Abstract
Dystrophin deficiency, which leads to severe and progressive muscle degeneration in patients with Duchenne muscular dystrophy (DMD), is caused by frameshifting mutations in the dystrophin gene. A relatively new therapeutic strategy is based on antisense oligonucleotides (AONs) that induce the specific skipping of a single exon, such that the reading frame is restored. This allows the synthesis of a largely functional dystrophin, associated with a milder Becker muscular dystrophy phenotype. We have previously successfully targeted 20 different DMD exons that would, theoretically, be beneficial for >75% of all patients. To further enlarge this proportion, we here studied the feasibility of double and multiexon skipping. Using a combination of AONs, double skipping of exon 43 and 44 was induced, and dystrophin synthesis was restored in myotubes from one patient affected by a nonsense mutation in exon 43. For another patient, with an exon 46–50 deletion, the therapeutic double skipping of exon 45 and 51 was achieved. Remarkably, in control myotubes, the latter combination of AONs caused the skipping of the entire stretch of exons from 45 through 51. This in-frame multiexon skipping would be therapeutic for a series of patients carrying different DMD-causing mutations. In fact, we here demonstrate its feasibility in myotubes from a patient with an exon 48–50 deletion. The application of multiexon skipping may provide a more uniform methodology for a larger group of patients with DMD.
Introduction
Antisense oligonucleotides (AONs) have recently become an attractive tool for the study and treatment of human disease. Initially, AONs were used for the sequence-specific inhibition of genes, either to elucidate developmental processes or to suppress malignant or aberrant gene expression (Dennis et al. 1998; Stevenson et al. 1999; Nasevicius and Ekker 2000; Corey and Abrams 2001; Dove 2002). In these studies, AONs mediated RNAse H degradation of dsRNA, or they blocked transcription or translation initiation. However, AONs are also capable of modulating the splicing of pre-mRNA (Sierakowska et al. 1996). Since it has been estimated that at least 15% of disease-causing point mutations result in RNA splicing defects (Krawczak et al. 1992; Cartegni et al. 2002; Buratti et al. 2003), this latter application may be highly relevant for future genetic therapies. For instance, RNAse H-resistant AONs have successfully been used to induce the skipping of pseudo-exons by blocking cryptic splice sites in the β-globin gene (Sierakowska et al. 1996) and the cystic fibrosis transmembrane conductance regulator gene (Friedman et al. 1999). Alternatively, AONs linked to 10 arginine-serine dipeptide repeats for the artificial recruitment of splicing enhancer factors have been applied in vitro to induce inclusion of mutated BRCA1 and SMN2 exons that otherwise would be skipped (Cartegni and Krainer 2003). AONs have also been effective in altering the ratio of alternative splicing, which was applied for cancer-related genes to direct malignant toward nonmalignant isoforms (Mercatante et al. 2001, 2002). Last, but not least, a promising, recently developed application of AONs is to induce the specific skipping of exons in order to correct the reading frame of a mutated transcript so that it can be translated into a partially functional protein. The DMD gene, which codes for dystrophin, is well suited for this latter application. The protein consists of an N-terminal domain that binds to actin filaments, a central rod domain, and a C-terminal cysteine-rich domain that binds to the dystrophin-glycoprotein complex (Hoffman et al. 1987; Koenig et al. 1988; Yoshida and Ozawa 1990). Mutations in the DMD gene that interrupt the reading frame result in a complete loss of dystrophin function, which causes the severe Duchenne muscular dystrophy (DMD [MIM 310200]) (Hoffman et al. 1988; Koenig et al. 1989; Ervasti et al. 1990). The milder Becker muscular dystrophy (BMD [MIM 300376]), on the other hand, is the result of mutations in the same gene that are not frameshifting and result in an internally deleted but partially functional dystrophin that has retained its N- and C-terminal ends (Koenig et al. 1989; Di Blasi et al. 1996). Over two-thirds of patients with DMD and BMD have a deletion of ⩾1 exon (den Dunnen et al. 1989). Remarkably, patients have been described who exhibit very mild BMD and who lack up to 67% of the central rod domain (England et al. 1990; Winnard et al. 1993; Mirabella et al. 1998). This suggests that, despite large deletions, a partially functional dystrophin can be generated, provided that the deletions render the transcript in frame.
These observations have led to the idea of using AONs to alter splicing so that the open reading frame is restored and the severe DMD phenotype is converted into a milder BMD phenotype. Several studies have shown therapeutic AON-induced single-exon skipping in cells derived from the mdx mouse model (Dunckley et al. 1998; Wilton et al. 1999; Mann et al. 2001, 2002; Lu et al. 2003) and various DMD patients (Takeshima et al. 2001; van Deutekom et al. 2001; Aartsma-Rus et al. 2002, 2003; De Angelis et al. 2002). To date, we have identified a series of AONs that can be used to induce the skipping of 20 different exons (exons 2, 8, 17, 19, 29, 40–46, 48–53, 55, and 59) (Aartsma-Rus et al. 2002). Of all patients with DMD, >75% would benefit from the skipping of these exons. So far, we have successfully applied single-exon skipping in cells derived from eight different patients with DMD (van Deutekom et al. 2001; Aartsma-Rus et al. 2003). In these studies, the skipping of exons flanking out-of-frame deletions or an in-frame exon containing a nonsense mutation restored the reading frame and induced the synthesis of BMD-like dystrophins in ∼75%–80% of treated cells. These novel dystrophins could be detected as early as 16 h after transfection; the dystrophins increased to significant levels within 4 d and were maintained for at least 7 d (Aartsma-Rus et al. 2003).
