Abstract
Peak bone mineral density (BMD) is a highly heritable trait and is a good predictor of the risk of osteoporosis and fracture in later life. Recent studies have sought to identify the genes underlying peak BMD. Linkage analysis in a sample of 464 premenopausal white sister pairs detected linkage of spine BMD to chromosome 1q (LOD 3.6). An independent sample of 254 white sister pairs has now been genotyped, and it also provides evidence of linkage to chromosome 1q (LOD 2.5) for spine BMD. Microsatellite markers were subsequently genotyped for a 4-cM map in the chromosome 1q region in all available white sister pairs (n=938), and a LOD score of 4.3 was obtained near the marker D1S445. Studies in the mouse have also detected evidence of linkage to BMD phenotypes in the region syntenic to our linkage finding on chromosome 1q. Thus, we have replicated a locus on 1q contributing to BMD at the spine and have found further support for the region in analyses employing an enlarged sample. Studies are now ongoing to identify the gene(s) contributing to peak spine BMD in women.
Introduction
Osteoporosis is major public health problem, producing disability and excess mortality through the development of fractures. It is a disease characterized by low bone mass and microarchitectural deterioration of bone tissue, leading to enhanced bone fragility and a consequent increase in fracture risk (World Health Organization Consensus Development Conference 1991). In the United States, as many as 54% of postmenopausal white women, or ∼17 million women, have low bone mass, and another 20%–30%, or 6–9 million, have osteoporosis (Melton et al. 1992; Melton 1995; Looker et al. 1997). Vertebral compression fractures are common osteoporotic fractures and lead to pain, deformity, and disability. Low bone mineral density (BMD) is a major contributing factor. BMD in later life is highly correlated with peak BMD in the 3rd or 4th decade of life. Sex and race are also important predictors of peak BMD, with men and African Americans having higher average peak BMD than women and whites, respectively (Bell 1997), as well as a concomitantly lower rate of osteoporosis and fracture (Bohannon et al. 1999).
There is a strong genetic component to peak spine BMD (Peacock et al. 2002). Indeed, a variety of study designs indicate that as much as 80% of the variability in peak BMD is attributable to genetic factors. Despite the substantial heritability of peak BMD, variability in this phenotype is still likely to be due to the effects of multiple, interacting genes. Several studies have also suggested greater phenotypic similarity among individuals of the same sex in a family (Jones and Nguyen 2000; Duncan et al. 2003; Van Pottelberg et al. 2003). Identification of the genes underlying spine BMD will be important in understanding the underlying mechanisms of bone formation and maintenance and may elucidate the mode of action of sex-specific QTLs. This, in turn, may provide molecular targets for future therapies for osteoporosis.
We previously performed an autosomal genome screen in 429 premenopausal white sister pairs and found evidence of linkage of peak spine BMD to markers on chromosome 1q (LOD 3.11) (Koller et al. 2000). Additional genotyping in an expanded sample of 464 white sister pairs increased the LOD score to 3.64 at marker D1S484 (Koller et al. 2000). The goals of the current study were to replicate our previous spine BMD linkage results by performing a 10-cM genome screen in an independent sample of 254 premenopausal white sister pairs, ascertained and evaluated using procedures identical to those used for our previously analyzed sample. In this independent sample, we have replicated linkage of spine BMD to chromosome 1q. Subsequently, all available white sister pairs (n=938) and their parents (n=316) were genotyped for a denser set of microsatellite markers in the chromosome 1q region, to further delineate the critical chromosomal interval. Our results provide significant evidence of the existence of a gene(s) on chromosome 1q that affects peak spine BMD.
Methods
Subjects
As in our previous studies, healthy, premenopausal sister pairs between the ages of 20 and 50 years were recruited through advertisement to participate in studies designed to identify genes contributing to bone-related phenotypes. Sibships of more than two individuals were actively recruited. When available, parents of the premenopausal sister pairs were requested to contribute a blood sample for DNA extraction. All studies were performed at the General Clinical Research Center at Indiana University Medical School, and all subjects gave written, informed consent prior to participation. The study was approved by the Indiana University institutional review board.
Sisters have been continually recruited since 1995, and molecular studies were performed as sufficient samples became available for analysis. As a result, a genome screen was previously completed in 429 sister pairs (Koller et al. 2000), and a genome screen was more recently completed in a subsequent sample of 254 independent sister pairs. Additional sisters and their parents have continued to be recruited (255 sister pairs). Therefore, to maximize the power of molecular studies to localize genetic effects, all available samples from these additional sister pairs and their parents were included in the follow-up genotyping on chromosome 1q (938 sister pairs and 316 parents).
