔN100 TBP due to the lack of the N-terminal 100 amino acids. The ΔC239 TBP (100 N-terminal amino acids), which has an estimated molecular mass of approximately 12 kDa, actually migrated much more slowly than anticipated due to the presence of the stretch of 38 Q residues in this polypeptide. 3Cpro treatment of ΔC239 generated the 6-kDa band (the N-terminal 18 aa linked to an additional 3.5-kDa sequence derived from the cloning vector) and an 18-kDa band (actual molecular mass of 10 kDa) (lane 6). These results together with mutagenesis studies are consistent with two 3Cpro-mediated cleavages within TBP: one at Q18-G19 and the other at Q104-S105 (Fig. 5B).
FIG. 5.
Comparison of complete 3Cpro digestion products of TBP, ΔN100 TBP, and ΔC239 TBP. (A) In vitro translated [35S]methionine-labeled wt TBP (lanes 1 and 2), ΔN100 (lanes 3 and 4), and ΔC239 (lanes 5 and 6) were digested with buffer (lanes 1, 3, and 5) or 3Cpro (lanes 2, 4, and 6) to completion. The radiolabeled products were analyzed by SDS-PAGE. (B) A schematic presentation of the predicted 3Cpro cleavage products generated from TBP and truncated TBP molecules. Arrowheads indicate Q18-G19 and Q104-S105 cleavage sites. The numbers within parentheses indicate the molecular weights of the products, while the numbers marked with asterisks indicate actual molecular weights of these products compared to how they migrate during gel electrophoresis.
Amino acid residues surrounding the Q104-S105 site modulate 3Cpro-mediated TBP cleavage.
Previous studies on the mechanism of 3Cpro-catalyzed cleavage of viral precursor polypeptides have shown that in addition to the scissile Q-G bond, surrounding amino acids are important for proteolytic cleavage by 3Cpro. In particular, the enterovirus 3Cpro prefers alanine (or aliphatic amino acids) in the P4 position, whereas cardiovirus 3C seems to prefer proline in the P2 or P2′ position (25, 26, 27). Examination of the amino acid sequence preceding the Q104-S105 site showed that there was a stretch of seven aliphatic residues in positions P3 through P9 (V102, A101, A100, A99, A98, V97, and A96) (Fig. 6A). To determine which of these surrounding residues were important for TBP cleavage, we introduced point mutations throughout this stretch. There was no significant effect on 3Cpro-mediated cleavage (that generates the 27-kDa product) of TBP mutants containing the Q103A, V102A, A101V, A99L, A98L, V97L, and A96L substitutions compared to the wt TBP (Fig. 6B and C, and Table 1). However, in the A101G substitution mutant, formation of the 27-kDa band was reduced ∼90% compared with the wt TBP (Fig. 6C, lane 2, and Table 1). Substitution of both A100 and A101 by glycine totally blocked the formation of the 27-kDa product, and almost all of mutant TBP was converted to a product that comigrated with ΔN18 TBP (Fig. 6D, lanes 1 and 2, and Table 1). Similar results were obtained when these two alanine residues (A100 and A101) were replaced by leucine (Fig. 6D, lanes 5 and 6). Double amino acid substitutions in positions 102 to 103 (VQ to LL) did not have a significant effect on the formation of the 27-kDa product (lanes 9 and 10). However, an AV (101 and 102) to LL substitution did have a significant effect (∼65%) on the generation of the 27-kDa product. These results suggest that A100 (P4) and A101 (P5) are important for the 3Cpro cleavage at the Q104-S105 site in TBP.
FIG. 6.
Amino acid residues preceding the Q104-S105 bond important for 3Cpro-induced cleavage of TBP. (A) The primary sequence of amino residues 95 to 111 of TBP is shown. The Q104-S105 bond is underlined. The upward arrowhead indicates the P4 residue. (B) Single amino acid substitution between residues 96 to 99 does not influence TBP cleavage at the Q104-S105 site. TBP mutants with the single amino acid substitutions A96L, V97L, A98L, and A99L were digested with buffer (lanes 1, 3, 5, and 7) or 3Cpro (lanes 2, 4, 6, and 8). (C) Substitution of A101 (P4 position) with G, but not with V, significantly inhibits 3Cpro-mediated cleavage that generates the 27- and 24-kDa products. Single amino acid substitution mutants A101G, A101V, V102A, Q103A, and wt TBP were digested with buffer (lanes 1, 3, 5, 7, and 9) or 3Cpro (lanes 2, 4, 6, 8, and 10) prior to analysis by SDS-PAGE. (D) Double amino acid substitution at A100-A101 completely blocks the 3Cpro-mediated cleavage that generates 27- and 24-kDa products. TBP double amino acid substitution mutants A100G-A101G, A100L-A101L, A101L-V102L, V102L-Q103L, and wt TBP were digested with buffer alone (lanes 1,3, 5, 7, 9, and 11) or 3Cpro (lanes 2, 4, 6, 8, 10, and 12).
