Abstract
Deletion of the entire AZFc locus on the human Y chromosome leads to male infertility. The functional roles of the individual gene families mapped to AZFc are, however, still poorly understood, since the analysis of the region is complicated by its repeated structure. We have therefore used single-nucleotide variants (SNVs) across ∼3 Mb of the AZFc sequence to identify 17 AZFc haplotypes and have examined them for deletion of individual AZFc gene copies. We found five individuals who lacked SNVs from a large segment of DNA containing the DAZ3/DAZ4 and BPY2.2/BPY2.3 gene doublets in distal AZFc. Southern blot analyses showed that the lack of these SNVs was due to deletion of the underlying DNA segment. Typing 118 binary Y markers showed that all five individuals belonged to Y haplogroup N, and 15 of 15 independently ascertained men in haplogroup N carried a similar deletion. Haplogroup N is known to be common and widespread in Europe and Asia, and there is no indication of reduced fertility in men with this Y chromosome. We therefore conclude that a common variant of the human Y chromosome lacks the DAZ3/DAZ4 and BPY2.2/BPY2.3 doublets in distal AZFc and thus that these genes cannot be required for male fertility; the gene content of the AZFc locus is likely to be genetically redundant. Furthermore, the observed deletions cannot be derived from the GenBank reference sequence by a single recombination event; an origin by homologous recombination from such a sequence organization must be preceded by an inversion event. These data confirm the expectation that the human Y chromosome sequence and gene complement may differ substantially between individuals and more variations are to be expected in different Y chromosomal haplogroups.
AZFc deletions in distal Yq11 are the most frequent known genetic cause of human male infertility, leading to azoospermia or severe oligozoospermia (MIM 400024, MIM 415000). They occur in different populations with a frequency of 10%–20% of infertile men (Vogt 1998; Krausz and McElreavey 1999; Krausz et al. 2000; Simoni 2001). AZFc contains eight gene families expressed only in testis tissue, three of which code for proteins, but the importance of each individual gene for fertility is not understood. The underlying AZFc sequence is composed mainly of large repeated-sequence blocks called “amplicons” (Kuroda-Kawaguchi et al. 2001) organized into palindromic structures showing high (>99.9%) sequence identity between the arms (fig. 1). Such structures may undergo frequent inversion and gene-conversion events (Skaletsky et al. 2003), as well as duplications and deletions (Repping et al. 2003). We therefore set out to investigate the structure and gene content of this region in normal individuals, using SNVs (single-nucleotide variants) and STSs extending along 3 Mb of the AZFc sequence, supplemented by Southern blot analyses.
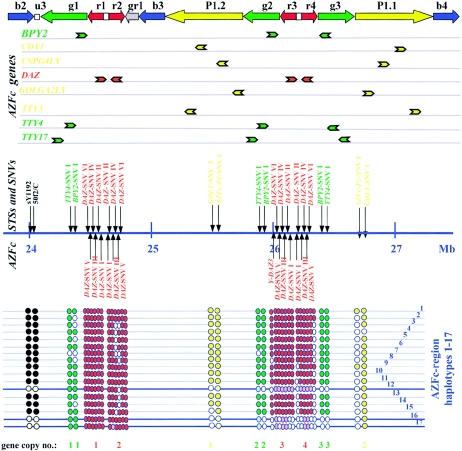
Figure 1.
Schematic view of the AZFc locus, showing its gene content and the locations of the STSs and SNVs used in this study. The AZFc amplicon structure is drawn according to the color code of Kuroda-Kawaguchi et al. (2001). AZFc genes are listed below the amplicon structure, with their 5′→3′ polarities indicated by the directions of the arrows. The STSs and SNVs used in this study are shown below the gene map. Their GenBank accession numbers and molecular data are listed in the work of Fernandes et al. (2002) and in table 1, respectively. All AZFc markers and genes were mapped onto the Ensembl Y-chromosome-sequence between 23.79 and 27.49 Mb. With the complete b2 and b4 amplicons included, a length of ∼3.7 Mb for the AZFc sequence was estimated. The presence or absence of an STS or SNV in the genomic DNA samples analyzed could be grouped into 17 patterns (AZFc deletion haplotypes 1–17). These are symbolized below the STS/SNV map by rows of filled or empty circles, with the gene copies numbered at the bottom of the figure. Thick lines represent deletion haplotypes belonging to haplogroup N. The pale colors of some circles indicates that these multilocus SNVs were inferred to be absent because all unambiguously typed flanking and intervening markers were absent.
