Abstract
The aim of this study was to elucidate protein-protein interactions between tegument proteins of herpes simplex virus type 1 (HSV-1). To do so, we have cloned and expressed in the LexA yeast (Saccharomyces cerevisiae) two-hybrid system, 13 of the 21 currently known tegument proteins of HSV-1. These included the tegument proteins essential for replication in cell lines, UL17, UL36, UL37, UL48, and UL49, and the nonessential tegument proteins US11, UL11, UL14, UL16, UL21, UL41, UL46, and UL47. A total of 104 combinations were screened in the yeast two-hybrid assay, with 9 interactions identified. These included: UL11-UL16, UL36-UL37, UL36-UL48, UL46-UL48, UL47-UL48, and UL48-UL49. The remaining interactions consisted of self-associations that were observed for US11, UL37, and UL49. The interactions UL36-UL37, UL36-UL48, UL37-UL37, UL46-UL48, and UL47-UL48 have not been previously reported for HSV-1. The interaction of UL46-UL48 was verified using an in vitro pull-down assay. The interactions of UL36-UL37 and UL37-UL37 were verified with a coimmunoprecipitation assay. Knowledge of HSV-1 tegument protein-protein interactions will provide insights into the pathways of tegument assembly, and the identified interactions are potential targets for new antiviral drugs.
The herpes simplex virion has four components: an electron-dense core containing the double-stranded DNA genome (152 kb), the capsid, the tegument, and an outer envelope containing glycoprotein (33). The icosadeltahedral capsid encloses the DNA core and consists of 162 capsomeres, 7 different viral proteins (VPs), VP5 (UL19), VP19C (UL38), VP21 (UL26), VP22a (UL26.5), VP23 (UL18), VP24 (UL26), and VP26 (UL35), and the products of the UL6 and UL25 genes (33, 44). VP5 is the major protein and, together with VP19C, VP23, and VP26, is present on the surface. The surrounding tegument contains about 20 proteins, including VP1/2 (UL36), VP11/12 (UL46), VP13/14 (UL47), VP16 (UL48), VP22 (UL49), ICP0, ICP4, and the virion host shutoff protein (UL41) plus the products of genes US2, US3, US10, US11, UL11, UL13, UL14, UL16, UL17, UL21, UL37, UL51, and UL56 (18, 28, 29). The envelope contains at least 8 of the 11 different glycoproteins: gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, and gM (33).
The structure of the capsid has been well defined by cryoelectron microscopy (cryo-EM) after assembly in vitro (43, 44). In addition, interactions between capsid proteins have been determined by yeast (Saccharomyces cerevisiae) two-hybrid analysis (1). In contrast, the structure of the herpes simplex virus type 1 (HSV-1) tegument is largely unknown. There are at least 20 proteins within the tegument. Clues to the structure of tegument have come from a number of studies. By cryo-EM, UL36 is present in the deepest layers of the tegument attached to the vertices of the capsid (42) and interacts with the capsid protein VP5 (27). Recently the use of cryoelectron tomography has revealed the tegument as frequently forming an asymmetric cap between the capsid and outer envelope (16). The identification of cytoplasmic HSV “light particles” (26), which consist of the authentic viral membrane with a full complement of embedded glycoproteins and subjacent tegument, indicates that most of the tegument and glycoprotein can self-assemble within the cytoplasm (37). Most of the tegument proteins are present in HSV-1 light particles, including UL36 (37) and UL37 (25). In contrast, UL36 and UL37 are not required for the assembly of pseudorabies virus (PRV; a closely related alphaherpesvirus) light particles (11). Evidence for UL48 in both the inner and outer tegument layers has come from detergent solubilization/salt extractions of purified virions (35, 40). Recently, UL48 has been shown to interact with the cytoplasmic tail of glycoprotein gH (15). In PRV, there is evidence for UL49 interacting with glycoproteins E/I and M (13) and for UL36 interacting with UL37 (20, 21), suggesting superficial and deep positions within the tegument, respectively. Studies with PRV mutants suggest that UL37, UL46, UL47, and UL48 are important structural components of the tegument (11, 12, 21, 23, 28, 29). Those tegument proteins that are present in high amounts (1,000 to 2,000 copies per virion), such as UL46, UL47, UL48, and UL49, are likely to play a structural role (17, 40). Proteins such as ICP0 and ICP4, which are relatively low in abundance (100 to 150 copies per virion), probably perform purely regulatory roles and are carried in the tegument rather than being structural (39). From previous individual reports, the following interactions between HSV tegument proteins have been identified: UL48 and UL49 (10), UL41 and UL48 (34, 36), ICP0 and ICP4 (39), and UL11 and UL16 (24).