N-terminal 100-amino-acid deletion drastically reduces the transcription restoration activity of TBP in PV-infected extracts.
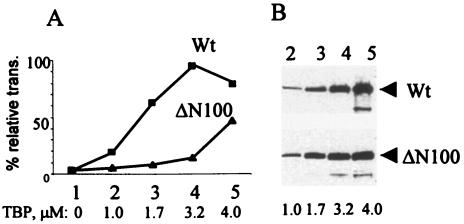
To determine whether truncation of the N-terminal domain of TBP interferes with its transcriptional activity, we used the ΔN100 TBP in a transcription-restoration assay that measures the ability of recombinant TBP to rescue inhibition of transcription in PV-infected extracts. The bacterially expressed recombinant ΔN100 TBP was purified, and its ability to restore basal transcription from a TATA promoter in PV-infected nuclear extract was compared with that of the wt rTBP. In previous studies we have shown that the inhibition of TATA-mediated transcription from a core promoter in PV-infected cell extracts can be completely restored by TFIID as well as purified recombinant TBP (19, 37). As can be seen in Fig. 7, the ΔN100 TBP was severely defective in restoring transcription from the TATA promoter compared with the wt TBP. Although it could partially restore transcription at very high concentrations (Fig. 7A), the specific activity of the truncated TBP was 8- to 10-fold lower than the wt TBP in this assay. Thus, the 3Cpro-mediated cleavage of TBP at the Q104-S105 site is likely to reduce TBP transcriptional activity severely, an observation consistent with our previous findings (37).
FIG. 7.
ΔN100 TBP is highly defective in restoring RNA Pol II-mediated transcription in PV-infected extracts. (A) TATA box-dependent in vitro transcription from a template DNA containing Ad MLP was performed in poliovirus-infected (4 h postinfection) cell extracts in the absence and presence of purified ΔN100 TBP or wt TBP. The primer-extended product was quantified by densitometric scanning, and the percentage of relative transcription with respect to 4-h mock-infected control was plotted against the concentration of TBP in the reaction. (B) Western blots of various concentrations of wt and ΔN100 TBP used in the transcription assay are shown.
Constitutive expression in HeLa cells of recombinant mutant TBP resistant to viral protease results in incomplete shutoff of transcription and yields virus with a small-plaque phenotype.
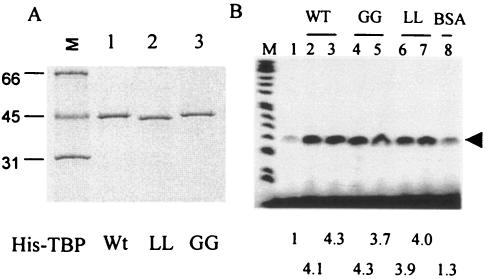
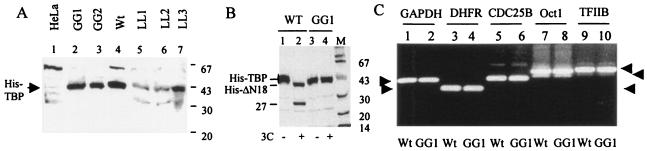
To determine whether efficient replication of PV requires host cell transcription shutoff, HeLa cell lines constitutively expressing mutant TBP, which is resistant to cleavage by the poliovirus-encoded 3C protease, was prepared. Previous results from our laboratory as well as results presented here have shown that altogether TBP can be cleaved at three sites in PV-infected cells: Q18-G19, Y34-G35, and Q104-S105. The cleavages at the Q18-G19 and Q104-S105 sites are mediated by 3Cpro, while the Y34-G35 bond is cleaved by 2Apro (Fig. 1A). We have shown previously that 2Apro-mediated cleavage of TBP at the Y34-G35 bond does not contribute to the shutoff of transcription by RNA Pol II in vitro (39). Results presented here indicate that 3Cpro-mediated cleavage of the Q104-S105, but not the Q18-G19, bond is most likely responsible for shutoff of transcription by RNA Pol II. Although cleavage of TBP at the Q18-G19 and Y34-G35 sites does not appear to reduce its transcriptional activity in vitro, it is possible that TBP cleaved at these sites could become destabilized and degraded quickly in vivo. For this reason, we decided to make mutant TBP cell lines in which all three viral protease-cleavable sites in TBP are altered. We cloned and expressed two TBP mutants for this purpose containing the following mutations: Q18A-G19A, Y34A-G35A, and A100G-A101G (called GG rTBP), and Q18A-G19A, Y34A-G35A, and A100L-A101L (called LL rTBP). The bacterially expressed His-tagged proteins were purified and examined by SDS-PAGE. Both GG rTBP and LL rTBP were stable (Fig. 8A), and their transcriptional activity was comparable to that of wt TBP in the in vitro transcription restoration assay (Fig. 8B). Stable HeLa cell lines constitutively expressing the wt and mutant TBP were then prepared as outlined in Materials and Methods. The level of TBP expression in wt and two GG lines was comparable (Fig. 9A, lanes 2 to 4). In particular, the level of TBP expression was very similar between the wt and GG1 rTBP cells. However, expression of TBP in LL rTBP cells was relatively poor (lanes 5 to 7). There was no significant difference between the wt rTBP- and GG1 rTBP-expressing cells with respect to the rate of cell growth, morphology, and overall RNA and protein synthesis (data not shown). Additionally, the levels of mRNA encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH), dihydrofolate reductase, CDC25B, Oct-1, and TFIIB were very similar between the cell lines expressing wt and GG1 rTBP, as determined by semiquantitative reverse transcription-PCR (RT-PCR) analysis (Fig. 9C). It should be noted that for each mRNA, the data from one concentration from the linear range of RT-PCR analysis are shown. The GG1 rTBP from the cell line was completely resistant to 3Cpro at a concentration that efficiently cleaves wt rTBP in vitro (Fig. 9B).