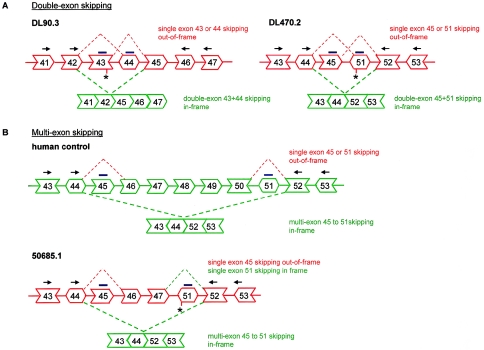
Here, we significantly extend the therapeutic applicability of this technique by demonstrating double and multiexon skipping (fig. 1). Following the simultaneous skipping of two exons (double-exon skipping), the reading frame was restored for one patient with a nonsense mutation in an out-of-frame exon and for another patient with a deletion that could not be bypassed by the skipping of only one exon. Furthermore, through the skipping of an entire stretch of consecutive exons (multiexon skipping), a BMD-like deletion was generated with the potential to restore up to 14% of all known DMD mutations.
Figure 1.
Schematic overview of double and multiexon skipping in three patients with and one human control. AONs are indicated by blue lines. Primers for RT-PCR analysis are indicated by arrows. A, Patient DL90.3 has a nonsense mutation in exon 43 (indicated by an asterisk). The resulting out-of-frame transcript is indicated in red. In contrast to single-exon skipping of exon 43 or 44, double-exon skipping of both exons restores the reading frame (in-frame transcript indicated in green). When myotubes derived from this patient are targeted by AONs specific for exons 43 and 44, single-exon skipping of exon 43 and exon 44 (indicated by red dotted lines) can be expected in addition to the anticipated double-exon skipping. Patient DL470.2 carries an exon 46–50 deletion, which results in a frameshift and a stop codon in exon 51. Single-exon skipping of exon 45 or exon 51 is not frame restoring, whereas double-exon skipping of both exons 45 and 51 is. B, Multiexon skipping of exons 45–51 preserves the reading frame in a control transcript (individual KM109). Patient 50685.1 has a deletion of exons 48–50, resulting in a stop codon in exon 51. Multiexon skipping of exons 45, 46, 47, and 51 restores the reading frame for this patient. In addition to the skipping of exons 45–51, single-exon skipping of exon 45 and exon 51 can also be expected.
Material and Methods
AONs and Primers
Exon-internal AONs targeting exons 44 (h44AON1) and 51 (h51AON2) were described elsewhere (Aartsma-Rus et al. 2002). The AONs targeting exons 43 and 45 were specifically designed for this study (h43AON5: CUGUAGCUUCACCCUUUCC; h45AON5: GCCCAAUGCCAUCCUGG). BLAST analysis of the AONs did not reveal perfect homology to other sequences in the human genome (maximum homology 94%, minimum E value 0.3). All AONs contain a 5′ fluorescein group (6-FAM), a full-length phosphorothioate backbone, and 2′-O-methyl–modified ribose molecules (Eurogentec). To avoid interference with the fluorescent signals of the secondary antibodies, unlabeled AONs were used for immunohistochemical analyses. Primers for RT-PCR analysis (fig. 1) were synthesized by Eurogentec (sequences available upon request).
Myogenic Cell Cultures and AON Transfections
Primary myoblasts from a human control and from two patients with DMD (DL470.2 [exon 46–50 deletion] and 50685.1 [exon 48–50 deletion]) were isolated from a muscle biopsy and cultured as described elsewhere (Aartsma-Rus et al. 2002). Myotubes were obtained from confluent myoblast cultures, after 7–14 d of serum deprivation. These were transfected with mixtures of 200 nM of each AON. Polyethylenemine (PEI) was used as transfection reagent, according to the manufacturer’s instructions (ExGen 500 [Fermentas]). Separate AON-PEI dilutions were made for each AON, with 3.5 μl PEI applied per μg of transfected AON.
For patient DL90.3, who has a point mutation in exon 43, only fibroblasts were available. Following infection (multiplicity of infection [MOI] 50–100) with an adenoviral vector containing the MyoD gene (Ad50MyoD), the fibroblasts were forced into myogenesis, according to protocols described elsewhere (Murry et al. 1996; Roest et al. 1996; Aartsma-Rus et al. 2002; Havenga et al. 2002). Transfection conditions were identical to those described above.
RNA Isolation and RT-PCR Analysis of the Skip Products
RNA isolation, RT-PCR, and sequence analysis were performed as described elsewhere (Aartsma-Rus et al. 2002). See figure 1 for the location of the primers. For quantification of the skip products, nested PCRs were performed using 24 cycles. PCR products were analyzed using the DNA 1000 LabChip kit and the 2100 Bioanalyzer (Agilent Technologies).