The assays for analyses of specific variants within the DAZ genes (MIM 400003, MIM 400026, MIM 400027) were described recently (Fernandes et al. 2002); three STSs (DAZ-RRM3 and sY152 in DAZ1 and DAZ4; Y-DAZ3 in DAZ3) and six DAZ SNVs (I–VI). Typing the SNVs involves coamplification of two or more similar loci, followed by restriction enzyme digestion to reveal the absence (A allele) or presence (B allele) of a given restriction enzyme site. Since the locations of the A and B-SNV alleles are known from the sequence, loss of the A and/or B pattern indicates a loss of specific DAZ gene copies. Some markers outside the DAZ gene locus were already available: 50f2/C or DYS7/C (GenBank accession number Y07728) and sY1192 (GenBank accession number G67166), both of which lie in the proximally located u3 block, and AZFc-P1, an SNV marker that distinguishes the yellow P1.1 amplicon from the P1.2 amplicon (table 1). In addition, we used the AZFc sequence data published in GenBank to look for SNVs in the seven other gene families in this region: BPY2 (basic protein Y2 [MIM 400013]), CDY1 (chromo-domain Y [MIM 400016]), CSPG4LY (chondrotin-sulfate proteoglycan-4-like Y), GOLGA2LY (Golgi-antigene 2–like Y), TTY3, TTY4, and TTY17 (testis-transcript Y 3, 4, and 17, respectively). We found no suitable variants in the CDY1, CSPG4LY, TTY3, and TTY17 genes located in the yellow and green amplicons P1.1/P1.2 and g1–3, respectively. However, SNVs were found, for the genes BPY2 and TTY4, located in the green amplicons (g1–3) and, for GOLGA2LY, located in the yellow (P1.1/ P1.2) amplicons, and assays were established (table 1). These markers, together with the DAZ-STS/SNVs, provided assays for 37 positions in eight loci spread over ∼3,000 kb in the AZFc region (fig. 1).
Table 1.
AZFc-SNVs and STSs
| SNV or STS | AZFc Site | GenBank Accession Number | SNV Restriction Enzyme | AZFc Amplicon | Ensembl Y DNA Sequence Position(bp) | SNVAllele | RestrictedFragment(bp) |
| SNV: | |||||||
| P1.1 | 26.709.824–26.710.291 | A | 467 | ||||
| AZFc-P1/I | P1.1/P1.2 | G73351 | DdeI | P1.2 | 25.556.438–25.556.845 | B | 284+183 |
| P1.1 | 26.763.718–26.764.249 | A | 531 | ||||
| GOLY/I | GOLGA2LY genes (two copies) | BV012731 | HhaI | P1.2 | 25.501.889–25.502.420 | B | 289+242 |
| g2 | 25.930.869–25.931.339 | A | 470 | ||||
| g3 | 26.335.746–26.336.216 | A | |||||
| BPY2/I | BPY2 genes (three copies) | BV012732 | EcoRV | g1 | 24.388.898–24.389.368 | B | 289+181 |
| g3 | 26.368.272–26.368.812 | A | 541 | ||||
| g1 | 24.356.372–24.356.912 | B | 323+218 | ||||
| TTY4/I |
TTY4 genes (three copies) |
BV012733 |
HaeIII |
g2 |
25.898.343–25.898.883 |
B |
|
| AZFc Site |
GenBankAccessionNumber |
BAC Clonein GenBank |
BACSequencePosition |
Ensembl Y DNA Sequence Position(bp) |
AnnealingTemperature(°C) |
PCR Product(bp) |
|
| STS: | |||||||
| sY1192 | u3 | G67166 | BAC5C5; AC010080 | 52109–52362 | 24.019.703–24.020.304 | 64 | 255 |
| 50f2/C | u3 | Y07728 | BAC5C5; AC010080 | 65711–66830 | 24.033.305–24.034.424 | 65 | 1119 |
With this marker set, we analyzed the AZFc region in 31 individuals (In1-In31). Sixteen different “AZFc haplotypes” were present, 14 of which showed loss of variants in our set (table 2). In most cases, one or a small number of variants were missing, and the missing loci were not located adjacent to one another on the physical map (fig. 1). These were interpreted as likely to have arisen in multiple ways such as mutations in the restriction enzyme or primer binding sites, gene conversion events, or small deletions, but do not seem to involve the loss of large segments of DNA. In contrast, two AZFc region haplotypes (12 and 16), containing a total of five individuals (16%), stood out. These haplotypes lacked the two STSs from u3, as well as all SNVs from a large contiguous region in distal AZFc extending from P1.2 and g2 through DAZ3 and DAZ4 to g3 (fig. 1), suggesting loss of a large AZFc segment in distal Yq11.