FIG. 8.
Purification and transcriptional activity of GG rTBP and LL rTBP. (A) Recombinant His-tagged wt TBP, GG rTBP, and LL rTBP were expressed in bacteria and purified by Ni++ affinity chromatography. Silver-stained gel of the purified proteins is shown. (B) The transcriptional activity at 3 and 5 μM of each of the wt (lanes 2 and 3), GG (lanes 4 and 5), and LL (lanes 6 and 7) rTBP was measured using a template containing the Ad MLP (lanes 2 to 7) in PV-infected cell extract as previously described (37). Bovine serum albumin (5 μM) was used as a negative control (lane 8). Lane 1 shows transcriptional activity of the infected extract in the absence of any added protein. The numbers at the bottom indicate the increase in stimulation (n-fold) over control as determined by densitometric quantification of the transcript.
FIG. 9.
Cell lines that constitutively express wt and mutant TBP resistant to cleavage by 3Cpro. (A) Detection of mutant TBP in various cell lines. Five clonally selected HeLa cell lines that constitutively express His-TBP were isolated as described in Materials and Methods. Each line was examined for expression of recombinant TBP by Western analysis using an antibody to the His tag. Two of these lines (GG1 and GG2) contained His-TBP in which the two alanines at positions 100 and 101 were replaced by glycine. The two alanine residues in three other lines (LL1, LL2, and LL3) were replaced by L. In both mutants the 18Q-19G and the 34Y-35G bonds were mutated to 18A-19A and 34A-35A, respectively. The wild-type line expresses wt His-TBP. In lane 1, HeLa cell extracts containing the endogenous TBP, not detected by anti-His antibody, was used. (B) GG1 rTBP is resistant to 3Cpro-mediated cleavage. Cell extracts from wild-type and GG1 rTBP (GG1) lines were digested with buffer (lanes 1 and 3) or purified 3Cpro (lanes 2 and 4) prior to Western analysis using anti-His antibody. (C) Five cellular mRNA levels as indicated were examined by RT-PCR from wt rTBP (odd-numbered lanes) and GG1 rTBP (even-numbered lanes) cells.
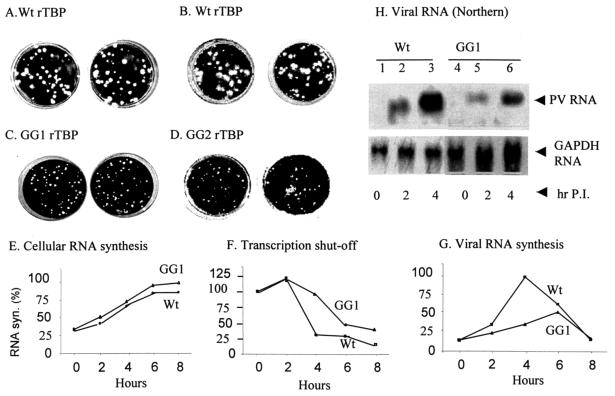
To determine whether replication of poliovirus is affected in HeLa cells that constitutively express GG1 rTBP, clonally selected cell lines expressing wt rTBP and GG1 rTBP (or GG2 rTBP) were infected with poliovirus type 1 (Mahoney strain) and assayed for formation of viral plaques as described in Materials and Methods. As can be seen in Fig. 10, HeLa cells expressing the wt rTBP showed relatively large plaques characteristic of wt PV infection (panels A and B, in duplicate). In contrast, infection of HeLa cells expressing GG1 rTBP and GG2 rTBP resulted in the formation of significantly smaller plaques compared with those expressing the wt rTBP (panels C and D). The number of plaques, however, did not change significantly between the wt rTBP and GG1 rTBP cells. The cell-free lysates from two separate PV-infected GG1 rTBP (and wt rTBP) cultures were used to infect naïve HeLa cells. The viral yields were found to be lowered approximately 20- to 30-fold with the virus harvested from GG1 rTBP cells compared with yields from the wt rTBP cells in two separate experiments (Table 2). These results suggest that the efficiency of virus replication is significantly affected in HeLa cells that express the form of TBP not cleaved by the viral proteases and that shutoff of transcription may contribute to efficient viral replication in infected cells.