To analyze the splicing of the entire DMD gene, RT reactions were performed with 1 μg RNA, random hexamer primers, and SuperScript III (Invitrogen). PCR analyses were performed using the protein-truncation test (PTT) primers described elsewhere (Roest et al. 1993). Since some of the PCR primers were located in exons that were deleted for patient DL470.2, we specifically designed additional primers in exons 41, 42, 45, 53, and 54 (sequences available on request).
Analysis of the Dystrophin Protein
Immunohistochemical and Western blot analyses were performed as described elsewhere (Aartsma-Rus et al. 2002). Myosin polyclonal antibody L53 (a gift from Dr. M. van den Hoff, Amsterdam Medical Center, The Netherlands) was used to detect myosin. MANDYS1 (a gift from Dr. G. Morris, North East Wales Institute, United Kingdom), NCL-DYS2 (Novacastra Laboratories), and NCL-DYS1 (Novacastra Laboratories) were used to detect dystrophin. For the Western blot analysis, dystrophin levels were quantified using LumiAnalyst 3.0 (Roche).
Results
Double-Exon Skipping in Two Patients with DMD
The skipping of only a single exon is not sufficient to restore the reading frame for every mutation. In a significant fraction of mutations, it is necessary to skip two exons simultaneously (fig. 1A). For instance, patient DL90.3 carries a nonsense mutation in exon 43. Considering that this single exon is out of frame, the skipping of exon 43 removes the nonsense mutation but does not restore the reading frame. Since the combination of exons 43 and 44 is in frame, we aimed at the simultaneous skipping of both exons. Patient DL470.2 is affected by a deletion of exons 46–50. Frame restoration requires the joint skipping of the two exons flanking the deletion (fig. 1A).
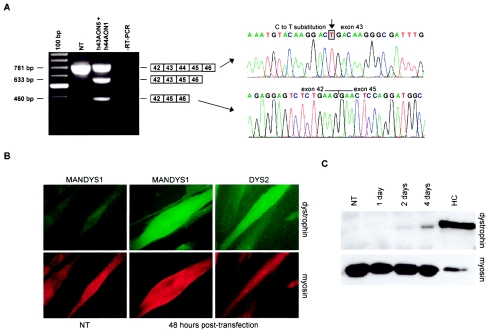
Myotube cultures from these patients were transfected with a mixture of either exon 43– and 44–specific AONs (patient DL90.3) or exon 45– and 51–specific AONs (patient DL470.2). Transfection efficiencies were typically >80%, as indicated by the number of cells with specific nuclear uptake of fluorescent AONs. RT-PCR analysis at 24 or 48 h after transfection indeed demonstrated the feasibility of specific double-exon skipping in both patients (30% and 75% of total transcript fragments for patients DL90.3 and DL470.2, respectively), which was confirmed by sequence analysis (figs. 2A and 3A). As expected, in addition to double-exon skipping, we also detected single-exon skipping in patient DL90.3 (exon 44; 27% of total transcripts) and patient DL470.2 (exon 51; 12% of total transcripts).
Figure 2.
Double-exon skipping in patient DL90.3, who carries a nonsense mutation in the out-of-frame exon 43. A, RT-PCR analysis of dystrophin mRNA fragments in AON-treated myotube cultures showed a shorter, novel transcript not present in nontransfected (NT) myotube cultures. Sequence analysis confirmed the precise skipping of both exon 43 and exon 44. Along with the double skip, we also detected a single skip of exon 44 but not a single-exon skip of exon 43. Note that weak additional fragments, slightly shorter than the wild-type products, were present. These were derived from heteroduplex formation. 100 bp = size marker; RT-PCR = negative control. B, Immunohistochemical analysis of the AON-treated myotube cultures. Cells were stained for myosin to identify fully differentiated myotubes (lower panel). Monoclonal antibodies MANDYS1 and Dys2 were used to detect dystrophin 2 d after transfection. No dystrophin signals could be detected in untreated cells stained with MANDYS1 (left panel) or Dys2 (not shown), whereas clear dystrophin signals could be detected in treated cells for both dystrophin antibodies. Magnification × 63. C, Western blot analysis of the AON-treated myotube cultures. The monoclonal antibody Dys1 was used to detect dystrophin. Protein extracts isolated from MyoD-transduced human control (HC) fibroblasts were used as a positive control. To avoid overexposure, this sample was diluted 1:5. To confirm equal loading of protein samples, blots were additionally stained with an antibody against myosin. No dystrophin was detected in NT myotube cultures. Clear dystrophin signals were observed in AON-treated myotube cultures after 2 d, which were increased at 4 d.
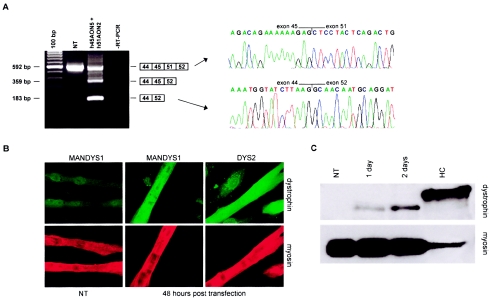
Figure 3.