In this study, we have defined a number of interactions among the tegument proteins of HSV-1 using a yeast two-hybrid approach. We have confirmed a number of the identified interactions by expression of individual tegument proteins in bacterial or mammalian cells.
MATERIALS AND METHODS
Expression constructs.
The cloning of HSV-1 tegument genes into the yeast two-hybrid vectors displayBait and displayTarget (Display Systems Biotech) has been previously described (9). Additional tegument genes cloned in the present study included UL11, UL14, UL16, UL21, and UL49. UL36 fragments corresponding to amino acids 124 to 511, 512 to 767, and 1 to 767 were amplified from displayBait/UL36(1-1874) (9) and inserted into EcoRI/XhoI-digested displayBait or EcoRI-digested displayTarget. For bacterial expression, the UL46 gene was excised from displayBait and inserted into EcoRI-digested pGEX-5X-1 (Amersham). Cloning of UL48 into pET28a (Novagen) to generate an expression plasmid which encodes untagged UL48 has been previously described (7). For expression in mammalian cells, the UL36 fragment corresponding to amino acids 512 to 767 and UL37 were inserted into EcoRI/XhoI- or EcoRI-digested pCMV-HA and/or pCMV-myc (Clontech) (both vectors were modified so that the EcoRI site was in the same reading frame as displayBait and displayTarget). Human kinesin KIF5B fragment 814 to 963 was obtained from a displayBait construct (5) and inserted into EcoRI/XhoI-digested pCMV-HA and pCMV-myc.
Yeast two-hybrid assay.
The use of the LexA-based yeast two-hybrid assay to determine protein-protein interactions has been previously described (5). The protocols for qualitative assessment of protein-protein interactions, quantification of each positive interaction using a β-galactosidase assay, and determination of protein expression in yeast were as described previously (5).
In vitro pull-down assay.
The glutathione S-transferase (GST)-tagged UL46 and untagged UL48 constructs were expressed and harvested as previously described (7). Conditions for binding, wash, and elution were as previously described (5). GST fusion proteins were immobilized on glutathione-Sepharose beads prior to incubation with either HSV-1-infected Hep-2 cell lysates, prepared as previously described (7), or bacterial lysates containing untagged UL48.
Coimmunoprecipitation assay.
HeLa cells were grown at 37°C (5% CO2) in Dulbecco's modified Eagle medium supplemented with 10% (vol/vol) fetal calf serum (JRH Bioscience), 100 U/ml penicillin, and 100 μg/ml streptomycin (Sigma). HeLa cells were grown to 80% confluence in six-well plates and were transiently transfected using GeneJuice transfection reagent (Novagen). Cells were harvested at 48 h posttransfection and lysed in M-PER buffer (Pierce) containing mammalian protease inhibitors (Sigma). Lysates were processed with the Profound mammalian c-myc tag coimmunoprecipitation kit (Pierce) according to the manufacturer's instructions. Protein complexes were eluted by heating in 2× nonreducing sample buffer (Pierce).
Analysis of protein complexes.
Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and, for analysis of expression in yeast or for in vitro pull-downs, were processed by immunoblotting as previously described (6). For coimmunoprecipitation experiments, immunoblots were washed with phosphate-buffered saline (PBS) before incubatoin for 1 h with Odyssey blocking buffer (Licor-Biosciences). Blots were then incubated for 1 h with primary antibody diluted in Odyssey blocking buffer containing 0.1% (vol/vol) Tween 20. Blots were then washed four times for 5 min with PBS containing 0.1% (vol/vol) Tween 20 before incubation for 1 h with secondary antibody diluted as described above. Blots were then washed as described above before being rinsed and stored protected from light in PBS. Detection was performed with an Odyssey infrared imaging system (Licor-Biosciences). The primary antibodies used included mouse monoclonal and rabbit polyclonal antibodies against hemagglutinin (HA), mouse monoclonal antibodies against LexA and UL48 (all from Santa Cruz Biotechnology), rabbit polyclonal antibody against HSV-1 (Dako), and mouse monoclonal against c-myc (Clontech). Secondary antibodies for the Odyssey system, both used at a 1:2,000 dilution, included goat anti-mouse Alexa Fluor 680-conjugated immunoglobulin G (IgG; Molecular Probes) and goat anti-rabbit IRdye800-conjugated IgG (Rockland, Inc.).
