Abstract
We show that human adenovirus inhibits RNA interference (RNAi) at late times of infection by suppressing the activity of two key enzyme systems involved, Dicer and RNA-induced silencing complex (RISC). To define the mechanisms by which adenovirus blocks RNAi, we used a panel of mutant adenoviruses defective in virus-associated (VA) RNA expression. The results show that the virus-associated RNAs, VA RNAI and VA RNAII, function as suppressors of RNAi by interfering with the activity of Dicer. The VA RNAs bind Dicer and function as competitive substrates squelching Dicer. Further, we show that VA RNAI and VA RNAII are processed by Dicer, both in vitro and during a lytic infection, and that the resulting short interfering RNAs (siRNAs) are incorporated into active RISC. Dicer cleaves the terminal stem of both VA RNAI and VA RNAII. However, whereas both strands of the VA RNAI-specific siRNA are incorporated into RISC, the 3′ strand of the VA RNAII-specific siRNA is selectively incorporated during a lytic infection. In summary, our work shows that adenovirus suppresses RNAi during a lytic infection and gives insight into the mechanisms of RNAi suppression by VA RNA.
When double-stranded RNA (dsRNA) is introduced into a cell, it will induce specific degradation of mRNA with the homologous sequence (17) through a mechanism named RNA interference (RNAi). The mechanism for RNAi involves an initial processing of the trigger dsRNA into short interfering RNAs (siRNAs) of 21 to 23 nucleotides by an RNase III type enzyme, called Dicer. The siRNAs are subsequently incorporated into the RNA-induced silencing complex (RISC) enzyme complex, which targets and degrades mRNA with the homologous sequence (reviewed in reference 12).
Many viruses, including those with a DNA genome, will produce dsRNA as a replication intermediate or as a side product of symmetrical transcription of both strands of the viral genome. Therefore, RNAi has the potential to act as a defense against virus infections. In plants, it is well documented that RNAi has an important function as an antiviral defense mechanism (30, 32). Since viruses are masters of adopting strategies to subvert cellular defense mechanisms, it is not surprising that many plant DNA and RNA viruses have evolved proteins that act as suppressors of RNA silencing (reviewed in references 25 and 45).
Assuming that RNAi plays an important function in humans, one would envisage that human viruses also have developed counter-mechanisms to overcome this silencing. However, unlike plants and invertebrates, vertebrates have the interferon system, which responds to dsRNA by inducing the synthesis of a large group of proteins that have a general inhibitory effect on virus multiplication. The best-characterized interferon-induced genes encode the protein kinase R (PKR) and the 2′,5′-oligo(A) synthetase enzymes, both of which are activated in response to dsRNA (reviewed in reference 22). Activated PKR causes an inhibition of protein synthesis by phosphorylation of eukaryotic initiation factor 2 (eIF2), and 2′,5′-oligo(A) synthetase induces general RNA degradation via activation of RNase L, leading to ultimate cell death via apoptosis. Thus, mammals with their adaptive immune system already have defense mechanisms that respond to dsRNA. However, the discovery that RNAi is triggered by siRNAs (9, 15), which are too short to efficiently activate the interferon response pathway (reviewed in reference 39), suggested that RNAi also may play a role in the cellular defense against infection by human viruses.
It is well established that most mammalian viruses have evolved defense strategies to suppress the negative effects that the interferon response pathway has on virus multiplication (reviewed in reference 18). This is of vital importance for the capacity of a virus to multiply successfully. Numerous viruses encode proteins or decoy RNAs that inhibit the activity of PKR by a surprisingly large range of different strategies (reviewed in references 19 and 21). For example, human adenovirus type 5 (Ad5) encodes two approximately 160-nucleotide-long nontranslated RNA polymerase III transcripts: the virus-associated (VA) RNAs, VA RNAI and VA RNAII (reviewed in reference 37). The VA RNAs are highly structured (10) and can adopt secondary structures that show similarity to microRNAs (reviewed in reference 6) in that they form imperfect stem-loop structures. Furthermore, VA RNAI has been shown to stimulate protein synthesis in infected cells (48) and in transient transfection assays (47) by blocking activation of the interferon-induced antiviral defense system (3, 27). VA RNAI binds to PKR and acts as a competitive inhibitor (reviewed in reference 19), thus preventing the dsRNA that is produced by symmetrical transcription of the viral DNA (36) from activating PKR.
Since the RNAi machinery is conserved in mammals, it appeared possible that human viruses, like plant viruses, have evolved strategies to suppress RNA silencing. Here we tested this hypothesis. We show that adenovirus blocks the RNAi machinery at late times of infection. The suppression of RNAi results from a virus-induced block of the two key enzymatic activities in RNAi, Dicer and RISC. We further show that adenovirus VA RNAI and VA RNAII have the capacity to suppress RNAi in transient transfection experiments. Mechanistically, the VA RNAs appear to block RNAi by acting as competitive substrates squelching Dicer. Further, we show that the VA RNAs are themselves processed into siRNA, which are incorporated into functional RISC. Collectively, our results suggest that the adenovirus VA RNAs antagonize the cellular defense pathways directed against both long (interferon-induced) and short (RNAi-induced) dsRNA by binding and inactivating two key enzymes, PKR and Dicer.
MATERIALS AND METHODS
Cell culture condition.
293 ATCC cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum and 1× antibiotic solution. C33A, cervix carcinoma cells, were grown in DMEM with pyruvate, supplemented with 10% fetal calf serum, minimal essential medium nonessential amino acids, 100 U/ml penicillin, and 100 U/ml streptomycin. All cell culture reagents were purchased from Gibco/BRL.
VA RNA mutant viruses.
All viruses used are derivatives of Ad5 and have previously been described. Mutant dl331(VA RNAI−) is a derivative of dl327 (48). dl703 is a variant of Ad5 in which the VA RNA region has been taken from Ad2 (8). Mutants dl704 (VA RNAII−), dl705 (VA RNAI−), and dl-sub720 (VA RNAI− VA RNAII−) are derivatives of dl703 (8). To simplify the nomenclature the dl-sub720 virus is referred to as dl720 throughout this report.
Plasmid construction.
Vectors expressing 29-base-pair short hairpin RNAs (shRNAs) directed against the b3a2 bcr/abl fusion sequence in pcDNA3.1zeo-sb3a2-4hEGFP (44) or the unrelated e1a2 bcr/abl fusion sequence were generated by cloning synthetic double-stranded b3a2 or e1a2 oligonucleotides (44) downstream of the human U6 snRNA promoter in vector pShuttle-U6 (S. Berenjian et al., unpublished data), generating plasmids pShuttleU6-b3a2 and pShuttle-U6-e1a2, respectively. Plasmids encoding Ad5 VA RNAI or VA RNAII under the transcriptional control of the H1 promoter were generated by inserting PCR fragments encoding VA RNAI or VA RNAII into the shRNA expression vector, psiRNA-hH1zeo (InvivoGen), generating plasmids H1-VA RNAI and H1-VA RNAII, respectively. Plasmids pVAI and pVAII have previously been described (47). To construct the luciferase-VA RNA reporter plasmids, pLuc-VAI 5′, pLuc-VAI 3′, pLuc-VAII 5′, and pLuc-VAII 3′, we used pGL3-HIV-Nef (51), which is a derivative of pGL3-Control (Promega). The pGL3-HIV-Nef construct contains human immunodeficiency virus type 1 (HIV-1) Nef sequences cloned into the XbaI site directly downstream of the firefly luciferase open reading frame. To allow for a replacement cloning strategy, EcoRI and PstI restriction sites were introduced upstream and downstream of the HIV-Nef insert, respectively. PCR fragments corresponding to either the 5′ or 3′ half of the Ad5 VA RNAI and VA RNAII genes were cloned in the antisense orientation in the modified pGL3-HIV-Nef plasmid using the EcoRI and PstI restriction sites. Further details about the cloning strategies are available on request.