Table 2.
Summary of AZFc-STS and SNV Analyses and Y Haplogroup Typing in 46 Genomic DNA Samples[Note]
| DNASample | DAZ-SNV I | DAZ-SNV II | DAZ-SNV III | DAZ-SNV IV | DAZ-SNV V | DAZ-SNV VI | Y-DAZ3 | 50f2/C | sY1192 | TTY4-SNV I | BPY2-SNV I | GOLY-SNV I | AZFc-P1-SNV I | AZFc del ht | Y Haplogroup | YCC Code |
| In1 | A+B | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | A+B | 1 | 104 | R1* |
| In2 | A+B | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A | A+B | B | 2 | 108 | R1a1* |
| In3 | A+B | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | A+B | 1 | 104 | R1* |
| In4 | A+B | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | A+B | 1 | 104 | R1* |
| In5 | A+B | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A | A+B | B | 2 | 108 | R1a1* |
| In6 | A+B | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | A+B | 1 | 104 | R1* |
| In7 | A+B | A+B | B | B | A+B | A | + | + | + | A+B | A | A+B | B | 3 | 52 | I* |
| In8 | A+B | A+B | B | B | A+B | A | + | + | + | A+B | A | A+B | B | 3 | 52 | I* |
| In9 | A+B | A+B | B | B | A+B | A | + | + | + | A+B | A | A+B | B | 3 | 52 | I* |
| In10 | A+B | A+B | B | A+B | A+B | A | + | + | + | A+B | A+B | B | B | 4 | 58 | J2* |
| In11 | A+B | A+B | B | A+B | A+B | A | + | + | + | A+B | A | A+B | B | 5 | 35 | E3b1* |
| In12 | A+B | A+B | B | A+B | A+B | A | + | + | + | A+B | A+B | A+B | B | 6 | 56 | J2f* |
| In13 | A+B | A+B | B | B | A+B | A | + | + | + | A | A | A+B | B | 7 | 58 | J2* |
| In14 | A+B | A+B | B | A+B | A+B | A | + | + | + | A+B | A | A+B | B | 5 | 32 | E3b3a* |
| In15 | A+B | A+B | A+B | B | A+B | A | − | + | + | A+B | A | A+B | B | 8 | 52 | I* |
| In16 | A+B | A+B | A+B | B | A+B | A | − | + | + | A+B | A | A+B | B | 8 | 35 | E3b1* |
| In17 | A+B | A+B | A+B | B | A+B | A | − | + | + | A+B | A | A+B | B | 8 | 32 | E3b3a* |
| In18 | A+B | A+B | A+B | B | A+B | A | − | + | + | A+B | A | A+B | B | 8 | 35 | E3b1* |
| In19 | A+B | A+B | A+B | B | A+B | A | − | + | + | A+B | A | A+B | B | 8 | 35 | E3b1* |
| In20 | A | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | A+B | 9 | 104 | R1* |
| In21 | A | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | A+B | 9 | 104 | R1* |
| In22 | A+B | A+B | A+B | A+B | A+B | A | + | + | + | A+B | A+B | A+B | B | 10 | 115 | Q3* |
| In23 | A | A+B | A+B | A+B | A+B | A+B | + | + | + | A+B | A+B | A+B | B | 11 | 108 | R1a1* |
| In24 | A | A+B | A+B | A+B | A+B | A | − | − | − | B | B | A | B | 12 | 92 | N3* |
| In25 | A | A+B | A+B | A+B | A+B | A | − | − | − | B | B | A | B | 12 | 92 | N3* |
| In26 | A | A+B | A+B | B | A+B | A | + | + | + | A+B | A+B | A+B | B | 13 | 76 | O3* |
| In27 | A | A+B | A+B | B | A+B | A | + | + | + | A+B | A | A+B | B | 14 | 52 | I* |
| In28 | A | A+B | B | A+B | A+B | A | + | + | + | A+B | A+B | A | B | 15 | 58 | J2* |