BMD
BMD was measured by dual energy X-ray absorptiometry (DPX-L [Lunar Corporation]) at lumbar vertebrae L2–L4. Sisters were measured on the same machine, usually at the same visit. Image analysis was performed using version 4.6/4.7 of the software. The coefficient of variation for the lumbar spine, measured in 20 women who had duplicate BMD measurements made after they were repositioned on the machine, was 0.52%. Height and weight were measured using a Harpenden Stadiometer and a Scale-Tronix weighing scale, which were regularly calibrated throughout the study.
DNA and Marker Genotyping
DNA was isolated using standard techniques and was stored in a DNA repository under a unique code. A 10-cM genome scan was performed at the Center for Inherited Disease Research by use of automated fluorescent microsatellite analysis. PCR products were sized on an ABI 3700 sequencer. The marker set was a modification of version 9 of the Cooperative Human Linkage Center (CHLC) marker set, with 392 markers at an average spacing of 9 cM and an average heterozygosity of 0.76. The error rate based on paired genotypes from blind duplicate samples was 0.1%. The overall missing-data rate was 5.7%. The marker genotype data from the genomewide screen was used to verify full-sibling relationships among subjects by use of the computer programs RELATIVE (Göring and Ott 1997) and RELPAIR (Boehnke and Cox 1997), and half-sibling pairs were eliminated from further analysis.
To maximize the power to further localize the genes contributing to spine BMD, additional genotyping by use of microsatellite repeat markers on chromosome 1q was performed on all available samples from white subjects (938 sister pairs). This enlarged sample included all individuals who were genotyped as part of either of the two genome screens (683 sister pairs), as well as the 35 sister pairs who were genotyped only in the selected chromosomal region (Koller et al. 2000) and those recruited after the initiation of the second genome screen (220 sister pairs). Since >80% of the subjects recruited for the sample were white and this subsample provided the greatest evidence of linkage to chromosome 1q, only white subjects were included in the denser microsatellite-marker genotyping. For recently recruited subjects who were not part of either genome screen, the full-sibling relationships were confirmed by analyzing 10 X-chromosome markers.
A total of 14 markers were genotyped in the chromosome 1q linkage region. All markers were selected from the Marshfield map on the basis of their reported heterozygosity and marker position, with a preference for tetranucleotide repeat markers, when available. The average intermarker distance in the chromosome 1q critical interval delimited by D1S2777 and D1S2823 was 4 cM, and the average marker heterozygosity was 76%. PCR products were sized on an ABI 3100 by use of the Genotyper program, version 3.6, with overreading by two independent observers. Chromosomal positions, marker order, and map positions were obtained from the Marshfield electronic database (Marshfield Center for Medical Genetics Web site).
Quantitative Linkage Analysis
Stepwise regression analysis was employed with the spine BMD phenotype, and height, weight, oral contraceptive use, pack-years of smoking, and age were used to identify significant covariates with spine BMD. Regression residuals, representing covariate-adjusted BMD values, were computed and used in all analyses. Multipoint quantitative linkage analysis was performed for BMD by use of the maximum-likelihood variance-estimation method, as implemented in the computer package Mapmaker/SIBS (Kruglyak and Lander 1995). Using all possible sibling pairs formed from families with more than two sisters, LOD scores were computed at 1-cM intervals along each autosome. Observed allele frequencies in the individuals genotyped for the genome screen were used. To confirm the robustness of linkage, analyses were also performed using only independent sibling pairs and implementing the more conservative Haseman-Elston regression approach (Kruglyak and Lander 1995).
Results
A sample of 254 independent white sister pairs was ascertained and evaluated to detect linkage to the phenotype of spine BMD. The mean age of the sister pairs was 32.6 years, and the mean difference in age between sisters was 3.8 years (table 1). Of the covariates studied for their effect on spine BMD, only age and weight approached significance (P<.10) in stepwise model fitting. Residuals from regression model fitting were used as age- and weight-adjusted spine BMD values in all subsequent analyses.
Table 1.