Infections and GFP reporter analysis.
The vector pcDNA3.1zeo-sb3a2-4hEGFP (b3a2-GFP, for short) encodes a chimeric bcr/abl-enhanced green fluorescent protein (EGFP) (44). To induce RNAi against EGFP, 293 cells were grown on 6-cm plates to 60% confluence and transfected with 0.5 μg of plasmid b3a2-GFP and 1 μg of pShuttle-U6-b3a2 or pShuttle-U6-e1a2 using the FuGENE6 transfection reagent (Roche), as described by the manufacturer. At 16 h posttransfection cells were infected with adenovirus wt900 (50) at a multiplicity of infection of 10 fluorescence-forming units per cell in DMEM containing 2% newborn calf serum. After 45 min the medium was replaced with fresh medium containing 10% serum. The noninfected control cells were treated identically except that virus was omitted. At 40 h posttransfection GFP-expressing cells were counted under a fluorescence microscope at a magnification of ×40. Ten fields from each plate were randomly selected and counted.
Luciferase reporter assay.
C33A cells were grown in 24-well plates to 60% confluence and transfected using the calcium phosphate method (11). To induce RNAi against luciferase, 0.1 μg of pGL3-Control (Promega) was cotransfected with 5 ng of plasmid, pShh1-Ff1 (referred to as shLuc), which encodes an shRNA directed against firefly luciferase (41). As a negative control plasmid pShh1-Ff1rev (referred to as shLucrev) was used, in which the shRNA is inserted in reverse orientation, resulting in synthesis of an improperly structured shRNA that does not induce silencing (41). To test the effect of the VA RNAs on RNAi, 0.1 or 0.5 μg of plasmid H1-VA RNAI or H1-VA RNAII was cotransfected with pGL3-Control and pSh11-Ff1. The total amount of DNA in each transfection mixture was brought to 1 μg by the addition of pBluescript DNA. Firefly luciferase levels were determined using a Dual-Luciferase Reporter assay system (Promega).
Dicer cleavage of VA RNA in vitro.
Double-stranded DNA fragments, with a T7 promoter upstream of the transcription initiation site of VA RNAI and VA RNAII, were generated by PCR amplification (primer sequences available upon request). PCR fragments were precipitated and used for in vitro transcription using a T7-MEGAshortscript kit (Ambion). After DNase treatment the RNAs were purified using NucAway spin columns (Ambion). Two micrograms of VA RNA was incubated overnight at 37°C with 1 unit of human recombinant Dicer (Stratagene) in a buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 2.5 mM MgCl2 in a final volume of 20 μl. To confirm a complete cleavage of the VA RNAs, 1 μl of the reaction mixture was separated on a 2% metaphor agarose gel (BMA) for fine separation of small nucleic acids. The diced VA RNAs (0.1 μg per μl) were further diluted to 0.02 and 0.004 μg per μl and used for cotransfection with the luciferase-VA RNA reporter plasmids, pLuc-VAI 5′, pLuc-VAI 3′, pLuc-VAII 5′, and pLuc-VAII 3′ in 24-well plates as described above.
Cytoplasmic extract preparation.
293 cells were cultured and infected as described above. Cells from three 10-cm culture plates were collected in ice-cold phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.3), washed once in phosphate-buffered saline, and resuspended in 5 volumes of hypotonic buffer (10 mM HEPES-KOH, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT]). The cells were allowed to swell on ice for 15 min and were disrupted by 20 to 30 strokes in a 23-gauge syringe needle. Nuclei were pelleted at 4,500 × g, and the supernatant was centrifuged at 15,000 × g for 60 min. The S15 extracts were supplemented with glycerol to 5%, quick-frozen in liquid nitrogen, and stored at −80°C. The protein concentration was determined by the Bradford assay and was typically 6 to 8 μg/μl.
Dicer assay.
The template for dsRNA synthesis, corresponding to positions 380 to 539 in the firefly luciferase gene of plasmid pGL2-Control (Promega), was amplified by PCR using primers introducing T7 or SP6 promoter sequences at the opposite ends. A 32P-labeled dsRNA was synthesized by in vitro transcription using T7 and SP6 RNA polymerase as previously described (43). Templates for synthesis of the hairpin and VA RNA substrates were amplified by PCR introducing a 5′ T7 promoter sequence. RNA was synthesized and purified as above, and the VA RNAs were analyzed on a nondenaturing polyacrylamide gel to confirm the absence of any dsRNA contaminants (38). Dicer was assayed as described previously (7). Briefly, a 10-μl reaction mixture contained 5 μl of cytoplasmic extract and 5 to 10 fmol of 32P-labeled dsRNA (approximately 50,000 dpm) in a final reaction mixture containing 20 mM HEPES, pH 7, 2 mM MgAc, 2 mM DTT, 1 mM ATP, and 0.75 mM MgCl2. Reaction mixtures were incubated for 2 h at 30°C, proteins were digested with proteinase K, and RNA was isolated and separated on a denaturing 15% polyacrylamide gel. The gel was dried and exposed to a phosphorimager screen (Fujix) or an X-ray film.
RISC assay.
The 484-nucleotide-long 32P-labeled capped p99 mRNA used in the experiments (see Fig. 3) was synthesized by T7 transcription on template pRSETA-p99 (28). RISC activity was assayed as previously described (49) with a synthetic 21-base-pair siRNA (Dharmacon) directed against positions 242 to 263 in the p99 mRNA. The 32P-labeled capped Luc-VA transcripts (see Fig. 8) were synthesized by T7 transcription from PCR templates generated by amplification using the forward primer T7-Luc-RISC-VAI (ATATATTAATACGACTCACTATAGGACCGCGAAAAAGTTG; promoter sequence in bold) or T7-Luc-RISC-VAII (ATATATTAATACGACTCACTATAGGAGGAGTTGTGTTTG; promoter sequence in bold) and the reverse primer Luc-RISC-(CCCTGAACCTGAAACAT) on the respective pLuc-VA plasmid. Each 10-μl RISC assay contained 5 μl of cytoplasmic extract, 25 to 50 fmol of 32P-labeled target mRNA (approximately 50,000 dpm), and 1 pmol of siRNA (optional) in a final reaction mixture containing 15 mM HEPES-KOH, pH 7.4, 50 mM KCl, 1 mM MgCl2, 1 mM ATP, 0.2 mM GTP, 10 μg/ml RNasin (Promega), 30 μg/ml creatine kinase, 25 mM creatine phosphate, 0.5 mM DTT, and 2.5% glycerol. Reaction mixtures were incubated for 2 h at 30°C, proteins were digested with proteinase K, and RNA was isolated and separated on a denaturing 8% polyacrylamide gel. The gel was dried and exposed to a phosphorimager screen (Fujix) or an X-ray film.