FIG. 10.
Constitutive expression in HeLa cells of recombinant mutant TBP resistant to 3Cpro cleavage yields virus with a small-plaque phenotype. (A to D) HeLa cells expressing wild-type His-rTBP (A and B), GG1 His-rTBP (C), or GG2 His-rTBP (D) were infected with poliovirus type 1 (Mahoney strain) in duplicates and assayed for plaque formation as described in Materials and Methods. (E to H) Transcription shutoff is delayed and viral RNA synthesis is significantly inhibited in GG1 His-rTBP cells. HeLa cell lines expressing either wt rTBP or GG1 rTBP were infected with poliovirus type 1 (Mahoney strain) as described in Materials and Methods. Mock- and PV-infected cells were labeled with 5μCi of [3H]uridine per 0.3 × 106 cells in the absence and presence of 5 μg/ml actinomycin D, and cellular (E) and viral RNA synthesis (G) were measured as described in the text. Transcription shutoff (F) was determined by using the values of acid-insoluble [3H]uridine incorporation according to the following formula: Inf − [(Inf + actD) − (Uninf + actD)]/[(Uninf) − (Uninf + actD)]. The experiments shown in panels E to G were performed in duplicate, and the average values are plotted. (H) Northern analysis of viral RNA was performed from total RNA isolated from wt and GG1 rTBP cells at 0 (lanes 1 and 4), 2 (lanes 2 and 5), and 4 (lanes 3 and 6) h postinfection. The lower panel shows GAPDH RNA used as a loading control.
TABLE 2.
Viral yields in single cycle infection
| Virus stock prepared from cell line | Titer (PFU/cell)a |
|---|---|
| Experiment 1 (6 h) | |
| Wt rTBP | 723 |
| GG1 rTBP | 36 |
| Experiment 2 (12 h) | |
| Wt rTBP | 1,639 |
| GG1 rTBP | 53 |
wt rTBP and GG1 rTBP cells were infected with wt poliovirus at an MOI of 10 at 37°C. Cells were harvested at 6 and 12 h postinfection, and virus stocks were prepared by cell lysis. Viral yields were determined by plaque assay on naive HeLa cells as described in Materials and Methods.
To determine whether the overall pattern of shutoff of host cell transcription by poliovirus is altered in HeLa cell lines that express the protease-resistant form of TBP, we compared the kinetics of shutoff by poliovirus of cellular RNA synthesis between cells expressing wt rTBP and GG1 rTBP. In the absence of virus infection, the rate of cellular RNA synthesis, as determined by acid-insoluble [3H]uridine incorporation, was comparable between Wt rTBP- and GG1 rTBP-expressing cells (Fig. 10E). These results were consistent with our semiquantitative RT-PCR analysis of at least five individual cellular mRNAs having stabilities ranging from those having relatively long half-lives to moderate half-lives (Fig. 9C). Thus, cellular RNA synthesis was not significantly affected by expression of wt or mutant rTBP in the cell lines. HeLa cell lines expressing wt and GG1 rTBP were either mock-infected or infected with poliovirus in the absence and presence of actinomycin D, and the extent of transcription shutoff was determined by applying the values of acid-insoluble [3H]uridine incorporation into the following formula: Inf − [(Inf + actD) − (Uninf + actD)]/[(Uninf) − (Uninf + actD)] (2, 3), where Inf indicates [3H]uridine incorporation (cpm) in infected cells in the absence and presence of actinomycin D and Uninf indicates [3H]uridine incorporation (cpm) in uninfected cells in the absence and presence of actinomycin D. These values were then plotted as percentages of RNA synthesis. As can be seen in Fig. 10F, host cell transcription shutoff in GG1 rTBP cells was significantly delayed compared to the wt rTBP cells. At 4 h postinfection, almost 75% of cellular RNA synthesis was inhibited by poliovirus in wt rTBP cells, while ∼10% inhibition was observed in GG1 rTBP-expressing cells. Even at 8 h postinfection, cellular RNA synthesis was inhibited by ∼60% in GG1 rTBP cells compared to ∼85% in wt rTBP-expressing cells. The overall viral RNA synthesis was reduced in GG1 rTBP cells approximately 3.5-fold and appeared to peak at a later time point compared with that in wt rTBP cells as determined by the [3H]uridine incorporation assay (Fig. 10G). Northern analysis showed approximately 3.2-fold inhibition of viral RNA synthesis in GG1 rTBP compared to wt rTBP cells at 4 h postinfection (Fig. 10H). These results suggest that the lack of proper transcription shutoff could influence the efficiency of viral RNA replication in infected cells.