| In29 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| In30 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| In31 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| Only Y haplogroup N: | ||||||||||||||||
| In32 | A | A+B | A+B | A+B | B | A | − | − | + | B | B | A | B | 16 | 87 | N*(xN3) |
| In33 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In34 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In35 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In36 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In37 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| In38 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| In39 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| In40 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 92 | N3* |
| In41 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In42 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In43 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In44 | A | A+B | A+B | A+B | B | A | − | − | − | B | B | A | B | 16 | 87 | N*(xN3) |
| In45 | A | A | A+B | A+B | B | A | − | − | − | B | B | A | B | 17 | 92 | N3* |
| In46 | A | A | A+B | A+B | B | A | − | − | − | B | B | A | B | 17 | 92 | N3* |
Note.— Data for individuals who had a DAZ3/DAZ4 deletion are shown in boldface italics.
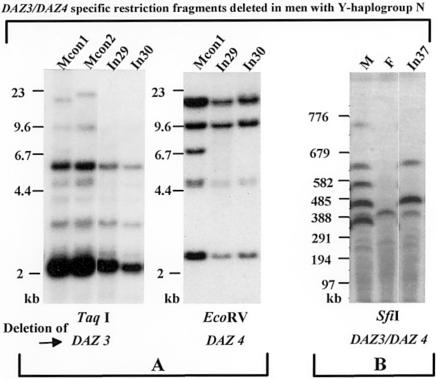
Local gene conversions or other changes involving small numbers of nucleotides in the DAZ locus can be distinguished from large DAZ deletions by Southern blot experiments using probes of DYS1-DNA from the DAZ gene (Fernandes et al. 2002). Despite the similarity of the DAZ gene sequences, EcoRV or TaqI digests produce patterns in which fragments specific for the DAZ gene copies can be distinguished. Similar analyses on a larger scale can be also performed after genomic SfiI restriction digestion. Scoring the DAZ-BAC-contig in GenBank for rare-cutter restriction sites identified genomic SfiI fragments of ∼450 kb and ∼350 kb, carrying the DAZ1/DAZ2 gene doublet and DAZ3/DAZ4 gene doublet, respectively (Floridia et al. 2000). We therefore performed such experiments to investigate whether men lacking the SNVs carry a deletion, and the results are shown in figure 2. A deletion of the DAZ3 and DAZ4 genes of In29 and In30 is indicated by absence of the DAZ3-specific 19.6-kb TaqI fragment and of the DAZ4-specific 7.3-kb EcoRV fragment (fig. 2A). Deletion of the DYS1-SfiI fragment specific for the DAZ3/DAZ4 gene doublet (∼350 kb) is seen in figure 2B for sample In37. Southern blot hybridization therefore shows that substantial DNA segments are missing, and we conclude that the absence of the P1.2-g2-DAZ3-DAZ4-g3 markers in the Y chromosome of these individuals is due to a large DNA deletion in this part of the AZFc region. We therefore refer to these chromosomes as carrying a DAZ3/DAZ4 deletion.
Figure 2.