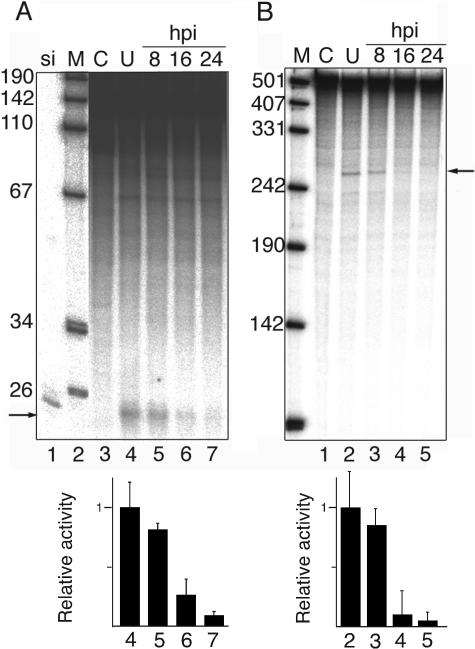
FIG. 3.
Time course of RNAi inactivation. Cytoplasmic extracts were prepared from uninfected 293 cells (U) or cells collected at 8, 16, or 24 h postinfection (hpi) with adenovirus (wt900) M, DNA size marker; C, uninfected extract. The reaction was stopped at time zero. (A) Dicer activity assayed against a 1,600-base-pair dsRNA. si, 32P-labeled 21-base-pair siRNA marker. The arrow indicates the position of 21-base-pair siRNA products. (B) RISC activity in the same extracts assayed against a 484-nucleotide, uniformly labeled mRNA in the presence of a synthetic siRNA. The arrow indicates the position of the 252-nucleotide 5′ cleavage product. Quantitative results based on three independent experiments are shown below respective autoradiograms.
FIG. 8.
siRNAs derived from the terminal stem of VA RNAI and VA RNAII are incorporated into active RISC in adenovirus-infected cells. (A) Cytoplasmic extracts prepared from uninfected 293 cells or cells infected with dl703 (wild type) or VA RNA mutant viruses were assayed for RISC activity against synthetic Luc-VA transcripts with a target region complementary to the 5′ or 3′ half of VA RNAI (upper panel) or VA RNAII (lower panel). The arrows indicate the span of the VA RNA target regions. The predicted positions of the cleavage products generated by Dicer cleavage at the terminal stem of VA RNAI and VA RNAII are indicated by a dot. (B) Secondary structure of VA RNAI and VA RNAII (adapted from references 1 and 37). Note that the VA RNAII structure is a computer model and has not been verified experimentally. The position where Dicer interacts with the VA RNAs, as deduced from the fragments generated in panel A, is indicated. The structure of VA RNAI contains an approximately 20-base-pair apical stem that could theoretically be cleaved by Dicer. Such a cleavage would have generated a 5′ fragment of 162 nucleotides and a 3′ fragment of 219 nucleotides. Cleavage of VA RNAII at the hypothetical apical stem would have generated fragments in the size range of 132 to 141 (5′ transcript) and 185 to 193 (3′ transcript) nucleotides, dependent on the exact position of Dicer interaction.
In vitro kinase assay.
A recombinant ASF/SF2 protein (20) (0.15 μg) was incubated with 0.25 μg of 293-S15 cytoplasmic extracts at room temperature for 40 min in a kinase buffer containing 20 mM Tris (pH 7.4), 130 mM KCl, 15 mM MgCl2, 5 mM DTT, 0.1 mM ATP, and 0.25 μCi of [γ-32P]ATP. Products were resolved on a 12% reducing sodium dodecyl sulfate (SDS)-polyacrylamide gel, dried, and subjected to autoradiography.
Northern blot analysis.
Total cytoplasmic RNA was prepared by lysis with IsoB-NP-40 (10 mM Tris-HCl [pH 7.9], 150 mM NaCl, 1.5 mM MgCl2, 1% NP-40), followed by two rounds of phenol-chloroform-isoamylalcohol extraction and one extraction with chloroform-isoamylalcohol (5). Two micrograms of cytoplasmic RNA was separated on a 1% agarose gel containing 2.2 M formaldehyde, transferred to a nitrocellulose filter, and hybridized with L3- and L5-specific DNA probes (29) 32P labeled by random priming as described (5). For siRNA analysis total RNA was prepared from uninfected or infected 293 cells using the Trizol reagent (Invitrogen). Thirty micrograms of RNA was separated on denaturing 15% polyacrylamide gels and transferred to Hybond XL membranes (Amersham Biosciences) by electroblotting (200 mA at +4°C overnight). PCR products encoding VA RNAI and VA RNAII were 32P labeled by random priming (5). Membranes were blocked by preincubation in a solution containing 100 mg/ml dextran sulfate, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 37.5% formamide, 1× Denhardts solution, 12.5 mM Tris-HCl, pH 7.5, and 5 μg/ml single-stranded DNA for 4 h. The probe was added, and the membranes were incubated overnight at +30°C, washed two times at 30°C (5 min each time) in a solution containing 2× SSC-0.1% SDS, washed three times in 1× SSC-0.1% SDS, and exposed to a phosphorimager. 32P-labeled siRNAs and a DNA ladder were used as size markers.
RESULTS
Suppression of RNAi in adenovirus-infected cells.
To determine whether human adenovirus inhibits RNAi, we cotransfected 293 cells with a bcr/abl-GFP reporter construct (44) and a vector expressing a 29-base-pair short hairpin RNA (41) directed against the bcr/abl-GFP fusion portion. As shown in Fig. 1, the homologous sh-b3a2 hairpin RNA reduced the number of fluorescent cells expressing GFP significantly (lane 2) compared to an identical vector expressing a nonhomologous sh-e1a2 hairpin (lane 1). Infection of transfected cells with adenovirus completely annulled the inhibitory effect of the sh-b3a2 hairpin on GFP expression (lane 4), indicating that adenovirus inactivates the RNAi machinery. Adenovirus infection of cells transfected with the sh-e1a2 control hairpin RNA did not lead to a dramatic increase in GFP expression (lane 3), suggesting that the adenovirus-induced increase in GFP expression (lane 4) was not mediated by a general translational effect of VA RNA via inactivation of PKR. This type of experiment is technically difficult since RNAi induced by siRNA transfection generally requires much longer time to be fully established compared to the rapid replication cycle of adenovirus. Similar problems have been reported by others (35). However, the results, based on six independent experiments, showed a statistically significant effect of the shRNA in uninfected cells, whereas no effect was seen in infected cells, and thus provide an indication that an adenovirus infection results in a suppression of RNAi.
FIG. 1.