Double-exon skipping in patient DL470.2, who carries a deletion of exons 46–50. A, RT-PCR analysis of dystrophin mRNA fragments of AON-treated myotube cultures showed a shorter, novel transcript not present in NT myotube cultures. The precise skipping of both exon 45 and exon 51 was confirmed by sequence analysis. Along with the double skip, we also detected a single skip of exon 51 but no single skip of exon 45. Because of heteroduplex formation, we observed weak additional fragments, slightly shorter than the wild-type products. 100 bp = size marker; RT-PCR = negative control. B, Immunohistochemical analysis of the AON-treated myotube cultures. Cells were stained for myosin to identify fully differentiated myotubes (lower panel). Monoclonal antibodies MANDYS1 and Dys2 were used to detect dystrophin 2 d after transfection. In treated cells, clear dystrophin signals could be detected for both antibodies, in contrast to untreated cells stained with MANDYS1 (left panel) or Dys2 (not shown). Magnification × 63. C, Western blot analysis of the AON-treated myotube cultures. Monoclonal antibody Dys1 was used to detect dystrophin. Protein extracts isolated from human control myotubes were used as a positive control, which was diluted 1:10 to avoid overexposure. Blots were additionally stained with an antibody against myosin to confirm equal loading of all samples. No dystrophin was detected in NT myotube cultures, whereas clear dystrophin signals were observed in AON-treated myotube cultures after 1 d, which increased after 2 d. Note that the dystrophin from patient DL470.2 is shorter than the control dystrophin. This correlates with the size of the deletion.
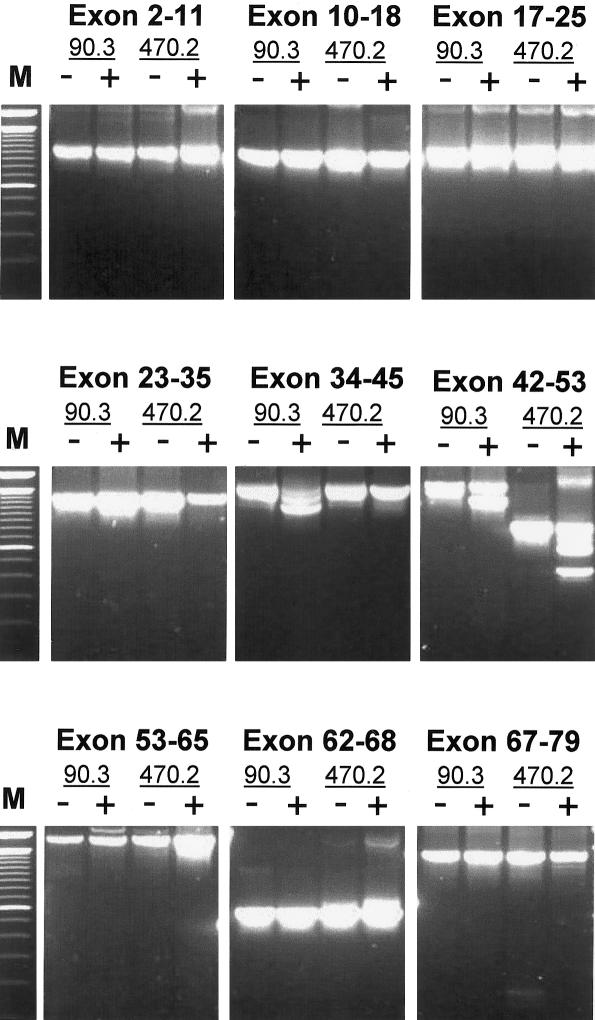
To verify that no larger or other regions of the DMD transcript were affected by the AON treatments, we performed RT-PCR analysis on the entire DMD gene, with consecutive sets of exons. In both treated and untreated myotubes, we did not detect additional aberrant splicing patterns in the DMD gene (fig. 4).
Figure 4.
RT-PCR analysis of the entire DMD transcript from RNA, isolated from untreated (−) and treated (+) myotubes from both patients. The anticipated specific-exon skipping patterns are present in the fragments containing exons 34–45 (patient DL90.3) and 42–53 (patients DL90.3 and DL470.2). Note that because of the deletion of exons 46–50, the wild-type fragment for this patient is shorter than that of patient DL90.3. No unexpected splicing anomalies were detected in other parts of the DMD gene, confirming the sequence specificity of the AONs. “M” is the 100-bp size marker.
Immunohistochemical analysis using two different antibodies directed against internal (MANDYS1) and C-terminal (Dys2) parts of dystrophin showed that the in-frame transcripts derived from double-exon skipping produced BMD-like dystrophins. On average, 70% of the myosin-positive myotubes showed dystrophin expression in response to AON transfection (figs. 2B and 3B). Western blot analysis confirmed the presence of dystrophin for patient DL90.3 (after 4 d) and patient DL470.2 (after 2 d), at levels of 3.3% and 1.8%, respectively, when compared to control myotubes (figs. 2C and 3C).