RESULTS
Yeast two-hybrid analysis of the interaction of HSV-1 tegument proteins.
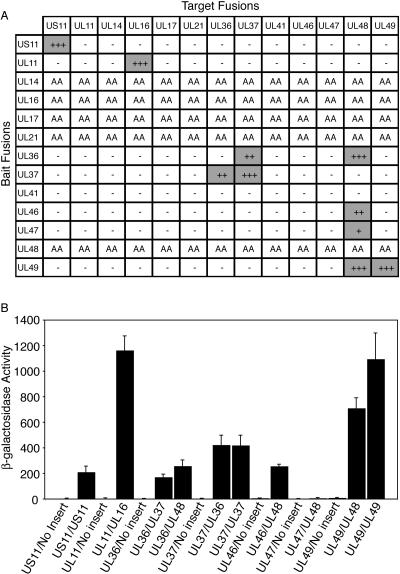
In this study, we have cloned and expressed, in the LexA yeast two-hybrid system, 13 of the 21 known tegument proteins of HSV-1 (29). Those cloned and expressed include US11, UL11, UL14, UL16, UL17, UL21, UL36, UL37, UL41, UL46, UL47, UL48, and UL49. All genes cloned were full-length constructs except UL36, which was cloned in the bait vector as a fragment corresponding to acids 1 to 1874 (9) and in the target vector as a fragment corresponding to amino acids 1 to 767. A matrix approach, testing 104 combinations in the yeast two-hybrid assay was then undertaken. Combinations involving bait constructs of UL14, UL16, UL17, or UL48 could not be assessed due to autoactivation observed when tested against target vector containing no insert (not shown). A qualitative plate-based assay was used to initially identify positive interactions. This assay identifies positive interactions through expression of the reporter genes LEU2 and lacZ. This yielded 9 positive interactions that included US11-US11, UL11-UL16, UL36-UL37, UL36-UL48, UL37-UL37, UL46-UL48, UL47-UL48, UL48-UL49, and UL49-UL49 (Fig. 1A). Quantitative β-galactosidase activity values were then obtained for each positive interaction using a liquid assay (Fig. 1B). Only in the case of UL47-UL48 was there a lack of β-galactosidase activity above the background (Fig. 1B). Whether this particular interaction is a true positive in the Y2H assay, therefore, requires further investigation. The expression of each construct used in the yeast two-hybrid assay was confirmed by immunoblotting (not shown) using antibodies against the LexA fusion tag (bait constructs) and the HA fusion tag (Target constructs).
FIG. 1.
Yeast two-hybrid analysis of the interaction of HSV-1 tegument proteins. (A) Summary of data obtained with yeast cotransformed with various combinations of bait and target fusion constructs. Interactions were initially assessed using a qualitative in vivo plate assay (at day 3), with readout being expression of the reporter genes LEU2 and lacZ. Positive interactions (shaded) were observed as blue colonies, with + indicating the lightest blue and +++ indicating the darkest blue. No growth (−) indicates a negative interaction. Autoactivation (AA) was observed for a number of bait constructs. (B) Summary of quantitative liquid β-galactosidase assay. The activity was calculated from the following equation: β-galactosidase activity = 1,000 × A420/(t × V × OD660), where t is time (in minutes) of incubation, V is volume of cells (in ml) used in the assay, and OD660 is the optical density at 660 nm. The values obtained for β-galactosidase activity are the averages of measurements from at least three separate colonies. Negative controls consisted of displayBait/viral gene insert and displayTarget/no insert. UL36 in displayBait encodes amino acids 1 to 1874, while UL36 in displayTarget encodes amino acids 1 to 767.
Of the 9 identified interactions, only that of UL36-UL37 could be tested and confirmed in both orientations in the yeast two-hybrid assay. The other interactions were either self-associations or involved one partner which autoactivates when inserted in the bait vector, e.g., UL16 and UL48 (Fig. 1A). Four of the interactions have been previously identified in HSV-1 using other assays. The self-association of HSV-1 tegument protein US11 has previously been demonstrated (4). Interaction of HSV-1 tegument protein UL11 with UL16 using an in vitro pull-down assay has recently been reported (24). The self-association of UL49 and interaction with UL48 has also been previously reported (10).