An adenovirus infection suppresses RNAi. 293 cells were cotransfected with a b3a2-GFP reporter plasmid and a vector expressing a 29-base-pair nonhomologous e1a2 or homologous b3a2 shRNA. At 16 h posttransfection the samples were infected with wild-type adenovirus (wt900; lanes 3 and 4) or mock infected (lanes 1 and 2). Fluorescent cells were counted under the microscope at 40 h posttransfection. The figure shows a typical result with mean and standard deviation from 10 counted fields. The effect of b3a2 shRNA in uninfected cells is variable but statistically significant (36 ± 18%; n = 6; P = 0.05). In contrast, no significant effect of b3a2 shRNA is seen in adenovirus-infected cells (8 ± 13%; n = 9; P = 0.05).
Adenovirus VA RNAI and VA RNAII function as suppressors of RNAi in transiently transfected cells.
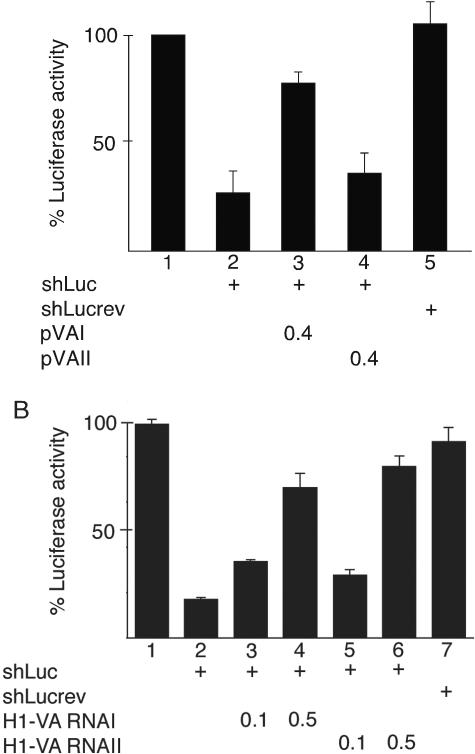
The adenovirus VA RNAs are highly structured with relatively long dsRNA regions (reviewed in reference 37) and interact with a large number of cellular dsRNA binding proteins (34). Therefore, the VA RNAs were the prime candidates to function as viral suppressors of RNAi. To test this hypothesis, C33A cells were cotransfected with a firefly luciferase reporter, a vector expressing a luciferase-specific shRNA (shLuc), and plasmid pVAI or pVAII expressing VA RNAI or VA RNAII. As shown in Fig. 2 (panels A and B) cotransfection of the shLuc plasmid resulted in a dramatic reduction in luciferase expression (lanes 1 and 2). This inhibition was specific since cotransfection of a plasmid expressing a control shRNA (shLucrev) had no significant inhibitory effect on firefly luciferase expression (Fig. 2A, lane 5, and B, lane 7). Further, cotransfection of the shLuc plasmid with a Renilla luciferase reporter construct did not affect luciferase expression (data not shown). Interestingly, cotransfection of plasmid pVAI suppressed the shRNA-mediated reduction of luciferase expression (Fig. 2A, lane 3), whereas cotransfection of plasmid pVAII had only a marginal effect on luciferase reporter activity (Fig. 2A, lane 4). This result was not entirely unexpected since the VA RNAI promoter is much stronger than the VA RNAII promoter and this RNA is produced in substantially larger quantities during a lytic infection (8, 46). To determine whether VA RNAII has an intrinsic capacity to block RNAi, we placed the VA RNA genes under the transcriptional control of the strong H1 promoter, generating plasmids H1-VA RNAI and H1-VA RNAII, respectively. As shown in Fig. 2B, cotransfecting increasing amounts of a plasmid expressing H1-VA RNAI (lanes 3 and 4) or H1-VA RNAII (lanes 5 and 6) resulted in a dose-dependent recovery of luciferase expression. It is noteworthy that, under these conditions, VA RNAI and VA RNAII were equally efficient in relieving shRNA-mediated suppression of luciferase expression. Collectively, these results are consistent with the conclusion that both VA RNAI and VA RNAII have the capacity to function as suppressors of RNAi.
FIG. 2.
VA RNA restores expression of a silenced luciferase reporter. C33A cells were cotransfected with a luciferase reporter plasmid, a vector expressing an shRNA against luciferase (shLuc), or a control shRNA (shLucrev). The effect of VA RNA expression on the silencing of the luciferase reporter was measured by cotransfection of 0.4 μg of plasmid pVAI or pVAII (A) or 0.1 or 0.5 μg of plasmid H1-VA RNAI or H1-VA RNAII (B). The quantitative results are based on three independent experiments.
Adenovirus blocks the activity of Dicer and RISC.
Although the observed loss of RNAi in adenovirus-infected or VA RNA-transfected cells suggests that one or several enzymes in the RNAi pathway are blocked, it does not rule out the possibility that it is a secondary effect, for example, as a result of inhibited RNA export through a competition for the exportin-5 receptor (35). To confirm that there is, indeed, an inactivation of the RNAi machinery, we measured the activity of Dicer and RISC in cytoplasmic 293 cell extracts prepared at different time points after infection. Dicer activity was assayed using a 32P-labeled dsRNA template (Fig. 3A), and RISC activity was measured by cleavage of a 32P-labeled mRNA incubated with a complementary synthetic siRNA (Fig. 3B). The activity of both enzymes was reduced in virus-infected extracts, with an almost complete inhibition at 16 h postinfection. This result was not unique to the 293 cell line since a similar inhibition of Dicer and RISC was observed in extracts from adenovirus-infected HeLa Williams and HeLa spinner cells (data not shown).
Theoretically, the reduction in Dicer and RISC activity at late times of infection could result from virus-induced cell death. To demonstrate that the extracts are enzymatically active, we measured protein kinase activity directed against the SR protein ASF/SF2 (20). We selected ASF/SF2 as a substrate since it is well established that ASF/SF2 is phosphorylated by SRPK1 (23), a cytoplasmic protein kinase that should be present in the S15 extract used to measure Dicer and RISC activity. As shown in Fig. 4A, the recombinant ASF/SF2 protein was phosphorylated as efficiently, if not slightly better, in infected extracts compared to uninfected extracts. Furthermore, studies on adenovirus alternative RNA splicing are routinely performed in our laboratory with nuclear extracts prepared at 20 to 22 h postinfection, and these extracts are fully competent in virus-specific splicing (reviewed in reference 2). Taken together, these results suggest that the inhibition of Dicer and RISC in cell extracts at late times of adenovirus infection is selective.
FIG. 4.

The suppression of Dicer and RISC extracts at late times of adenovirus infection is specific. (A) An adenovirus infection does not inhibit protein kinase activity against a recombinant ASF/SF2 protein. Cytoplasmic extracts prepared from uninfected 293 cells (U) or cells infected with wild-type adenovirus for 8, 16, or 24 h were tested for their capacity to phosphorylate ASF/SF2 in vitro. Products were resolved on a 12% reducing SDS-polyacrylamide gel and subjected to autoradiography. (B) Restoration of Dicer activity in infected extracts by increasing the concentration of the dsRNA substrate. Dicer activity in cytoplasmic extracts prepared from uninfected or adenovirus-infected (16 h postinfection [hpi]) 293 cells was assayed against a 32P-labeled GL2 dsRNA. The figure shows the Dicer activity in infected extracts relative to uninfected extract at concentrations of 5, 20, and 100 nM dsRNA. The mean values and standard deviations from three independent experiments are shown. For further details see Fig. S1 in the supplemental material.