Multiexon Skipping
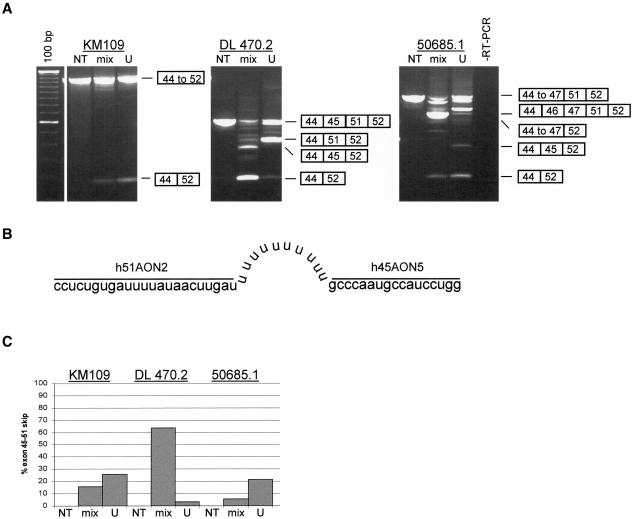
The splicing of exon 44 directly to exon 52 (as induced in patient DL470.2) generates an in-frame transcript. Inducing the skipping of this entire stretch of exons—that is, multiexon skipping—would generate a BMD-like deletion (exons 45–51) spanning a range of smaller, internal DMD mutations (fig. 1B). Multiexon skipping was first tested in human control myotubes (individual KM109) treated with a mixture of 200 nM of both the exon 45– and 51–specific AONs (fig. 5A). We observed a novel, shorter transcript, corresponding to a size that would result from the targeted skipping of exons 45–51. Indeed, sequence analysis revealed that exon 44 was directly spliced to exon 52 (data not shown). We then applied it to myotubes derived from a patient with DMD (50685.1) carrying an exon 48–50 deletion (fig. 1B). The AON-induced skipping of all exons from 45 to 51 yielded the intended in-frame transcript (fig. 5A).
Figure 5.
Double and multiexon skipping in myotubes from the human control (individual KM109) and from patients DL470.2 (deletion of exons 46–50) and 50685.1 (deletion of exons 48–50). A, RT-PCR analysis of dystrophin mRNA fragments in myotube cultures treated with either a mixture of h45AON5 and h51AON2 (mix) or a U-linked combination of these AONs (U). In all samples treated with either the mixture of AONs or the U-linked AON, a shorter transcript fragment was detected that contained exon 44 precisely spliced to exon 52 (confirmed by sequence analysis; data not shown). This was not present in NT myotubes. The novel in-frame transcript arose from double-exon skipping in patient DL470.2 (the targeted exons 45 and 51 are flanking the deletion), but in both the control and patient 50685.1 the transcript was a result of multiexon skipping. Also observed were additional shorter fragments, which were caused by single-exon skipping (exon 45 or 51) or partial multiexon skipping (exons 46–51). Note that in some lanes, other fragments, slightly shorter than the wild-type products, were present. This was a result of heteroduplex formation. 100 bp = size marker; RT-PCR = negative control. B, Schematic drawing of the U-linked AON. The exon 51–specific AON (h51AON2) is linked to the exon 45–specific AON (h45AON5) by 10 uracil nucleotides. C, All fragments were quantified using the DNA 1000 LabChip and the Bioanalyzer (Agilent). The percentage of double or multiexon skipping of exons 45–51 was determined by the ratio of this fragment to the total of transcript fragments. In contrast to patient DL470.2, the U-linked AON was more efficient for patients KM109 and 50685.1, when compared to the mixture of individual AONs.
Double and Multiexon Skipping Using a U-Linked Combination of AONs
The skipping of more than one exon from one pre-mRNA molecule requires both AONs to be present in the same nucleus, targeting the same molecule. To specifically enhance this probability, we designed a combined AON containing both the exon 45 and 51 AONs (h45AON5 and h51AON2), linked by 10 uracil nucleotides (fig. 5B). After transfection of this “U-linked AON” into myotubes from the human control (individual KM109) and the two patients with DMD (DL470.2 and 50685.1), RT-PCR analysis demonstrated its efficacy to generate the in-frame transcript with exon 44 spliced to exon 52 (fig. 5A). This multiexon skipping occurred specifically and precisely at the exon boundaries, as confirmed by sequence analysis (data not shown). Quantification showed that the U-linked AON was more efficient than the mixture of AONs in myotubes from patient 50685.1 and the human control but not in myotubes from patient DL470.2 (fig. 5C).
Discussion
Antisense-induced skipping of a single exon has shown therapeutic potential to correct the reading frame and induce the synthesis of BMD-like dystrophins in cultured muscle cells from patients with DMD (Takeshima et al. 2001; van Deutekom et al. 2001; De Angelis et al. 2002; Aartsma-Rus et al. 2003). Here, we demonstrate the feasibility of targeted antisense-induced multiexon skipping for therapeutic purposes. The spontaneous skipping of multiple exons frequently occurs in nature, albeit at low levels. This has been detected both in patients with DMD and in unaffected individuals (Sironi et al. 2002). Furthermore, this phenomenon has been suggested to be the underlying mechanism for the occurrence of dystrophin-positive “revertant” fibers in the mdx mouse model and in patients with DMD (Sherratt et al. 1993; Thanh et al. 1995; Lu et al. 2000). Multiexon skipping has also been observed in DMD gene-therapy studies aimed at the targeted skipping of the mutated exon 23 in mdx mouse muscle cells (Dunckley et al. 1998; Wilton et al. 1999; Bertoni et al. 2003). Both AONs and chimeric DNA-RNA oligonucleotides directed at the 3′ splice site of this exon generated shorter in-frame or out-of-frame transcripts in which additional exons adjacent to exon 23 were skipped. In this study, we specifically focused on inducing multiexon skipping. By use of combinations of AONs, the double skipping of exons 43–44 and 45–51 was induced in patient-derived myotube cultures. Immunohistochemical analysis of single myotubes indicated that this allowed the synthesis of dystrophins in up to 70% of myotubes. This percentage is not significantly lower than those obtained in our previous single-exon–skipping studies (75%–80%) (Aartsma-Rus et al. 2003). This suggests that both the transfection and the skipping performance of two AONs simultaneously are not significantly less efficient than those of a single AON. However, in a number of the remaining dystrophin-negative myotubes only single-exon skipping occurred.