DISCUSSION
We present evidence here suggesting that poliovirus-induced shutoff of host cell transcription (by RNA polymerase II) is mediated by cleavage of transcription factor TBP at an alternate 3Cpro cleavage site, which is distinct from the most frequently cleaved site in the viral polyproteins by 3Cpro, namely a glutamine-glycine (Q-G) pair. Previous reports from our laboratory had identified the 18th Q-G bond as the most vulnerable site within TBP to be cleaved by 3Cpro both in vitro and during PV infection (9). However, a recombinant TBP mutant (ΔN18) that lacks the first 18 amino acids of TBP was found to be as active as the wt TBP in restoring RNA Pol II-mediated transcription in poliovirus-infected cell extracts. Moreover, time course experiments showed a lack of correlation between the onset of transcription shut-off and generation of the cleaved ΔN18 TBP as determined by Western analyses (Fig. 1).
Our previous studies have shown that the two other Q-G sites at positions 12 and 112 of TBP were not cleaved by 3Cpro (9). The PV 3Cpro has been shown to prefer glutamine at P1 and either glycine, alanine, or serine at P1′ for cleavage of viral precursor polypeptides (12, 17). In addition, aliphatic amino acids (such as alanine, valine, or isoleucine) at P4 are preferred for 3C/3CD-mediated cleavage between P1 and P1′ (4, 14). Initial amino- and carboxy-terminal as well as internal deletion studies of TBP indicated the presence of an additional 3Cpro cleavage site between amino acid residues 101 to 129 (Fig. 3). In vitro cleavage assays using purified 3Cpro and triple (data not shown) and double amino acid substitution mutants of TBP indicated that amino acid residues from 101 to 129 played important roles in 3Cpro-induced cleavage of TBP that resulted in generation of two polypeptides having approximate molecular masses of 27 and 24 kDa. These two polypeptides were detected when 35S-labeled TBP was incubated with PV- but not mock-infected extracts for 5 to 16 h (Fig. 1B). Moreover, the 27-kDa proteolysis product could be detected in PV-infected cell extracts by overexposing a Western blot developed with a polyclonal anti-TBP antibody (Fig. 2D). Finally, the 27-kDa polypeptide could be detected by incubating purified recombinant TBP with purified 3Cpro, suggesting a direct cleavage of TBP by 3Cpro (Fig. 2E). The 24-kDa polypeptide appears to be very unstable both in PV-infected extract and in the in vitro cleavage assay. Additionally, the 24-kDa peptide was not easily detectable by Western blotting.
Analysis of the primary sequence of TBP surrounding residues 101 to 129 revealed the presence of a number of alternate 3Cpro cleavage sites such as Q104-S105, Q109-A110, Q112-G113, Q117-A118, Q120-L121, and Q125-T126 (Fig. 4A). Double amino acid substitutions at these sites revealed that only mutagenesis of the Q104-S105 site blocked 3Cpro-induced generation of the 27- and 24-kDa products, although cleavage at the Q18-G19 site was still visible (Fig. 4B, lanes 1 to 4). Single amino acid substitutions of Q104 with proline and S105 with tyrosine almost totally blocked the formation of the 27- and 24-kDa bands (Fig. 3C, lanes 3 to 4 and 9 to 10). Thus, it is highly likely that Q104-S105 could constitute an alternative cleavage site of 3Cpro in TBP. This notion is also supported by mutational analysis of the amino acids preceding the Q104-S105 site. For example, generation of the 27-kDa product was inhibited to almost 90% of the control when the alanine at P4 was replaced by glycine (A101G) (Fig. 6C, lane 2, and Table 1). The 3Cpro activity was totally abolished when both alanines at P4 and P5 were replaced by either glycine (AA100-101GG) or lysine (AA100-101LL), suggesting that the amino acid residue at P4/P5 could contribute to efficient cleavage at the Q104-S105 site (Fig. 6D).