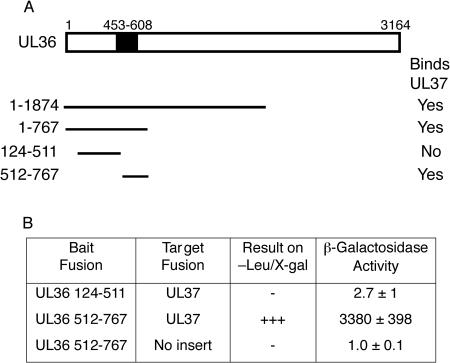
The remaining 5 interactions, UL36-UL37, UL36-UL48, UL37-UL37, UL46-UL48, and UL47-UL48, have not been documented previously for HSV-1. In the case of UL36-UL37, the interaction has been reported using homologous PRV tegument proteins and the LexA yeast two-hybrid assay (20). It should be noted that both HSV-1 UL36 fragments 1 to 1874 and 1 to 767 used in this study contain the region (amino acids 453 to 608) which is homologous to the UL37-interacting region identified in PRV UL36 (20). The additional UL36 fragments 124 to 511 and 512 to 767 were also tested for UL37 binding (Fig. 2). The fact that UL36 fragment 512 to 767 is strongly positive for UL37 binding implies that the minimal UL37-binding site in HSV-1 UL36 corresponds to this region (Fig. 2).
FIG. 2.
Yeast two-hybrid analysis of the interaction of UL36 and UL37. (A) Summary of full-length UL37 binding to fragments of UL36. The region 453 to 608, which is homologous to the previously determined UL37-binding site in PRV, is indicated (20). (B) Summary of quantitative liquid β-galactosidase assay for UL36 fragments 124 to 511 and 512 to 767. The activity was calculated from the following equation: β-galactosidase activity = 1,000 × A420/(t × V × OD660), where t is time (in minutes) of incubation, V is the volume of cells (in ml) used in the assay, and OD660 is the optical density at 660 nm. The values obtained for β-galactosidase activity are the averages of measurements from at least three separate colonies. X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; −, no growth or color change at day 3; +++, darkest blue colonies at day 3.
The interaction of tegument proteins UL46 and UL48 has been suggested from colocalization and copurification studies with HSV-2 (19). Modulation of HSV-1 UL48 transcriptional activity by both UL46 and UL47 supports a direct interaction (40). No report to date has documented the self-association of UL37 in any alphaherpesviruses.
The known interaction of UL41 with UL48, which was previously demonstrated using the GAL4 yeast two-hybrid assay (36), could not be reproduced in the LexA yeast two-hybrid assay (Fig. 1A). One of the reasons for this may be the difference between the two yeast systems with respect to the DNA-binding and activation domain fusion partners, each of which would have differing effects on the correct folding of the respective fused viral protein. Reversing the LexA bait and target fusion partners was not possible, since UL48 strongly autoactivates in bait. A recently reported interaction between UL16 and UL21 in PRV (22) could not be confirmed for HSV-1, as both autoactivate in the yeast two-hybrid assay (Fig. 1A).
Nevertheless, we have been able to reproduce four previously observed interactions for HSV-1 tegument proteins. This strongly supports the chosen LexA yeast two-hybrid assay as a valid system for determining binary interactions between HSV-1 structural proteins. Five new interactions were also identified with this screen.
In vitro pull-down assay.
To further validate the yeast two-hybrid approach, lysates from HSV-infected mammalian cells were added to GST-UL46. There was evidence not only of the expected pairwise interaction (UL46-UL48) but also of apparent secondary interactions with other viral proteins (not shown). Whether the two viral proteins are interacting directly or through an intermediate viral protein was unclear. Therefore, an approach based on expression of individual viral proteins was subsequently used to confirm interactions.