Western blot analysis of protein extracts from patient-derived total myotube cultures showed relatively low levels (⩽3%) of dystrophin. Considering the high levels (70%) observed in the immunohistochemical analysis, there seems to be a discrepancy. However, this can be explained by the fact that in the immunohistochemical analysis we focus on single myosin-positive myotubes, whereas the samples in the Western blot analysis also include a significant number of dystrophin-negative cells. Furthermore, the control protein sample was derived from a culture that exhibited a two-fold higher degree of myogenicity (i.e., ∼40% in the patient cultures vs. >90% in the control sample) and that accordingly contained a higher number of dystrophin-producing cells. Finally, the control protein sample in the Western blot analysis was derived from myotubes expressing dystrophin over the entire period of differentiation (2 wk), whereas in the patient-derived myotubes the dystrophin synthesis was only just induced at the time of analysis (at most, 4 d). Since myotubes are viable for only several days after transfection, longer expression periods were unachievable.
Different ratios of exon 45– and 51–specific AONs were evaluated, and the highest levels of double-exon skipping were obtained with a 1:1 ratio (data not shown). Both AONs were apparently equally efficient at entering the nucleus. Besides double-exon skipping, we also observed single-exon skipping of exon 44 (patient DL90.3) and exon 51 (patient DL470.2), in particular. This suggests that the local secondary pre-mRNA structure was probably more affected by AONs targeting these exons, which subsequently rendered exons 43 and 45 inaccessible.
To increase the probability not only that both AONs are taken up by the same nucleus but also that they hybridize to the same RNA molecule, we designed an AON that consisted of two AONs linked with 10 uracil nucleotides. Compared to the mixture of the two AONs, the U-linked AON indeed induced higher levels of multiexon skipping in myotubes from a human control and a patient carrying an exon 48–50 deletion. In contrast, for yet-unknown reasons it was less efficient in inducing double-exon skipping in myotubes from a patient carrying an exon 46–50 deletion. In follow-up studies, we assessed the influence of the length of the U-linker. We did not observe significant differences (data not shown). There are two possible explanations for the mechanism of multiexon skipping. First, the entire region of both exons and introns between exon 45 and 51 may have been spliced out because of an overall AON-induced alteration in the secondary structure of the pre-mRNA. Second, the splicing of the individual exons 45 through 51 may have occurred earlier than that of exon 44 to exon 45 (which is not unlikely, given that intron 44 is 270 kb long). In that case, the splicing machinery regarded exons 45 to 51 as one large exon and omitted it because of the AONs that hybridized to exons 45 and 51. The latter explanation suggests that multiexon skipping may only be effective in those areas of the DMD gene in which downstream introns are spliced prior to upstream introns. We are currently verifying this by using different combinations of (U-linked) AONs in other regions of the gene.
When only single-exon skipping is taken into account, the antisense-based reading frame correction therapy would theoretically be beneficial for >75% of all patients with DMD. Multiexon skipping would significantly extend this percentage to most DMD mutations, except those that affect the functionally critical domains of the N-terminus or the cysteine-rich C-terminal domain, or those that involve the promoter region, the first exon, or translocations. However, these latter mutations are found in <8% of all patients with DMD reported in our database (den Dunnen 1996). Multiexon skipping not only increases the number of patients that would benefit from this approach but, more importantly, also decreases its mutation specificity. The multiexon skipping of exons 45–51 shown in this study would be frame restoring for 14% of all the deletions and 6% of all small mutations reported in the DMD mutation database (den Dunnen 1996). Furthermore, it offers the therapeutic potential of generating relatively large in-frame deletions known to be associated with mild BMD phenotypes (England et al. 1990; Winnard et al. 1993; Mirabella et al. 1998). Its eventual therapeutic application will largely depend on the efficiencies in vivo. Lu and colleagues (2003) recently demonstrated AON-induced exon 23–skipping in mdx muscle by using the Pluronic copolymer F127 as a delivery reagent. Close-to-normal levels of a nearly full-length dystrophin were observed in many myofibers, which improved muscle function. Although the effect was optimal at 2–4 wk, dystrophin was still detectable at 3 mo after injection (Lu et al. 2003). In our experience, the in vitro and in vivo effects of AONs in muscle cells correlate relatively well, which provides a promising basis for our current studies on multiexon skipping in mice.
Acknowledgments
We would like to thank Frank Baas and Ruud Wolterman for providing patient cell cultures and Stefan White and Eveline Mank for assistance with the Agilent Bioanalyzer. This work was financially supported by the Duchenne Parent Project (The Netherlands), the Princess Beatrix Fund (The Netherlands), and the European Union (QLG2-CT-1999-00920).