Finally, analysis of full-length, ΔN100, and ΔC339 TBP as well as the products generated by complete digestion of these molecules by 3Cpro suggested two 3Cpro-mediated cleavages: one at Q18-G19 and the other at Q104-S105 site (Fig. 5). Amino acid composition of the 24-kDa polypeptide indicated that this polypeptide had an actual molecular mass of 12 kDa. The aberrant migration of this polypeptide with an apparent higher molecular mass was most probably due to the presence of 38 consecutive glutamine residues in this molecule. These results along with the mutational analysis strongly suggest that, in addition to cleavage at the Q18-G19 site, TBP is also cleaved at an alternate site, Q104-S105.
Mammalian TBP consists of a 181-amino-acid core that is common to all eukaryotes, fused to a vertebrate-specific 141-amino-acid long N-terminal domain (Fig. 1). Previous studies have suggested the importance of the conserved C-terminal 181 amino acids as well as some acidic amino acids just N-terminal to the conserved core of yeast TBP in transcription and normal cell growth (16, 28, 41). However, recent studies have underscored the importance of the TBP N-terminal domain in transcriptional regulation both in yeast and mammalian systems (15, 21). Moreover, deletion of 55 and 96 amino acids from the N-terminal domain of TBP leads to inactivation of TATA-mediated transcription from the U6 small nuclear RNA promoter (24). Our functional studies on the transcription-restoration activity of ΔN100 TBP in poliovirus-infected cell extracts clearly suggest that a cleavage of TBP at Q104-S105 could lead to significant inactivation of TBP transcriptional activity, as observed during infection of HeLa cells with poliovirus (Fig. 7), underscoring the importance of the N-terminal domain of TBP in RNA Pol II transcription. The specific activity of TBP, as determined by in vitro reconstitution assay, was reduced six- to eightfold as a result of deletion of the N-terminal 100 amino acids (data not shown). Thus, both transcription-restoration and in vitro reconstitution assays suggest that the loss of the N-terminal 104 amino acids of TBP could lead to significant inhibition of RNA polymerase II transcription.
The cleavage at the Q104-S105 site appears to be inefficient compared with that at the Q18-G19 site. This could be due to the fact that TBP in the SL-1 (Pol I) and TFIIIB (Pol III) complexes is cleaved only at the Q18-G19 site, whereas TBP in the TFIID (Pol II) complex is cleaved at the Q104-S105 site. Additionally, the 27- and 24-kDa products generated by cleavage of TBP in the TFIID complex could act in a dominant-negative manner, leading to significant inhibition of Pol II transcription in PV-infected cells. It is well known that TBP is present in limiting quantities in mammalian cells and is shared by all three polymerase systems. Thus, small of amounts of the cleavage products could be sufficient to block wt TBP that is actively engaged in transcription of mRNA.
The identification of the Q104-S105 3Cpro-cleavage site in TBP that could lead to transcriptional inactivation of TBP presented us with the opportunity to examine the role of transcription shutoff in viral replication in vivo. Two cell lines expressing mutant TBP, termed GG1 rTBP and GG2 rTBP, showed a small-plaque phenotype compared to the cell line expressing wt rTBP in a poliovirus plaque assay (Fig. 10). The titer of the cell-free lysates recovered from GG1 rTBP cells was found to be 20- to 30-fold lower than those recovered from wt rTBP cells (Fig. 10). This suggested that viral replication was significantly compromised in cells expressing the mutant noncleavable forms of TBP. Both the wt and mutant TBP-containing cells grew with similar kinetics and did not show any significant differences in overall cellular RNA (Fig. 10E) and protein synthesis (data not shown), suggesting that a defect in cellular transcription shutoff could lead to slower viral replication. Indeed, transcription shutoff was incomplete or delayed in GG1 rTBP cells compared to wt rTBP cells (Fig. 10F). The highest difference (three- to fourfold) in the magnitude of transcription shutoff between GG1 and wt rTBP cells was evident at 4 h postinfection. Since TBP is an essential component of Pol I factor, SL-1, and Pol III factor (TFIIIB) (7, 33), the defect in cellular transcription shutoff observed at 4 h postinfection is likely to include incomplete shutoff of transcription of both RNA polymerase I and III in addition to a defect in Pol II transcription shutoff. The decline of cellular transcription in GG1 rTBP cells at later time points of infection could be due to inactivation of activated Pol II transcription factors Oct-1, p53, and CREB (35, 38, 40) as well as upstream binding factor (Pol I factor) and TFIIIC (Pol III factors), whose activities are independent of TBP (5, 36). In addition, inhibition of TBP-independent RNA Pol II transcription (23) could also contribute to the decline of transcription seen in PV-infected GG1 rTBP cells.