A, EcoRV and TaqI DAZ deletion analyses. According to Fernandes et al. (2002), the DAZ3/DAZ4 deletion in the genomic DNA samples of In29 and In30 is shown by the absence of the DAZ3-specific 19.6-kb TaqI fragment and the DAZ4-specific 7.3-kb EcoRV fragment. Some genomic DYS1-TaqI fragments (9.6 kb, 5 kb, 4 kb) that are only visible in the male controls are common in all DAZ genes but are reduced in intensity in the In39 and In30 samples because of the DAZ3/DAZ4 deletion. The length of the 19.6-kb TaqI fragment in the DAZ3 gene is known to be polymorphic, as is shown here for the male control samples Mcon1 (with a 19.6-kb TaqI fragment) and Mcon2 (with a 24-kb TaqI fragment) in the DAZ3 gene. B, SfiI DAZ-blot deletion pattern of the genomic DNA sample of In37. The SfiI site at the proximal end of each fragment is partially resistant to cleavage (Floridia et al. 2000), so pulsed-field gel electrophoresis produces a pattern in which hybridization is also seen to partially cleaved products containing the adjacent ∼196-kb fragment: ∼649 kb and ∼543 kb, respectively. Both SfiI fragments specific for the DAZ3/DAZ4 gene doublet (∼350 and ∼550 kb) are deleted in In37, confirming the complete loss of these DAZ genes. The genomic male (M) and female (F) control samples display the normal SfiI fragments associated with the two DAZ gene doublets DAZ1/DAZ2 and DAZ3/DAZ4 (M control) and with the autosomal DAZL1 gene on chromosome 3 (∼380 kb SfiI fragment) in M and F control samples because of cross-hybridization. Molecular lengths of the λ-HindIII (A) and λ-multimer (B) size markers are given on the left.
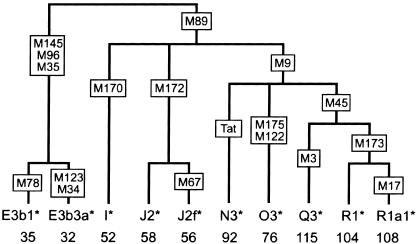
In parallel, the same DNA samples were typed with 118 Y chromosomal binary markers to establish their haplogroup (Paracchini et al. 2002). Results are given according to the Underhill haplogroup numbering (h1-h116) (Underhill et al. 2000) and the current haplogroup nomenclature of the Y Chromosome Consortium (Y Chromosome Consortium 2002) (table 2; fig. 3). Only 10 of the 116 haplogroups found worldwide were present in our samples, as would be expected from their predominant origin from Europe. Of the 10 haplogroups, 8 are common in Europe; the other 2 (O3* and Q3*), found in one individual each, are common in the countries of origin of these individuals (China and Costa Rica, respectively). There were striking (although not perfect) correspondences between the AZFc haplotypes and the Y-SNP phylogeny (table 2), and some of the AZFc variants could be placed on the Y phylogeny as unique mutations, whereas others were recurrent. The DAZ3/DAZ4 deletion chromosomes all belong to haplogroup N3* and thus suggest a single origin for this AZFc deletion.
Figure 3.
Phylogeny of the Y chromosomes analyzed in this work. Selected Y-SNPs are shown in boxes, and the haplogroup names according to the YCC (2002) or Underhill et al. (2000) are shown at the bottom. Only the haplogroups present in the samples analyzed are included. The phylogeny represents the branching order, but not timescale. In accordance with the YCC convention, haplogroups that may not be monophyletic (paragroups) are labeled with an asterisk (*).
To further investigate the relationship between this deletion polymorphism and the phylogeny, we carried out the reciprocal study, testing 15 individuals (In32–In46), who had initially been ascertained as belonging to the N lineage, for their AZFc deletion haplotype (table 2). There was some variation in DAZ-SNV II, leading to an additional AZF-region haplotype (17 in fig. 1), but, most strikingly, all but one of the individuals in the reciprocal study had lost both u3 and the same P1.2-g2-DAZ3-DAZ4-g3 markers lacking from the five haplogroup N individuals in the first sample set. The amplification of sY1192 in the one exceptional individual is most likely due to cross reaction with the related Yp sequence block (Skaletsky et al. 2003). We therefore conclude that a common Y lineage, haplogroup N, which lacks numerous AZFc markers, carries a deletion of two segments of the GenBank AZFc reference sequence.