Characteristics of Participants in Study
|
No. of Sibling Pairs |
Mean ± SD |
|||||||
| Sample | No. of Participants | No. of Families | Independent | All Possible | Age of Participant (years) | Difference in Sibling Age (years) | Weight of Participant (kg) | Lumbar (L2–L4) Spine BMD (g/cm2) |
| Independent | 365 | 164 | 201 | 254 | 32.6 ± 7.1 | 3.8 ± 2.4 | 68.4 ± 15.2 | 1.28 ± 0.13 |
| Expanded | 1,342 | 602 | 740 | 938 | 33.2 ± 7.1 | 3.7 ± 2.4 | 69.4 ± 16.0 | 1.28 ± 0.14 |
A genome screen in the independent sample of 254 white sister pairs did not identify any chromosomal region with a LOD score >3.6, the typically employed threshold for genomewide significance (Lander and Kruglyak 1995). However, linkage to chromosome 1q with a LOD score of 2.5 was detected in the same chromosomal region identified in our first linkage study (Koller et al. 2000), with a peak slightly distal to the original peak. Thus, linkage to the chromosome 1q region was replicated by our independent sample on the basis of the criteria (LOD > 1.2) of Lander and Krugylak (1995). No other chromosomal region had a LOD score >1.3 in our sample of 254 white sister pairs.
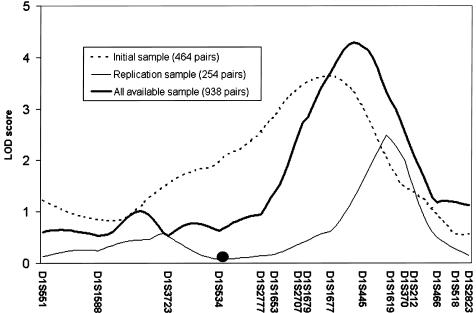
Additional genotyping of microsatellite markers was performed in the chromosome 1q region that was reported in both of our independent linkage samples. To maximize power, all available white sister pairs and their parents were genotyped. The analysis of these 938 sister pairs and their 316 parents resulted in a LOD score of 4.3 at position 181 cM on the Marshfield map (fig. 1). Since spine BMD is calculated from the bone mineral content (BMC) of the vertebra divided by its area, we also performed linkage analysis with BMC and area to determine if either of these phenotypes was responsible for our observed linkage with spine BMD. The multipoint LOD score for vertebral BMC was 2.8, with the peak in approximately the same position as the peak for BMD. There was no evidence for linkage with bone area, suggesting that the evidence for linkage to peak spine BMD in our sample does not result from an effect on bone size.
Figure 1.
Multipoint linkage results for chromosome 1 in the sample of white subjects in the study by Koller et al. (2000) (dashed line), the independent sample of white subjects (thin line), and the expanded sample of white subjects (thick line). The position of the centromere is indicated by a dark circle.
Discussion
In our first linkage study (Koller et al. 2000), we detected significant evidence of linkage of spine BMD to chromosome 1q21-23 with a LOD score of 3.6. In the current work, we performed a genomewide linkage scan in an independent sample of 254 white sister pairs, to identify chromosomal regions that harbor genes that affect peak spine BMD in premenopausal women. In this replication study, only one chromosomal region produced a LOD score >1.3 when analyzing the highly heritable spine BMD phenotype. This LOD score of 2.5 occurred at chromosome 1q22-23. Although the LOD-score graph of the current study substantially overlaps with the results of our original linkage study (Koller et al. 2000), the two figures are not superimposable. However, given the imprecision of linkage methods for localizing the contributory gene(s) (Roberts et al. 1999), we have strong evidence that we have replicated our significant linkage finding in an independent sample.
As noted above, spine BMD is calculated from the BMC of the vertebra divided by its area. Therefore, it was possible that our linkage findings resulted from differences in vertebral size between individuals rather than variation in spine BMD. To exclude this possibility, we performed the same analysis but designated BMC and vertebral area as phenotypes. Our results indicate that the observed linkage findings result from a gene(s) affecting spine BMD rather than bone size.
We performed further analyses with microsatellite markers on chromosome 1q in an expanded group of 938 premenopausal white women, a group that included all white women from both genome screens in addition to newly ascertained samples. We have focused our molecular studies on only the white families for several reasons. First, although our initial analyses (Koller et al. 2000) found that the inclusion of African American sister pairs increased our LOD score on chromosome 1q, further marker genotyping in additional samples suggest that the 131 African American families in our study do not provide strong evidence of linkage to this region of chromosome 1q. Second, numerous studies of complex disease-related phenotypes have found that different susceptibility genes may be segregating in samples of various ancestry (Cox 2001; Fullerton et al. 2002). Therefore, it is quite possible that some of the genes contributing to the variation in spine BMD in the white sister pairs may not manifest a strong effect in samples from other racial groups. This hypothesis is further supported by the well-known racial differences in BMD (Bell et al. 1991). Thus, to maximize our power to further localize the gene(s) contributing to variation in peak spine BMD, we elected to limit our analyses to only the white sister pairs, who represent the single largest racial group in our sample.