Viral RNA synthesis, as measured by both actinomycin D-resistant, acid-insoluble [3H]uridine incorporation and Northern analysis, was significantly inhibited and delayed in GG1 rTBP cells compared with wt rTBP cells (Fig. 10). It appears, therefore, that lack of efficient transcription shutoff can lead to significantly lower levels of viral RNA synthesis. The exact cause for this is not known but could, at least in part, be due to competition between the viral and cellular RNA polymerases for the ribonucleoside triphosphate pool in infected cells, as previously suggested by Baltimore and Franklin (1). Since poliovirus overproduces viral RNA and proteins, it is not clear whether a 3.5-fold reduction in viral RNA synthesis could cause a 20- to 30-fold decrease in the viral yield. While translation of viral proteins in GG1 rTBP cells was reduced proportionately to viral RNA synthesis, there were no obvious polyprotein processing defects in these cells (data not shown). However, we cannot rule out the possibility that viral processes (such as assembly) other than translation, processing, and RNA synthesis could contribute to lower viral yields in infected GG1 rTBP cells. It is also possible that subtle intracellular changes created by expression of the protease-resistant rTBP mutant could influence viral replication directly or indirectly.
In summary, we have presented evidence here which suggests that cleavage of the TATA-binding protein at an alternative 3Cpro cleavage site, Q104-S105, could lead to shutoff of host cell RNA polymerase II transcription by poliovirus. Analogous to viral precursor polypeptide processing, cleavage at the Q104-S105 dipeptide is facilitated by the presence of an aliphatic amino acid (alanine) at the P4 position. We have also shown that PV-mediated transcription shutoff is inefficient (or incomplete) in stable cell lines that express a mutant, noncleavable form of TBP compared to those expressing the wt TBP. Interestingly, viral replication is significantly affected in HeLa cells that constitutively express a mutant TBP resistant to 3Cpro cleavage. These results suggest for the first time that the shutoff of host cell transcription could play an important role in efficient replication of poliovirus in cultured cells.
Acknowledgments
This work was supported by NIH grant AI27451 to A.D.
The authors are grateful to Padmaja Yalamanchili for her suggestion of 3Cpro cleavage of TBP at an alternative site.
REFERENCES
- 1.Baltimore, D., and R. M. Franklin. 1962. The effect of mengovirus infection on the activity of the DNA-dependent RNA polymerase of L cells. Proc. Natl. Acad. Sci. USA 48:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltimore, D., R. M. Franklin, and J. Callender. 1963. Mengovirus-induced inhibition of host ribonucleic acid and protein. Biochim. Biophys. Acta 76:425-430. [PubMed] [Google Scholar]
- 3.Baltimore, D., and R. M. Franklin. 1963. A new ribonucleic acid polymerase appearing after mengovirus infection of L cells. J. Biol. Chem. 238:3395-3400. [PubMed] [Google Scholar]
- 4.Blair, W. S., and B. L. Semler. 1991. Role for the P4 amino acid residue in substrate utilization by the poliovirus 3CD proteinase. J. Virol. 65:6111-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, M. E., T. Hammerle, E. Wimmer, and A. Dasgupta. 1991. Poliovirus proteinase 3C converts an active form of transcription factor IIIC to an inactive form: a mechanism for inhibition of host cell polymerase III transcription by poliovirus. EMBO J. 10:2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, M. E., P. M. Lieberman, A. J. Berk, and A. Dasgupta. 1993. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol. Cell. Biol. 13:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comai, L., N. Tenese, and R. Tjian. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL-1. Cell 68:965-976. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, N., A. Fires, N. Samuels, P. A. Sharp, and D. Baltimore, D. 1981. Inhibition of transcription factor activity by poliovirus. Cell 27:555-561. [DOI] [PubMed] [Google Scholar]
- 9.Das, S., and A. Dasgupta. 1993. Identification of the cleavage site and determinants required for poliovirus 3Cpro-catalyzed cleavage of human TBP. J. Virol. 67:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta, A., P. Yalamancholi, M. Clark, S. Kliewer L. Fradkin, S. Rubinstein, S. Das, Y. Shen, M. K. Weidman, R. Banerjee, U. Datta, M. Igo, P. Kundu, B. Barat, and A. J. Berk. 2003. Effects of picornavirus proteinases on host cell transcription, p. 321-335. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 11.Dewalt, P. G., and B. L. Semler. 1987. Site-directed mutagenesis of proteinase 3C results in poliovirus deficient in synthesis of viral RNA polymerase. J. Virol. 61:2162-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewalt, P. G., M. A. Lawson, R. J. Colonno, and B. L. Semler. 1989. Chimeric picornavirus polyproteins demonstrate a common 3C proteinase substrate specificity. J. Virol. 63:3444-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hampsey, M. 1998. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62:465-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, K. S., S. R. Reddigari, M. J. Nicklin, T. Hammerle, and E. Wimmer. 