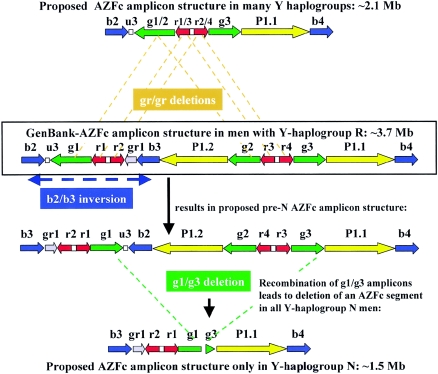
Intrachromosomal deletions commonly arise by homologous recombination between repetitive sequence blocks lying in the same orientation (Chen et al. 1997). For example, complete AZFc deletions were accounted for by intrachromosomal recombination between the b2 and b4 amplicons (Kuroda-Kawaguchi et al. 2001), and, similarly, deletions of the DAZ1/DAZ2 gene doublet were accounted for by intrachromosomal recombination between the g1 and g2 amplicons (Fernandes et al. 2002). We therefore investigated whether the lineage N DAZ3/DAZ4 deletions could be explained in a similar way. There were, however, no obvious candidates for directly repeated amplicons flanking the two deleted regions of the N lineage Y chromosome (fig. 1). If the deletion substrate in the pre-N-lineage Y chromosome resembled the GenBank sequence, two independent deletions must have occurred, neither based on long regions of homology but one removing u3 and the other the P1.2-g2-DAZ3-DAZ4-g3 region. The deletion, however, was an ancient event (see below) and did not occur in a modern sequence. The deletion substrate could therefore differ from the GenBank AZFc sequence with an organization, in the Y chromosomes of the pre-N lineage, that carries u3 adjacent to the P1.2-g2-DAZ3-DAZ4-g3 region, so that a single deletion event would remove all of the sequence missing in the N lineage by an intrachromosomal recombination event between homologous amplicons similar to that seen for complete AZFc deletions with the GenBank AZFc sequence.
The GenBank sequence, which is largely derived from the RPCI-11 donor (Kuroda-Kawaguchi et al. 2001), belongs to Y chromosome haplogroup R because it carries the derived state of the marker M207 (position 139.206 in the RPCI-11 BAC clone 386L3 [GenBank accession number AC006376]). The finding that the AZFc-P1 genotype A+B, present in the RPCI-11 donor, is restricted to haplogroup R1* (table 2) also supports this assignment. Haplogroups N and R are not closely related, and their most recent common ancestor probably lived ∼36,000 ± 6,000 years ago (Hammer and Zegura 2002), so substantial differences in structure, including inversions, could have accumulated, especially in the large palindromic structure in distal AZFc, where homologous recombination could lead to inversions of the intervening DNA (Zhou et al. 2001). A rearrangement of the GenBank AZFc structure could place u3 adjacent to the P1.2-g2-DAZ3-DAZ4-g3 region: inversion between the b2 and b3 amplicons (fig. 4). Then a g1/g3 recombination could delete, in a single step, all the sequences found to be absent from lineage N chromosomes (fig. 4). If this holds true, we must consider that the GenBank AZFc amplicon structure of the R-lineage Y chromosome is only one of many possible arrangements, and the arrangement proposed here for the AZFc structure in the N-lineage Y chromosome is only one other example. Indeed, the large P1 palindrome in distal AZFc was proposed to originate by duplication and inversion of an ancestral AZFc sequence similar to that proposed here for the pre-N lineage AZFc sequence (Kuroda-Kawaguchi et al. 2001).
Figure 4.
Schematic representation of putative genomic rearrangements in the amplicon structure of the AZFc sequence, in men from Y haplogroup R (GenBank reference sequence), that could lead to an AZFc amplicon structure with u3 adjacent to the P1.2-g2-DAZ3-DAZ4-g3 AZFc segment. After the proposed inversion, mediated by b2/b3, the g1 and g3 amplicons flank the contiguous u3-P1.2-g2-DAZ4-DAZ3 AZFc segment with direct polarity. Recombination between the g1/g3 amplicons in this proposed pre-N AZFc sequence would then lead to deletion of an AZFc segment in men with Y haplogroup N as indicated. This AZFc structure is different from that proposed by Repping et al. (2003) for structures with gr/gr deletions found in 14 different Y haplogroups but not in N (illustrated above the GenBank reference sequence). Recombinations of gr/gr deletions can take place between g1/g2 amplicons, r1/r3 amplicons, and r2/r4 amplicons, respectively, as indicated by the three brownish lines, but not between the g1/g3 amplicons, as found in men from Y haplogroup N. For further details, see text.