Interestingly, the addition of increasing amounts of target dsRNA to infected extracts restored Dicer activity (Fig. 4B). This result is important for two reasons. First, it shows that the Dicer-inhibitory activity in the infected extract is mediated by a limiting factor in the extract that can be out titrated. Second, this result supports the conclusion that the infected cell extracts are not metabolically dead extracts since the activity of Dicer can simply be reactivated by spiking the extract with an excess of dsRNA.
The VA RNAs interfere with the activity of Dicer in infected extracts.
Having a specific assay allowed us to test the effect of the individual VA RNAs on the activity of Dicer during a lytic adenovirus infection (Fig. 5). For this purpose, we used a collection of adenovirus mutant viruses deficient in VA RNA expression (8). In these experiments 293 cells were infected with the wild type or the mutant viruses, and the activity of Dicer was assayed in extracts prepared at late times of infection. As shown in Fig. 5A and quantitated in Fig. 5B, the wild-type virus (dl703) efficiently suppressed Dicer (lane 2), whereas the mutant dl720, which is defective both in VA RNAI and VA RNAII expression, failed to block Dicer (lane 5). This result indicates that the VA RNAs are required for suppressing the activity of Dicer during a lytic infection. Interestingly, a mutant virus defective in VA RNAII expression (dl704) was almost as efficient as the wild-type virus in suppressing Dicer (compare lanes 2 and 3). In contrast, mutant virus dl705, which is defective in VA RNAI expression, showed a markedly reduced capacity to suppress the activity of Dicer compared to the wild-type virus (compare lanes 2 to 4), suggesting that VA RNAII is a slightly less efficient Dicer suppressor compared to VA RNAI. Most likely this difference results from the fact that significantly more VA RNAI is produced during a lytic adenovirus infection (46). As shown in Fig. 2, the inherent activity of VA RNAI and VA RNAII to inhibit RNAi appears to be similar under conditions where expression of these RNAs are boosted by the strong H1 promoter.
FIG. 5.
VA RNA expression is necessary for suppression of Dicer. (A) Cytoplasmic extracts prepared at 22 h postinfection from wild-type or VA RNA mutant virus-infected 293 cells were assayed for their activity of Dicer as described in Materials and Methods. (B) Quantitative results based on three independent experiments are shown.
The VA RNAs are substrates for Dicer.
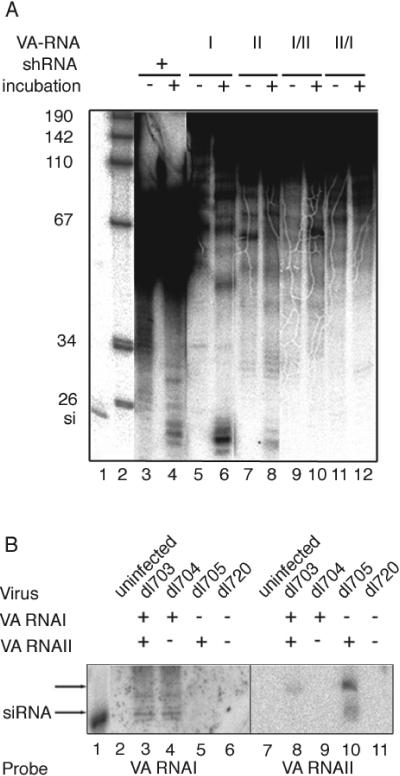
Since the VA RNAs appear to suppress RNAi (Fig. 2) by acting as competitive substrates sequestering Dicer (Fig. 4B), we speculated that the VA RNAs might themselves be cleaved by Dicer. To test this hypothesis we incubated in vitro transcribed 32P-labeled VA RNAs in cytoplasmic extracts prepared from uninfected cells. As shown in Fig. 6A, VA RNAI was efficiently cleaved to small RNAs (lane 6) with the same length as a synthetic 21-nucleotide siRNA (lane 1) or an siRNA cleaved from a 29-base-pair shRNA (lane 4). A quantitative analysis of the results suggests that Dicer activity against VA RNAI is approximately fourfold higher than the activity against the 29-base-pair shRNA, which in turn means that VA RNAI is cleaved with a similar efficiency as a dsRNA. VA RNAII was also cleaved to siRNA, although with much reduced efficiency compared to VA RNAI (compare lanes 6 and 8). The processing of the VA RNAs to siRNA appears to require VA RNA structure since hybrid VA RNAs, consisting of the 5′ half of VA RNAI fused to the 3′ half of VA RNAII (lanes 9 and 10), or vice versa, the 5′ half of VA RNAII fused to the 3′ half of VA RNAI (lanes 11 and 12), were not cleaved into siRNAs.
FIG. 6.
The VA RNAs are processed into siRNA both in vitro and in vivo. (A) 32P-labeled VA RNAI, VA RNAII, or VA RNA hybrid molecules (VA RNAI/II and VA RNAII/I) or a 29-base-pair shRNA were incubated in S100 extracts. After purification the reaction products were separated on a 15% denaturing polyacrylamide gel and visualized by autoradiography. −, reactions stopped at time zero. The control RNAs, VA RNA I/II and II/I, are unable to form normal secondary structures. (B) Total RNA prepared 24 h postinfection from dl703 (wild type) or VA RNA mutant virus-infected 293 cells was separated on a 15% denaturing polyacrylamide gel and transferred to a Hybond membrane. VA RNAI- and VA RNAII-specific probes were used to detect small RNAs. si, 32P-labeled 21-base-pair siRNA marker. Note that the predominant product from VA RNAII is slightly larger than the siRNA marker.
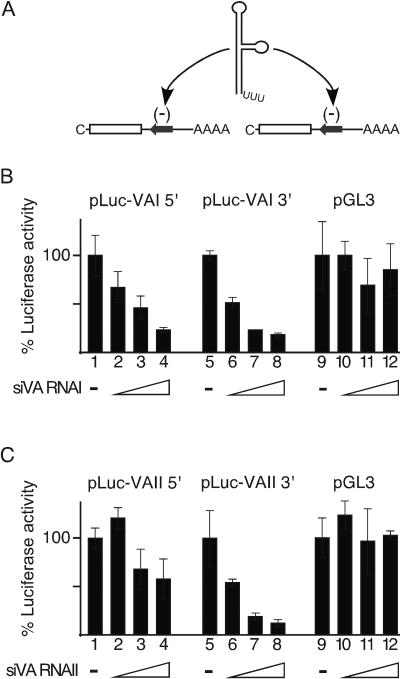
To confirm that the Dicer products formed are functional siRNAs, VA RNAI and VA RNAII were cleaved with a recombinant Dicer enzyme, and the resulting fragments were tested for their capacity to function as siRNAs in transiently transfected cells. For this experiment we constructed chimeric luciferase reporter constructs; either the 5′ half or the 3′ half of VA RNAI and VA RNAII was fused in an antisense orientation to the 3′ noncoding region of a luciferase reporter plasmid, generating plasmids pLuc-VAI 5′, pLuc-VAI 3′, pLuc-VAII 5′, and pLuc-VA 3′ (Fig. 7A). As shown in Fig. 7B, siRNAs derived from VA RNAI inhibited both Luc-VAI 5′ and Luc-VAI 3′ with a similar efficiency. In contrast, siRNAs derived by cleavage of VA RNAII inhibited Luc-VAII 3′ significantly better than Luc-VAII 5′ (Fig. 7C, compare lanes 2 to 4 with lanes 6 to 8), suggesting a strand bias for incorporation into functional RISC (see also below). The silencing effect was specific since siRNAs derived from the VA RNAs did not silence luciferase expression in cells transfected with the pGL3-control plasmid (Fig. 7B and C, lanes 9 to 12). Collectively, these results show that both VA RNA species can be cleaved by Dicer into biologically active siRNA.