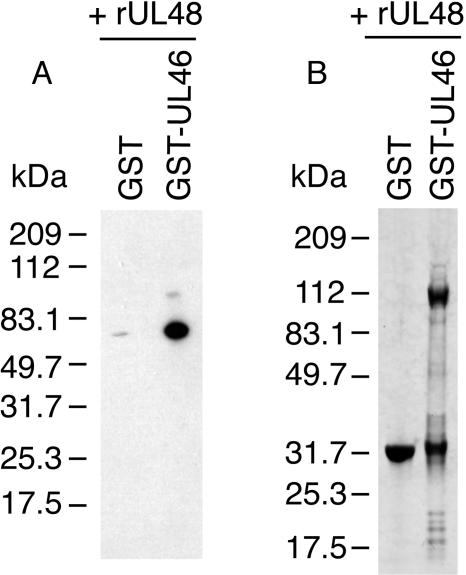
The same in vitro pull-down assay using bacterially expressed viral proteins was employed. The interaction between UL46 and UL48 was confirmed using this assay. Recombinant untagged UL48, in the absence of other viral proteins, bound only to GST-UL46 and not to GST (Fig. 3A). The presence of GST fusion proteins was also confirmed (Fig. 3B). Attempts to verify some of the other interactions with this assay were unsuccessful due to poor expression and/or low solubility of viral proteins such as UL36 and UL37 (not shown).
FIG. 3.
In vitro pull-down assay to assess the interaction of UL46 and UL48. Bacterial lysates containing recombinant untagged UL48 were added to GST or GST-UL46 on glutathione-Sepharose beads. Protein complexes were subsequently eluted and separated by SDS-PAGE (4 to 20%). (A) Immunoblotting with anti-UL48 confirms the presence of recombinant untagged UL48 coeluting preferentially with GST-UL46 in the absence of other viral proteins. (B) Coomassie blue staining to confirm the presence of GST proteins eluted from glutathione-Sepharose beads. The predicted masses of various protein products are as follows: GST, 26 kDa; GST-UL46, 104 kDa; untagged UL48, 54 kDa.
Coimmunoprecipitation assay.
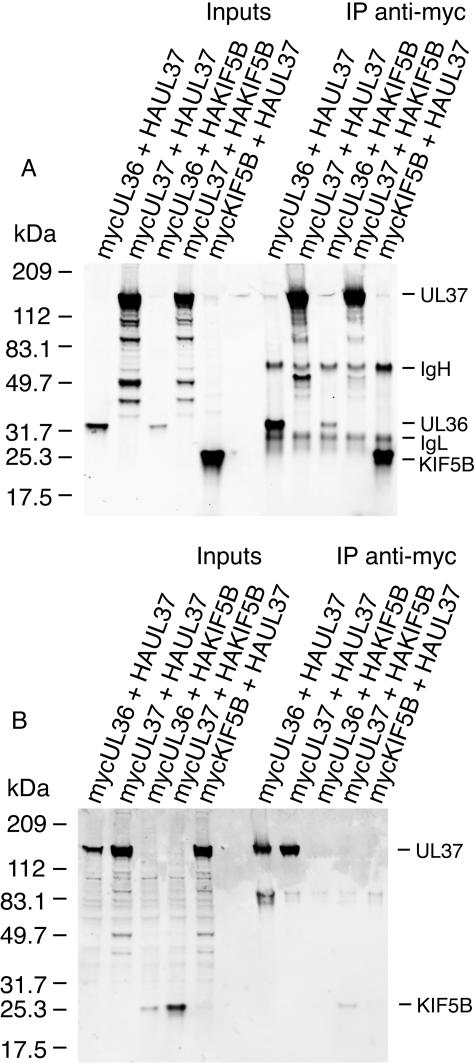
An alternative coimmunoprecipitation assay was subsequently employed to validate the yeast two-hybrid interactions. Two of the five new HSV-1 interactions (UL36-UL37 and UL37-UL37) identified from the yeast two-hybrid screen were chosen. A fragment of UL36 corresponding to amino acids 512 to 767 was expressed with a myc tag and found to interact with full-length HA-tagged UL37 (Fig. 4). UL36 fragment 512 to 767 was previously confirmed to interact with UL37 in the yeast two-hybrid assay (Fig. 2). Full-length myc-tagged UL37 was found to interact with full-length HA-tagged UL37 (Fig. 4). No significant binding of UL36 or UL37 was observed with either myc- or HA-tagged kinesin KIF5B, confirming the specificity of the coimmunoprecipitation assay (Fig. 4).
FIG. 4.