Y chromosomes carrying a large AZFc deletion variant removing the DAZ3 and DAZ4 genes may be common. In our sample, they corresponded precisely to haplogroup N chromosomes, including members of both the lineages N3* and N*(xN3). It therefore seems likely that all haplogroup N chromosomes carry this deletion. If so, we can use the known distribution of haplogroup N to deduce their frequency and distribution. This haplogroup is common throughout northern Europe and Asia, making up ∼12% of Y chromosomes in one worldwide survey and forming the majority in some populations such as the Finns (∼52%) and Yakuts (Sakha; ∼86%) (Zerjal et al. 1997). It forms a subset of the chromosomes previously identified as lacking 50f2/C (Jobling et al. 1996) but was erroneously considered to carry intact DAZ genes in this early study. The most recent common ancestor of haplogroup N chromosomes is estimated to have lived ∼8,800 ± 3,200 years ago (Hammer and Zegura 2002). It is therefore an ancient and successful lineage, and the lack of DAZ3 and DAZ4 has no detrimental effect on these men’s fertility. Other large Y deletion variants are for example a ∼3-Mb segment of Yp containing the AMELY and PRKY genes that also appears to be neutral, as it was absent from some normal males (Santos et al. 1998). Another partial AZFc deletion, designated the “gr/gr” deletion, seems, however, not to be neutral, since it was described as a significant risk factor for spermatogenic failure (Repping et al. 2003). This AZFc structure can arise by recombination between g1 and g2, but also between r1 and r3, or r2 and r4, respectively, in the AZFc GenBank sequence. No inversions are needed to arrange these amplicons into the same polarity (fig. 4). All gr/gr deletions therefore create an AZFc structure that is different from the one proposed here for men in Y haplogroup N. This is further illustrated by the gr/gr deletion’s retention of u3, but lack of the r1, r2, b3, and gr1 amplicons; in contrast, in Y haplogroup N, u3 is deleted and r1, r2, b3, and gr1 are present with reverse polarity (fig. 4). Interestingly, gr/gr deletions were not reported in men with Y-haplogroup N, although they were found in 14 other Y haplogroups (Repping et al. 2003). AZFc gr/gr deletions therefore appear to have arisen on many occasions and be continually eliminated by natural selection because of their modest negative impact on men’s fertility (Repping et al. 2003), whereas the g1/g3 deletion described here seems to be associated only with Y haplogroup N. The gr/gr deletion includes the g1/g2 deletions found in the AZFc structure of some men with oligozoospermia and deletion of the DAZ1/DAZ2 gene doublet (Fernandes et al. 2002), but it is not known whether it underlies the reduced sperm count associated with one haplogroup in Danish males (Krausz et al. 2001), so more similar deletions may be found in the future.
If the proposed g1/g3 recombination event is the origin of the AZFc deletion in the N-lineage Y chromosomes, the length of the AZFc sequence in these men is reduced by >50% (∼3.7 vs. ∼1.5 Mb) and with this two copies of the BPY2 gene family and also one copy each of the probably noncoding GOLGA2LY and TTY4 gene families are deleted (fig. 4). The functional contribution of the AZFc gene families to human spermatogenesis therefore appears to be genetically redundant and the roles of the individual genes may be most readily understood by investigating mutations in populations like the Finns, among whom many men carry fewer AZFc genes. Much further work is needed to understand the complete spectrum of normal and pathogenic variations in the structure of the Y-chromosomal AZFc region. The present study and forthcoming ones have implications for interpreting the results of Y STS deletion analyses in the infertility clinic, illustrating the care that must be taken when deletions are discovered and their phenotypic consequences for male fertility need to be predicted.