FIG. 7.
The VA RNAs are cleaved by a recombinant Dicer into functional siRNAs. (A) Schematic diagram showing the structure of the luciferase reporter mRNAs expressed from pLuc-VAI 5′, pLuc-VAI 3′, pLuc-VAII 5′, and pLuc-VAII 3′, respectively. The VA RNA genes were separated within the terminal loop into two halves and cloned in the antiparallel orientation in the 3′ untranslated region of the luciferase reporter mRNA. (B and C) 293T cells were cotransfected with increasing amounts of siRNAs derived by Dicer cleavage of VA RNAI or VA RNAII (4, 20, and 100 ng) and 0.1 μg of reporter plasmids pLuc-VAI 5′ (lanes 1 to 4), pLuc-VAI 3′ (lanes 5 to 8), or pGL3 (lanes 9 to 12). Luciferase expression was measured 48 h posttransfection.
To determine whether the VA RNAs are cleaved to siRNAs also during an adenovirus infection, cytoplasmic RNA derived from wild-type or VA RNA mutant virus-infected cells was separated on a high-percentage gel and probed for VA RNA-specific siRNAs by Northern blotting. As shown in Fig. 6B, VA RNAI-specific siRNAs were detected in dl703- and dl704-infected cells (lanes 3 and 4). As expected, such siRNAs were not produced in VA RNAI mutant virus-infected cells (lanes 5 and 6). VA RNAII produced small amounts of siRNAs of two size classes, approximately 21 and 27 nucleotides in length (lanes 8 and 10). In wild-type virus-infected cells the larger size class was weakly detected, whereas in VA RNAI mutant-infected cells, siRNAs of both the 27- and the 21-nucleotide length were observed. The dominance of siRNAs corresponding to VA RNAI in wild-type virus-infected cells most likely reflects the fact that approximately 20-fold more VA RNAI accumulates during a lytic adenovirus infection (46). The increase in VA RNAII-specific siRNA in dl705-infected cells most likely result from the expected increase in VA RNAII expression in a virus background lacking VA RNAI (8). Collectively, our results suggest that the VA RNAs function as substrates for Dicer cleavage both in vivo and in vitro.
The VA RNAs are incorporated into active RISC in cells at late times of adenovirus infection.
Since VA RNA are cleaved into siRNA both in vitro and in vivo (Fig. 6) and VA-derived siRNAs are biologically functional (Fig. 7), it became of interest to test whether VA RNA-specific siRNAs are incorporated into RISC during an adenovirus infection. We reasoned that if VA RNA-specific siRNAs become incorporated into RISC during the infection, extracts from these cells would cleave a synthetic mRNA harboring sequences complementary to VA RNA. Further, the position where RISC cleaves the VA RNA sequences would be indicative of where Dicer originally bound to the VA RNA. Also, by using targets complementary to the 5′ and 3′ halves of the VA RNAs, it should be possible to determine whether one or both strands of VA RNA-specific siRNAs were incorporated into RISC. To test this hypothesis, we analyzed RISC activity against substrate RNAs harboring sequences complementary to the 5′ and 3′ halves of VA RNAI and VA RNAII. As shown in Fig. 8A, cytoplasmic extracts prepared from wild-type virus-infected cells (dl703) or VA RNAII mutant-infected cells (dl704) were active in RISC, as shown by the generation of specific cleavage products directed against target RNA derived from both the 5′ (lanes 3 and 5) and 3′ (lanes 4 and 6) halves of VA RNAI. The size of the cleavage products corresponds to a predicted binding and cleavage of Dicer generating an siRNA from the terminal stem of VA RNAI (Fig. 8B). Interestingly, we do not detect RISC cleavage products corresponding to an siRNA generated from the apical stem of VA RNAI. Further, the finding of RISC activity targeting both the 5′ and 3′ halves of VA RNAI suggests that both strands of the VA RNAI-specific siRNA become incorporated into active RISC. Notably, the activity is significantly higher in extract from cells infected with the mutant virus deficient in VA RNAII expression (dl704), suggesting that VA RNAI and VA RNAII may compete for binding to Dicer and/or RISC.
A similar experiment was done to score for siRNAs derived from VA RNAII. However, it should be noted that the secondary structure of VA RNAII has not been experimentally tested. The structure shown in Fig. 8B is a computer-generated model (1). Incubation with target RNAs complementary to VA RNAII resulted in a cleavage product corresponding to an siRNA processed from the predicted terminal stem of VA RNAII (Fig. 8A, lower panel). However, in this case a cleavage product is only seen with the VA RNAII 3′ target RNA (lane 8), suggesting that there is a strand bias resulting in a strong preference for incorporation of the 3′ strand of the VA RNAII-derived siRNA into RISC. It is noteworthy that the VA RNA-specific cleavage product was hardly detectable in wild-type virus-infected cells (dl703, lane 4), whereas it became a prominent product in cells infected with a mutant unable to express VA RNAI (dl705, lane 8). This result was not surprising since VA RNAII is produced in much lower levels compared to VA RNAI in a wild-type infection (≈5%) ( 46). It has been shown that in the absence of VA RNAI, transcription of VA RNAII increases dramatically (8). Also, VA RNAII-specific siRNAs are only detected in dl705-infected cells (Fig. 6B).
As expected, extracts prepared from mutant virus infections deficient in VA RNAI expression (dl705 and dl720) or VA RNAII (dl704 and dl720) expression did not yield RISC activity against VA RNAI (Fig. 8A, lanes 7 to 10) or VA RNAII (Fig. 8A, lanes 5, 6, 9, and 10), respectively. Collectively, these results suggest that Dicer binds to the terminal stem of both VA RNAI and VA RNAII. Further, our results show that Dicer cleaves the VA RNAs to siRNAs in vivo and that siRNAs derived from the VA RNAs are incorporated into functional RISC during a lytic infection.