Coimmunoprecipitation assay. HeLa cells were cotransfected with expression plasmids encoding myc-tagged and HA-tagged proteins. Cells lysates were harvested at 48 h posttransfection and immunoprecipitated (IP) with mouse anti-myc monoclonal antibody. Protein complexes were separated by SDS-PAGE (4 to 20%). (A) Immunoblotting with mouse anti-myc monoclonal antibody; (B) immunoblotting with rabbit anti-HA polyclonal antibody. The positions of UL36, UL37, and KIF5B along with coeluting mouse anti-myc IgG heavy and light chains detected by the secondary anti-mouse conjugate used during immunoblotting are indicated. The predicted masses of various protein products are as follows: UL36 512 to 767, 30 kDa; UL37, 120 kDa; KIF5B 814 to 963, 19 kDa.
Therefore, 7 of the 9 novel interactions identified by the yeast two-hybrid screen have been confirmed by other assays, 3 in this study and 4 previously.
DISCUSSION
It has become widely accepted that the majority of the tegument is added to the capsid of HSV-1 in the cytoplasm (28-30). Capsids are first assembled in the nucleus before they traverse the inner and outer nuclear membrane in a primary envelopment and de-envelopment step. In nonneuronal cells, assembly is completed by secondary envelopment at the trans-Golgi compartment. In this assembly process, inner tegument proteins are added first to the capsid. Outer tegument proteins preassemble and interact with the cytoplasmic tails of viral glycoproteins at the Golgi. During secondary envelopment, inner tegument-coated capsid then links with outer tegument-glycoprotein to form the final viral particle, which is released by exocytosis (28, 29).
In this study, we have identified a number of binary interactions involving HSV-1 tegument proteins (Fig. 1). Whether these interactions are structural, regulatory, or other requires further elucidation. However, from the work of others and the present model for viral assembly, a number of the observed interactions are likely to be important for assembly. First, the interaction of UL36 and UL37 appears to have a definite structural role. The absence of either protein in HSV-1 or PRV significantly impairs or blocks further addition of tegument and subsequent viral maturation (2, 3, 20, 21). This fits with their apparent location in the virion, i.e., inner tegument adjacent to the capsid. UL36 has been shown to interact with the major capsid protein VP5 (product of UL19 gene) (27) and, by cryo-EM, appears to form the inner tegument layer (42). UL37 then interacts with VP5-bound UL36 to complete the inner tegument layer (20). In the case of PRV, a recent study shows that deletion of the UL37 binding site in UL36 does not abolish secondary envelopment (14). This suggests that the interaction, though important, is not essential for virion assembly. Whether the same applies to HSV-1 requires further investigation. Certainly this supports the existence of other links between the inner and outer tegument. One of these links may be provided by the interaction of UL36 and UL48 observed in this study. In support of this, previous studies with HSV-1 and PRV lacking UL48 suggest that it is important for linking capsids coated with UL36 and UL37 to the outer tegument and envelope (11, 31).
Whether the observed interactions among the outer tegument proteins (UL46, UL47, UL48, and UL49) have a structural role in HSV-1 assembly also requires further investigation. Certainly, deletion of either UL46 or UL47 in HSV-1 and PRV does not block viral assembly (23, 41), although deletion of UL47 does impair secondary envelopment in PRV (23). In fact, deletion of all the major tegument proteins, UL46, UL47, UL48, and UL49, in PRV (12) can be tolerated, supporting redundancy in tegument assembly. In the case of PRV, deletion of either UL46, UL47, or UL48 does not prevent incorporation of the remaining two proteins into the tegument, although deletion of UL47 may reduce the incorporation of UL48 into the tegument (11, 23). In HSV-1, deletion of UL47 leads to increased levels of incorporation of UL46 into virions, which also supports structural redundancy (40). Deletion of UL49 in HSV-1 also does not have a significant effect on viral assembly (32). Previous studies of HSV-1 have suggested that both UL46 and UL47 have a role in enhancing UL48-induced immediate-early viral gene expression (19, 40, 41). Nuclear localization has been demonstrated for UL47 (8, 30), supporting a role in regulation of UL48 activity. Using green fluorescent protein fusion proteins, UL46 was only found in the cytoplasm (38), arguing against a role for regulation of UL48 activity in the whole cell. However, lack of nuclear localization needs to be confirmed by other techniques.
A number of interactions have been demonstrated between tegument proteins and either membrane-associated tegument protein(s) or the cytoplasmic tails of viral glycoproteins. These include the interaction of UL16 with the membrane associated-tegument protein UL11 (24), which was confirmed in this study, UL49 with gE and gM (13), and UL48 with gH (15). Like the interactions between outer tegument proteins, there is some redundancy but each appears to play a role in secondary envelopment at the Golgi (28, 29).