1992. Purification and characterization of the poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J. Virol. 66:7481-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs, N. K., A. A. Bondareva, S. Barnett, M. R. Capecchi, and E. E. Schmidt. 2002. Removing the vertebrate-specific TBP N-terminus disrupts placental beta2m-dependent interactions with the maternal immune system. Cell 110:43-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, K., Y. Makino, T. Kishimoto, J. Yamauchi, S. Kato, M. Muramatsu, and T. Tamura. 1994. Multimerization of the mouse TATA-binding protein (TBP) driven by its C-terminal conserved domain. Nucleic Acids Res. 22:1179-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kean, K. M., N. Teterina, and M. Girard. 1990. Cleavage specificity of the poliovirus 3C protease is not restricted to Gln-Gly at the 3C/3D junction. J. Gen. Virol. 71:2553-2563. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 29:1547-1553. [DOI] [PubMed] [Google Scholar]
- 19.Kliewer, S., and A. Dasgupta. 1988. An RNA polymerase II transcription factor inactivated in poliovirus-infected cells copurifies with transcription factor TFIID. Mol. Cell. Biol. 8:3175-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krausslich, H. G., and E. Wimmer. 1988. Viral proteinases. Annu. Rev. Biochem. 57:701-754. [DOI] [PubMed] [Google Scholar]
- 21.Lee, M., and K. Struhl. 2001. Multiple functions of the nonconserved N-terminal domain of yeast TATA-binding protein. Genetics 158:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leong, E.-C., C. T. Cornell, and B. Semler. 2003. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-212. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 23.Martinov, I., S. Viville, and I. Davidson. 2002. RNA polymerase II transcription in murine cells lacking murine cells lacking the TATA-binding protein. Science 298:1036-1039. [DOI] [PubMed] [Google Scholar]
- 24.Mittal, V., and N. Hernandez. 1997. Role of the amino terminal region of the human TBP in U6 snRNA transcription. Science 275:1136-1140. [DOI] [PubMed] [Google Scholar]
- 25.Nicklin, M. J., K. S. Harris, P. V. Pallai, and E. Wimmer. 1988. Poliovirus proteinase 3C: large-scale expression, purification, and specific cleavage activity on natural and synthetic substrates in vitro. J. Virol. 62:4586-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmenberg, A. C., E. M. Kirby, M. R. Janda, N. L. Drake, G. M. Duke, K. F. Portratz, and M. S. Collet. 1984. The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res. 26:2969-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks, G. D., J. C. Baker, and A. C. Palmenberg. 1989. Proteolytic cleavage of encephalomyocarditis virus capsid region substrates by precursors to the 3C enzyme. J. Virol. 63:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon, D., S. Schroeder, C. K. Wang, T. Yamamoto, M. Horikoshi, R. G. Roeder, and P. A. Weil. 1991. The conserved carboxy-terminal domain of Saccharomyces cerevisiae TFIID is sufficient to support normal cell growth. Mol. Cell. Biol. 11:4809-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 30.Racaniello, V. R., and D. Baltimore. 1981. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc. Natl. Acad. Sci. USA 78:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication. .In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 32.Sharma, R., S. Raychaudhuri, and A. Dasgupta. 2004. Nuclear entry of poliovirus protease-polymerase precursor 3CD: implications for host cell transcription shut-off. Virology 320:195-205. [DOI] [PubMed] [Google Scholar]
- 33.Taggart, A. K. P., T. S. Fisher, and B. F. Pugh. 1992. The TATA-binding protein and associated factors are components of Pol III transcription factor TFIIIB. Cell 71:1015-1028. [DOI] [PubMed] [Google Scholar]
- 34.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP) associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 35.Weidman, M. K., P. Yalamanchili, B. Ng, W. Tsai, and A. Dasgupta. 2001. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology 291:260-271. [DOI] [PubMed] [Google Scholar]
- 36.Weidman, M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 95:75-85. [DOI] [PubMed] [Google Scholar]
- 37.Yalamanchili, P., K. Harris, E. Wimmer, and A. Dasgupta. 1996. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J. Virol. 70:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yalamanchili, P., M. K. Weidman, and A. Dasgupta. 1997. Cleavage of transcriptional activator Oct-1 by poliovirus-encoded protease 3Cpro. Virology 239:176-185. [DOI] [PubMed] [Google Scholar]
- 39.Yalamanchili, P., R. Banerjee, and A. Dasgupta. 1997. Poliovirus-encoded protease 2Apro cleaves the TATA-binding protein but does not inhibit host cell RNA polymerase II transcription in vitro. J. Virol. 71:6881-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yalamanchili, P., U. Datta, and A. Dasgupta. 1997b. Inhibition of host cell transcription by poliovirus: Cleavage of transcirption factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 71:120-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Q. A., M. C. Schmidt, and A. J. Berk. 1991. Requirement for acidic amino acid residues immediately N-terminal to the conserved domain of Saccharomyces cerevisiae TFIID. EMBO J. 10:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]