DISCUSSION
Here we show that human adenovirus, a double-stranded DNA virus, encodes two small nontranslated RNA polymerase III transcripts, VA RNAI and VA RNAII, which have the capacity to suppress RNAi. The idea that the VA RNAs might function as suppressors of RNAi originates from the observation that they form highly folded structures with imperfect stems (Fig. 8B) that resemble precursors to microRNA and therefore might sequester Dicer by acting as competing substrates or pseudo-substrates. The finding that VA RNAI and VA RNAII are indeed processed by Dicer into siRNAs both in vitro and during a lytic infection (Fig. 6) supports this model and shows that the VA RNAs can interact with Dicer. Further, the observation that suppression of the activity of Dicer in infected extracts can be overcome by increasing the substrate concentration (Fig. 4B) suggests that the VA RNAs act as competitive substrates. Since the VA RNAs are expressed at copious amounts at late times of an adenovirus infection (up to 108 copies/cell) (46) one would expect that they are produced in great excess over any aberrantly formed dsRNAs. Therefore, a simple competitive inhibition for binding to Dicer would be sufficient to explain the inhibitory effect of the VA RNAs on RNAi.
During the review process of this report, a study was published showing that, in transient transfection assays, VA RNAI inhibits RNAi and microRNA processing (35). The authors provided evidence suggesting that VA RNAI may achieve this by suppressing the nuclear export of shRNA by competing for binding to the exportin-5 nuclear transport factor. Also, they provided preliminary evidence that VA RNAI blocks Dicer by showing that a synthetic VA RNAI can suppress the activity of a recombinant Dicer in vitro (35). We have not addressed the significance of exportin-5 in VA RNA-mediated suppression of RNAi during a lytic virus infection. However, our experiments extend significantly on the study by Lu and Cullen (35) by providing strong evidence that the VA RNAs are, indeed, important for the suppression of Dicer in lytically infected cells (Fig. 5). Thus, we show that VA RNAs (i) suppress the activity of Dicer in extracts prepared from cells at late times of infection, (ii) bind Dicer through their terminal stems (Fig. 8), and (iii) are cleaved by Dicer both in vitro and in vivo (Fig. 6) into functional siRNAs (Fig. 7) that are incorporated in active RISC (Fig. 8). Taken together, the two studies provide strong support for the hypothesis that the VA RNAs exert their inhibitory effect on RNAi by suppressing the exportin-5-mediated export of dsRNA and cytoplasmic processing of dsRNA into siRNA.
The observation that the activity of RISC is also inhibited in virus-infected cells (Fig. 3B) is novel. The most simplistic model to explain this finding would be to postulate that RISC becomes saturated by VA RNA-specific siRNAs in vivo. Such VA RNA-specific RISC complexes could either suppress or, alternatively, redirect RISC in virus-infected cells toward VA RNA-specific sequences. Since the VA RNAs are encoded by intronic sequences, such VA RNA-specific RISC would not induce cleavage of cytoplasmic mRNA and, therefore, would not have any negative effects on virus multiplication. Also, siRNA target sequences that are located within secondary structures escape from cleavage by RISC (51). Although attractive, our preliminary experiments suggest that the VA RNAs are not required for inhibition of RISC. Thus, it appears more likely that another viral gene product yet to be identified is responsible for the inhibition of RISC. Our future work will be aimed at resolving this question.
Taken together, current data point to the possibility that adenovirus VA RNAI may serve two functions during adenovirus multiplication. It appears to antagonize the cellular defense pathways against both long (interferon) and short (RNAi) dsRNA by binding the two key enzymes in the respective pathways, PKR and Dicer. The two enzymes bind to nonoverlapping sequences. Thus, PKR binds to VA RNAI via interactions through the central domain and the apical stem (10), whereas Dicer appears to interact with the terminal stem of VA RNAI. Although VA RNAI and VA RNAII appear to have redundant activities in the suppression of RNAi, we suspect that during the natural life cycle of the virus, VA RNAI is of greater importance since it is produced in substantially larger quantities compared to VA RNAII (46). Such a speculation is also supported by the observation that predominantly VA RNAI-specific siRNA (Fig. 6B) and RISC (Fig. 8) are detectable in wild-type virus-infected cells.
It appears likely that many virus-encoded PKR inhibitory proteins or RNAs will also have a function as suppressors of RNAi. From this perspective it is interesting that a recent study showed that the interferon-antagonizing proteins of vaccinia virus (EL3) and influenza virus (NS1) function as suppressors of RNAi in transiently transfected Drosophila cells (33). It will be interesting to see whether these proteins also suppress RNAi during a lytic infection of human cells. Since both proteins presumably act by binding and sequestering dsRNA, thereby making it inaccessible for Dicer, they show similarity in function to the VA RNAs described here.
The finding that Dicer cleaves VA RNAI into siRNA both in vitro and during virus infection raises the interesting possibility that siRNAs generated from VA RNA could function as microRNAs and interfere with the expression of viral or cellular genes during infection. The Epstein-Barr virus was recently shown to encode such microRNAs (42). While a BLAST search with VA RNA sequences against the human genome generates no perfect matches, the possibility remains that imperfect matches to cellular mRNA sequences result in a translational block, as has been shown for microRNAs (13, 40). Identifying the targets of such an interaction may, however, be a formidable task since the sequence homology required to obtain repression apparently follows no simple rules and several different microRNAs and siRNAs can act cooperatively to obtain repression (14).
The finding that adenovirus inactivates the RNAi pathway may be interpreted to indicate that RNAi may play a role in the defense against viruses in mammals, as it does in plants (32) and invertebrates (31). However, it remains to be established how significant the RNAi machinery is in humans. It may be of importance that RNAi is only one of several regulatory pathways mediated by small RNAs. For example, small RNAs regulate translation and gene silencing through DNA methylation and heterochromatin formation (reviewed in reference 16). Perhaps the suppressive effect of VA RNA on Dicer interferes with one of these activities or another unrelated, yet-to-be-discovered cellular function that is mediated by small RNAs.
However, if suppression of RNAi is a general strategy used by mammalian viruses, therapeutic intervention of virus infections by siRNA treatment needs to take this suppressive mechanism into account. Also, the possible existence of viral suppressors of RNAi should have consequences on how adenoviral vectors, and potentially other viral vectors, are designed to create optimal vectors for siRNA delivery to target cells. Since the VA RNAs are also transcribed from nonreplicating adenoviral vectors, one would expect that VA RNA might negatively affect the efficiency of adenovirus-delivered shRNA. Further, it is possible that an adenoviral vector may alter cellular gene expression as a result of competition between VA RNA and cellular microRNAs as observed in virus-infected plants (26). From this point it is interesting that VA RNAI requires exportin-5 for nuclear export (24, 52) and inhibits export of premicroRNA precursors by competing for the exportin-5 transport factor (35). It is noteworthy that the joint pathways for RNAi and microRNA processing in vertebrates might pose a strong evolutionary constraint on the evolution of viral pathogens that cause persistent or latent infections. Since blocking microRNA processing would have serious effects on the expression of microRNA-regulated genes and may even kill many cells, including stem cells (reviewed in reference 4), a persistent/latent infection may require that the virus is hiding from RNAi rather than inhibiting it.
Supplementary Material
Acknowledgments
We thank Ernst-Jan Geutjes for help with the experimental work, Jan-Peter Kreivi for the p99 construct and the p99 siRNA, Walter de Vries for the kind gift of VA constructs, Bayar Thimmapaya for VA RNA mutant viruses, Patrick Paddison for the kind donation of the shRNA-luciferase construct, and Peter de Haan for stimulating discussions.