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for DMD and BMD)
References
- Aartsma-Rus A, Bremmer-Bout M, Janson A, den Dunnen J, van Ommen G, van Deutekom J (2002) Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscul Disord 12:S71 10.1016/S0960-8966(02)00086-X [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, Van Ommen GJ, et al (2003) Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet 12:907–914 10.1093/hmg/ddg100 [DOI] [PubMed] [Google Scholar]
- Bertoni C, Lau C, Rando TA (2003) Restoration of dystrophin expression in mdx muscle cells by chimeraplast-mediated exon skipping. Hum Mol Genet 12:1087–1099 10.1093/hmg/ddg133 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE, Pagani F (2003) Can a “patch” in a skipped exon make the pre-mRNA splicing machine run better? Trends Mol Med 9:229–232 10.1016/S1471-4914(03)00072-8 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR (2003) Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 10:120–125 10.1038/nsb887 [DOI] [PubMed] [Google Scholar]
- Corey DR, Abrams JM (2001) Morpholino antisense oligonucleotides: tools for investigating vertebrate development. Genome Biol 2:reviews1015.1–reviews1015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis FG, Sthandier O, Berarducci B, Toso S, Galluzzi G, Ricci E, Cossu G, Bozzini I (2002) Chimeric snRNA molecules carrying antisense sequences against the splice junctions of exon 51 of the dystrophin pre-mRNA induce exon skipping and restoration of a dystrophin synthesis in Delta 48–50 DMD cells. Proc Natl Acad Sci USA 99:9456–9461 10.1073/pnas.142302299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen J (1996) The Leiden Muscular Dystrophy pages. http://www.dmd.nl (accessed December 10, 2003)
- den Dunnen JT, Grootscholten PM, Bakker E, Blonden LA, Ginjaar HB, Wapenaar MC, van Paassen HM, van Broeckhoven C, Pearson PL, van Ommen GJ (1989) Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet 45:835–847 [PMC free article] [PubMed] [Google Scholar]
- Dennis JU, Dean NM, Bennett CF, Griffith JW, Lang CM, Welch DR (1998) Human melanoma metastasis is inhibited following ex vivo treatment with an antisense oligonucleotide to protein kinase C-alpha. Cancer Lett 128:65–70 10.1016/S0304-3835(98)00052-4 [DOI] [PubMed] [Google Scholar]
- Di Blasi C, Morandi L, Barresi R, Blasevich F, Cornelio F, Mora M (1996) Dystrophin-associated protein abnormalities in dystrophin-deficient muscle fibers from symptomatic and asymptomatic Duchenne/Becker muscular dystrophy carriers. Acta Neuropathol (Berl) 92:369–377 [DOI] [PubMed] [Google Scholar]
- Dove A (2002) Antisense and sensibility. Nat Biotechnol 20:121–124 10.1038/nbt0202-121 [DOI] [PubMed] [Google Scholar]
- Dunckley MG, Manoharan M, Villiet P, Eperon IC, Dickson G (1998) Modification of splicing in the dystrophin gene in cultured Mdx muscle cells by antisense oligoribonucleotides. Hum Mol Genet 7:1083–1090 10.1093/hmg/7.7.1083 [DOI] [PubMed] [Google Scholar]
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, Davies KE (1990) Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature 343:180–182 10.1038/343180a0 [DOI] [PubMed] [Google Scholar]
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP (1990) Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature 345:315–319 10.1038/345315a0 [DOI] [PubMed] [Google Scholar]
- Friedman KJ, Kole J, Cohn JA, Knowles MR, Silverman LM, Kole R (1999) Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J Biol Chem 274:36193–36199 10.1074/jbc.274.51.36193 [DOI] [PubMed] [Google Scholar]
- Havenga MJ, Lemckert AA, Ophorst OJ, van Meijer M, Germeraad WT, Grimbergen J, van Den Doel MA, Vogels R, van Deutekom JCT, Janson AAM, de Bruijn JD, Uytdehaag F, Quax PH, Logtenberg T, Mehtali M, Bout A (2002) Exploiting the natural diversity in adenovirus tropism for therapy and prevention of disease. J Virol 76:4612–4620 10.1128/JVI.76.9.4612-4620.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH, Jr., Kunkel LM (1987) Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51:919–928 [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, Loike JD, Harris JB, et al (1988) Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N Engl J Med 318:1363–1368 [DOI] [PubMed] [Google Scholar]
- Koenig M, Beggs AH, Moyer M, Scherpf S, Heindrich K, Bettecken T, Meng G, et al (1989) The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 45:498–506 [PMC free article] [PubMed] [Google Scholar]
- Koenig M, Monaco AP, Kunkel LM (1988) The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 53:219–226 [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90:41–54 [DOI] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, Fletcher S, Partridge TA, Wilton SD (2003) Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med 9:1009–1014 10.1038/nm897 [DOI] [PubMed] [Google Scholar]