A number of self-associations were also identified in this study. These involved the tegument proteins US11, UL37, and UL49 (Fig. 1). Whether the self-associations are simply dimers or higher-order oligomers requires further investigation. In the case of US11, higher-order oligomers have been previously demonstrated and may be important for the nonstructural functions of US11 (4). Recent work indicates that the tegument contains thin filaments, and it is possible that they are formed from the polymerization of tegument proteins (16).
In conclusion, we have identified a number of novel interactions among the tegument proteins of HSV-1. A number involve the essential tegument proteins UL36, UL37, and UL48. Further elucidation of the role of these interactions in viral assembly could form the basis for novel antiviral targets. The approach undertaken in this study now provides a significant platform for further biochemical and biological studies on viral assembly. Future studies will involve construction of viral mutants with appropriate mutations and/or deletions within individual tegument genes to study resulting effects on assembly.
Acknowledgments
This work was supported by grants 107374 and 253617 from the Australian National Health and Medical Research Council (to A.L.C. and R.J.D) and an initiating grant from the Westmead Millennium Foundation (to V.V.).
REFERENCES
- 1.Desai, P., and S. Person. 1996. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology 220:516-521. [DOI] [PubMed] [Google Scholar]
- 2.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz, J. J., D. Simonin, T. Masse, et al. 1993. The herpes simplex virus type 1 US11 gene product is a phosphorylated protein found to be non-specifically associated with both ribosomal subunits. J. Gen. Virol. 74:397-406. [DOI] [PubMed] [Google Scholar]
- 5.Diefenbach, R. J., E. Diefenbach, M. W. Douglas, and A. L. Cunningham. 2002. The heavy chain of conventional kinesin interacts with the SNARE proteins SNAP25 and SNAP23. Biochemistry 41:14906-14915. [DOI] [PubMed] [Google Scholar]
- 6.Diefenbach, R. J., J. P. Mackay, P. J. Armati, and A. L. Cunningham. 1998. The C-terminal region of the stalk domain of ubiquitous human kinesin heavy chain contains the binding site for kinesin light chain. Biochemistry 37:16663-16670. [DOI] [PubMed] [Google Scholar]
- 7.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, et al. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly, M., and G. Elliott. 2001. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas, M. W., R. J. Diefenbach, F. L. Homa, et al. 2004. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 279:28522-28530. [DOI] [PubMed] [Google Scholar]
- 10.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, et al. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1-12. [DOI] [PubMed] [Google Scholar]
- 16.Grunewald, K., P. Desai, D. C. Winkler, et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 17.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 19.Kato, K., T. Daikoku, F. Goshima, et al. 2000. Synthesis, subcellular localization and VP16 interaction of the herpes simplex virus type 2 UL46 gene product. Arch. Virol. 145:2149-2162. [DOI] [PubMed] [Google Scholar]
- 20.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., S. Bottcher, H. Granzow, M. Kopp, and T. C. Mettenleiter. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLauchlan, J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J. Gen. Virol. 78:189-194. [DOI] [PubMed] [Google Scholar]
- 26.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 27.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 28.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 30.Miranda-Saksena, M., R. A. Boadle, P. Armati, and A. L. Cunningham. 2002. In rat dorsal root ganglion neurons, herpes simplex virus type 1 tegument forms in the cytoplasm of the cell body. J. Virol. 76:9934-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 34.Schmelter, J., J. Knez, J. R. Smiley, and J. P. Capone. 1996. Identification and characterization of a small modular domain in the herpes simplex virus host shutoff protein sufficient for interaction with VP16. J. Virol. 70:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz, J. B., A. G. Albright, P. R. Kinchington, and F. J. Jenkins. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055-1065. [DOI] [PubMed] [Google Scholar]
- 36.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szilagyi, J. F., and C. Cunningham. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661-668. [DOI] [PubMed] [Google Scholar]
- 38.Willard, M. 2002. Rapid directional translocations in virus replication. J. Virol. 76:5220-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 68:8158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, Z. H., M. Dougherty, J. Jakana, et al. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Z. H., S. J. Macnab, J. Jakana, et al. 1998. Identification of the sites of interaction between the scaffold and outer shell in herpes simplex virus-1 capsids by difference electron imaging. Proc. Natl. Acad. Sci. USA 95:2778-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]