The work done in the Akusjärvi laboratory was supported by the Swedish Cancer Society and the Wallenberg Consortium North, whereas the work done in the Berkhout laboratory was sponsored by Viruvation BV, NWO-CW, and the ZonMw-Vici program. The collaboration between the two laboratories was supported by NORFA.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akusjärvi, G., M. B. Mathews, P. Andersson, B. Vennström, and U. Pettersson. 1980. Structure of genes for virus-associated RNAI and RNAII of adenovirus type 2. Proc. Natl. Acad. Sci. USA 77:2424-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akusjärvi, G., and J. Stevenin. 2003. Remodelling of the host cell RNA splicing machinery during an adenovirus infection. Curr. Top. Microbiol. Immunol. 272:253-286. [DOI] [PubMed] [Google Scholar]
- 3.Akusjärvi, G., C. Svensson, and O. Nygård. 1987. A mechanism by which adenovirus virus-associated RNAI controls translation in a transient expression assay. Mol. Cell. Biol. 7:549-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 6.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 8.Bhat, R. A., and B. Thimmappaya. 1984. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 12:7377-7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan. 2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, P. A., and M. B. Mathews. 1995. Interactions between the double-stranded RNA binding motif and RNA: definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA 1:7-20. [PMC free article] [PubMed] [Google Scholar]
- 11.Das, A. T., B. Klaver, and B. Berkhout. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 73:81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denli, A. M., and G. J. Hannon. 2003. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 28:196-201. [DOI] [PubMed] [Google Scholar]
- 13.Doench, J. G., C. P. Petersen, and P. A. Sharp. 2003. siRNAs can function as miRNAs. Genes Dev. 17:438-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18:504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 16.Finnegan, E. J., and M. A. Matzke. 2003. The small RNA world. J. Cell Sci. 116:4689-4693. [DOI] [PubMed] [Google Scholar]
- 17.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 18.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2003. Principles of virology: molecular biology, pathogenesis, and control of animal viruses. American Society for Microbiology, Washington, D.C.
- 19.Gale, M., Jr., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29-46. [DOI] [PubMed] [Google Scholar]
- 20.Ge, H., P. Zuo, and J. L. Manley. 1991. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell 66:373-382. [DOI] [PubMed] [Google Scholar]
- 21.Gil, J., and M. Esteban. 2000. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5:107-114. [DOI] [PubMed] [Google Scholar]
- 22.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 23.Gui, J. F., H. Tronchere, S. D. Chandler, and X. D. Fu. 1994. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. USA 91:10824-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gwizdek, C., B. Ossareh-Nazari, A. M. Brownawell, A. Doglio, E. Bertrand, I. G. Macara, and C. Dargemont. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278:5505-5508. [DOI] [PubMed] [Google Scholar]
- 25.Haasnoot, J. P. C., D. Cupac, and B. Berkhout. 2003. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 26.Kasschau, K. D., Z. Xie, E. Allen, C. Llave, E. J. Chapman, K. A. Krizan, and J. C. Carrington. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4:205-217. [DOI] [PubMed] [Google Scholar]
- 27.Kitajewski, J., R. J. Schneider, B. Safer, S. M. Munemitsu, C. E. Samuel, B. Thimmappaya, and T. Shenk. 1986. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell 45:195-200. [DOI] [PubMed] [Google Scholar]
- 28.Kreivi, J. P., L. Trinkle-Mulcahy, C. E. Lyon, N. A. Morrice, P. Cohen, and A. I. Lamond. 1997. Purification and characterisation of p99, a nuclear modulator of protein phosphatase 1 activity. FEBS Lett. 420:57-62. [DOI] [PubMed] [Google Scholar]
- 29.Larsson, S., C. Svensson, and G. Akusjärvi. 1992. Control of adenovirus major late gene expression at multiple levels. J. Mol. Biol. 225:287-298. [DOI] [PubMed] [Google Scholar]
- 30.Lecellier, C. H., and O. Voinnet. 2004. RNA silencing: no mercy for viruses? Immunol. Rev. 198:285-303. [DOI] [PubMed] [Google Scholar]
- 31.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 32.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 33.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao, H. J., R. Kobayashi, and M. B. Mathews. 1998. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. USA 95:8514-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, S., and B. R. Cullen. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J. Virol. 78:12868-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maran, A., and M. B. Mathews. 1988. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology 164:106-113. [DOI] [PubMed] [Google Scholar]
- 37.Mathews, M. B. 1995. Structure, function, and evolution of adenovirus virus-associated RNAs. Curr. Top. Microbiol. Immunol. 199:173-187. [DOI] [PubMed] [Google Scholar]
- 38.Mellits, K. H., T. Pe'ery, L. Manche, H. D. Robertson, and M. B. Mathews. 1990. Removal of double-stranded contaminants from RNA transcripts: synthesis of adenovirus VA RNAI from a T7 vector. Nucleic Acids Res. 18:5401-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moss, E. G., and J. M. Taylor. 2003. Small-interfering RNAs in the radar of the interferon system. Nat. Cell Biol. 5:771-772. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, P. H., and V. Ambros. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216:671-680. [DOI] [PubMed] [Google Scholar]
- 41.Paddison, P. J., A. A. Caudy, E. Bernstein, G. J. Hannon, and D. S. Conklin. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 43.Rotondo, G., and D. Frendewey. 2001. Pac1 ribonuclease of Schizosaccharomyces pombe. Methods Enzymol. 342:168-193. [DOI] [PubMed] [Google Scholar]
- 44.Scherr, M., K. Battmer, T. Winkler, O. Heidenreich, A. Ganser, and M. Eder. 2003. Specific inhibition of bcr-abl gene expression by small interfering RNA. Blood 101:1566-1569. [DOI] [PubMed] [Google Scholar]
- 45.Silhavy, D., and J. Burgyán. 2004. Effects and side effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76-82. [DOI] [PubMed] [Google Scholar]
- 46.Söderlund, H., U. Pettersson, B. Vennström, L. Philipson, and M. B. Mathews. 1976. A new species of virus-coded low molecular weight RNA from cells infected with adenovirus type 2. Cell 7:585-593. [DOI] [PubMed] [Google Scholar]
- 47.Svensson, C., and G. Akusjärvi. 1984. Adenovirus VA RNAI: a positive regulator of mRNA translation. Mol. Cell. Biol. 4:736-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thimmappaya, B., C. Weinberger, R. J. Schneider, and T. Shenk. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543-551. [DOI] [PubMed] [Google Scholar]
- 49.Tuschl, T., P. D. Zamore, R. Lehmann, D. P. Bartel, and P. A. Sharp. 1999. Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13:3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ulfendahl, P. J., S. Linder, J. P. Kreivi, K. Nordqvist, C. Svensson, H. Hultberg, and G. Akusjärvi. 1987. A novel adenovirus-2 E1A mRNA encoding a protein with transcription activation properties. EMBO J. 6:2037-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westerhout, E. M., M. Ooms, M. Vink, A. T. Das, and B. Berkhout. 2005. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 33:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi, R., Y. Qin, I. G. Macara, and B. R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.