- Lu QL, Morris GE, Wilton SD, Ly T, Artem'yeva OV, Strong P, Partridge TA (2000) Massive idiosyncratic exon skipping corrects the nonsense mutation in dystrophic mouse muscle and produces functional revertant fibers by clonal expansion. J Cell Biol 148:985–996 10.1083/jcb.148.5.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann CJ, Honeyman K, Cheng AJ, Ly T, Lloyd F, Fletcher S, Morgan JE, Partridge TA, Wilton SD (2001) Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci USA 98:42–47 10.1073/pnas.011408598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann CJ, Honeyman K, McClorey G, Fletcher S, Wilton SD (2002) Improved antisense oligonucleotide induced exon skipping in the mdx mouse model of muscular dystrophy. J Gene Med 4:644–654 10.1002/jgm.295 [DOI] [PubMed] [Google Scholar]
- Mercatante DR, Mohler JL, Kole R (2002) Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J Biol Chem 277:49374–49382 10.1074/jbc.M209236200 [DOI] [PubMed] [Google Scholar]
- Mercatante DR, Sazani P, Kole R (2001) Modification of alternative splicing by antisense oligonucleotides as a potential chemotherapy for cancer and other diseases. Curr Cancer Drug Targets 1:211–230 [DOI] [PubMed] [Google Scholar]
- Mirabella M, Galluzzi G, Manfredi G, Bertini E, Ricci E, De Leo R, Tonali P, Servidei S (1998) Giant dystrophin deletion associated with congenital cataract and mild muscular dystrophy. Neurology 51:592–595 [DOI] [PubMed] [Google Scholar]
- Murry CE, Kay MA, Bartosek T, Hauschka SD, Schwartz SM (1996) Muscle differentiation during repair of myocardial necrosis in rats via gene transfer with MyoD. J Clin Invest 98:2209–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC (2000) Effective targeted gene “knockdown” in zebrafish. Nat Genet 26:216–220 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Roest PA, Roberts RG, van der Tuijn AC, Heikoop JC, van Ommen GJ, den Dunnen JT (1993) Protein truncation test (PTT) to rapidly screen the DMD gene for translation terminating mutations. Neuromuscul Disord 3:391–394 10.1016/0960-8966(93)90083-V [DOI] [PubMed] [Google Scholar]
- Roest PA, van der Tuijn AC, Ginjaar HB, Hoeben RC, Hoger-Vorst FB, Bakker E, den Dunnen JT, van Ommen GJ (1996) Application of in vitro Myo-differentiation of non-muscle cells to enhance gene expression and facilitate analysis of muscle proteins. Neuromuscul Disord 6:195–202 10.1016/0960-8966(96)00006-5 [DOI] [PubMed] [Google Scholar]
- Sherratt TG, Vulliamy T, Dubowitz V, Sewry CA, Strong PN (1993) Exon skipping and translation in patients with frameshift deletions in the dystrophin gene. Am J Hum Genet 53:1007–1015 [PMC free article] [PubMed] [Google Scholar]
- Sierakowska H, Sambade MJ, Agrawal S, Kole R (1996) Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA 93:12840–12844 10.1073/pnas.93.23.12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Cagliani R, Pozzoli U, Bardoni A, Comi GP, Giorda R, Bresolin N (2002) The dystrophin gene is alternatively spliced throughout its coding sequence. FEBS Lett 517:163–166 10.1016/S0014-5793(02)02613-3 [DOI] [PubMed] [Google Scholar]
- Stevenson JP, Yao KS, Gallagher M, Friedland D, Mitchell EP, Cassella A, Monia B, Kwoh TJ, Yu R, Holmlund J, Dorr FA, O’Dwyer PJ (1999) Phase I clinical/pharmacokinetic and pharmacodynamic trial of the c-raf-1 antisense oligonucleotide ISIS 5132 (CGP 69846A). J Clin Oncol 17:2227–2236 [DOI] [PubMed] [Google Scholar]
- Takeshima Y, Wada H, Yagi M, Ishikawa Y, Minami R, Nakamura H, Matsuo M (2001) Oligonucleotides against a splicing enhancer sequence led to dystrophin production in muscle cells from a Duchenne muscular dystrophy patient. Brain Dev 23:788–790 10.1016/S0387-7604(01)00326-6 [DOI] [PubMed] [Google Scholar]
- Thanh LT, Nguyen Thi M, Hori S, Sewry CA, Dubowitz V, Morris GE (1995) Characterization of genetic deletions in Becker muscular dystrophy using monoclonal antibodies against a deletion-prone region of dystrophin. Am J Med Genet 58:177–186 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Bremmer-Bout M, Janson AA, Ginjaar IB, Baas F, den Dunnen JT, van Ommen GJ (2001) Antisense-induced exon skipping restores dystrophin expression in DMD patient derived muscle cells. Hum Mol Genet 10:1547–1554 10.1093/hmg/10.15.1547 [DOI] [PubMed] [Google Scholar]
- Wilton SD, Lloyd F, Carville K, Fletcher S, Honeyman K, Agrawal S, Kole R (1999) Specific removal of the nonsense mutation from the mdx dystrophin mRNA using antisense oligonucleotides. Neuromuscul Disord 9:330–338 10.1016/S0960-8966(99)00010-3 [DOI] [PubMed] [Google Scholar]
- Winnard AV, Klein CJ, Coovert DD, Prior T, Papp A, Snyder P, Bulman DE, et al (1993) Characterization of translational frame exception patients in Duchenne/Becker muscular dystrophy. Hum Mol Genet 2:737–744 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Ozawa E (1990) Glycoprotein complex anchoring dystrophin to sarcolemma. J Biochem (Tokyo) 108:748–752 [DOI] [PubMed] [Google Scholar]