Acknowledgments
We thank all the donors of the genomic DNA samples and are grateful to Karin Huellen and Alexandra Schadwinkel for their help in collecting and isolating them from the individual blood samples. We also wish to thank Mrs. Christine Mahrla for her help in preparing the final version of this manuscript. This study was supported by grants to S.F. from the Fundação para a Ciência e Tecnologia (FCT) (SFRH/BD/811/2000) and to P.H.V. from the Deutsche Forschungsgemeinschaft (DFG: Vo403/10-2). S.P and C.T.-S. were supported by the Cancer Research Campaign (now Cancer Research UK).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Ensembl genome browser, http://www.ensembl.org/ (for the AZFc sequence structure)
- GenBank STS database, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for data presented here first, accession numbers are as follows: GOLY-SNV I, accession number BV012731; BPY2-SNV I, accession number BV012732; TTY4-SNV I, accession number BV012733)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for AZF locus and the AZFc genes BPY2, CDY1, and DAZ)
References
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Fernandes S, Huellen K, Goncalves J, Dukal H, Zeisler J, Rajpert De Meyts E, Skakkebaek NE, Habermann B, Krause W, Sousa M, Barros A, Vogt PH (2002) High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod 8:286–298 10.1093/molehr/8.3.286 [DOI] [PubMed] [Google Scholar]
- Floridia G, Gimelli G, Zuffardi O, Earnshaw WC, Warburton PE, Tyler-Smith C (2000) A neocentromere in the DAZ region of the human Y chromosome. Chromosoma 109:318–327 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Zegura SL (2002) The human Y chromosome haplogroup tree: nomenclature and phylogeny of its major dicisions. Annu Rev Anthropol 31:303–321 10.1146/annurev.anthro.31.040402.085413 [DOI] [Google Scholar]
- Jobling MA, Samara V, Pandya A, Fretwell N, Bernasconi B, Mitchell RJ, Gerelsaikhan T, Dashnyam B, Sajantila A, Salo PJ, Nakahori Y, Disteche CM, Thangaraj K, Singh L, Crawford MH, Tyler-Smith C (1996) Recurrent duplication and deletion polymorphisms on the long arm of the Y chromosome in normal males. Hum Mol Genet 5:1767–1775 10.1093/hmg/5.11.1767 [DOI] [PubMed] [Google Scholar]
- Krausz C, McElreavey K (1999) Y chromosome and male infertility. Front Biosci 4:E1–E8 [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, McElreavey K (2000) Prognostic value of Y deletion analysis: what is the clinical prognostic value of Y chromosome microdeletion analysis? Hum Reprod 15:1431–1434 10.1093/humrep/15.7.1431 [DOI] [PubMed] [Google Scholar]
- Krausz C, Quintana-Murci L, Meyts ER, Jorgensen N, Jobling MA, Rosser ZH, Skakkebaek NE, McElreavey K (2001) Identification of a Y chromosome haplogroup associated with reduced sperm counts. Hum Mol Genet 10:1873–1877 10.1093/hmg/10.18.1873 [DOI] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, Page DC (2001) The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 29:279–286 10.1038/ng757 [DOI] [PubMed] [Google Scholar]
- Paracchini S, Arredi B, Chalk R, Tyler-Smith C (2002) Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res 30:e27 10.1093/nar/30.6.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S (2003) Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 35:247–251 10.1038/ng1250 [DOI] [PubMed] [Google Scholar]
- Santos FR, Pandya A, Tyler-Smith C (1998) Reliability of DNA-based sex tests. Nat Genet 18:103 [DOI] [PubMed] [Google Scholar]
- Simoni M (2001) Molecular diagnosis of Y chromosome microdeletions in Europe: state-of-the-art and quality control. Hum Reprod 16:402–409 10.1093/humrep/16.3.402 [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Vogt PH (1998) Human chromosome deletions in Yq11, AZF candidate genes and male infertility: history and update. Mol Hum Reprod 4:739–744 10.1093/molehr/4.8.739 [DOI] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Akgun E, Jasin M (2001) Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc Natl Acad Sci USA 98:8326–8333 10.1073/pnas.151008498 [DOI] [PMC free article] [PubMed] [Google Scholar]