Further marker genotyping in the chromosome 1q13-31 region was performed to achieve an average intermarker distance under the linkage peak of ∼4 cM. This resulted in some narrowing of the critical interval; however, the region remains broad, encompassing many potential candidate genes. This region of chromosome 1q contains several candidate genes, including the structural gene for osteocalcin, the interleukin-6 receptor gene, and a group of genes encoding the calcium-binding proteins. Linkage to chromosome 1q23.3-q24 has been reported for hypercalciuria, a common cause of kidney stones (Reed et al. 1999), which is frequently accompanied by reduced vertebral bone density (Pietschmann et al. 1992). Recently, polymorphisms in the soluble adenylate cyclase gene have been reported to contribute to hypercalciuria and low spine BMD (Reed et al. 2002).
Multiple studies in mouse models have linked BMD phenotypes to the syntenic region of human chromosome 1, which is mouse chromosome 1. Strong evidence of linkage to mouse chromosome 1 was observed for both spine and femur BMD in female C57BL/6J×C3H/HeJ F2 animals (Beamer et al. 2001; Koller et al., in press). Klein and colleagues reported linkage of whole-body BMD to mouse chromosome 1, initially using recombinant inbred lines derived from C57BL/6J and DBA/2 mice (Klein et al. 1998) and later confirmed in an F2 sample created from a cross of the same two progenitor lines (Klein et al. 2001). In addition, a study of a different cross employing C57BL/6J×CAST/EiJ F2 mice detected linkage to the same region of mouse chromosome 1 by use of the phenotype of peak femoral BMD, as measured by peripheral quantitative computed tomography (pQCT) (Beamer et al. 1999). Unfortunately, vertebral BMD was not measured in that study.
Although the linkage we found to chromosome 1q has also been observed in all mouse studies of similar BMD phenotypes, human studies of BMD have not consistently reported linkage to this region. A comparison of the various sample designs and phenotyping methods employed in human family studies highlights several important differences from our own unique study of peak spine BMD measured in premenopausal women. First, unlike our study and that of Spector and colleagues (Wilson et al. 2003), most study designs have included both males and females (Devoto et al. 1998; Niu et al. 1999; Karasik et al. 2002). On the basis of the recent observation of sex-specific QTLs in mouse models (Orwoll et al. 2001) and human studies suggesting greater phenotypic similarity among same-sex individuals (Jones and Nguyen 2000; Duncan et al. 2003; Van Pottelberg et al. 2003), it is quite likely that studies of both sexes may identify unique QTLs as compared with those samples that focus solely on women. Second, our sample includes only premenopausal women and therefore is designed to identify genes contributing to peak BMD. All other studies have sampled individuals across a wide age range, encompassing not only peak BMD but also bone loss as well as bone acquisition. The limited number of studies that have been performed to date have been quite inconclusive regarding the genetic contribution to bone loss (Christian et al. 1989; Kelly et al. 1993), and no study has yet demonstrated that the same genes contribute to peak BMD and bone loss. Therefore, the genes that will be identified in studies encompassing individuals from a wide age range are likely to be different from those linked to the highly heritable phenotype of peak BMD in premenopausal women.
In summary, we performed a genomewide linkage scan for spine BMD in an independent sample and found evidence for linkage to markers on chromosome 1q. Subsequently, we genotyped markers to a 4-cM density in an expanded sample of 938 white sister pairs. Our results indicate that this region harbors a gene(s) that influences peak BMD in premenopausal white women and that the gene(s) on chromosome 1q may affect BMD in both mice and humans. These studies represent an important step in identifying the genes underlying normal variation in BMD.
Acknowledgments
We gratefully acknowledge the sisters and parents who participated in this study, as well as the study coordinators, without whom this work could not have been accomplished. This work was supported by National Institutes of Health grants PO1 AG-18397, RO1 AR-43476, MO1 RR-00750, K24 AR-02095, AR-4370, and T32 HD-07373.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Center for Inherited Disease Research, http://www.cidr.jhmi.edu
- Marshfield Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/ (for chromosomal positions, marker order, and map positions)
References
- Beamer WG, Shultz KL, Churchill GA, Frankel WN, Baylink DJ, Rosen CJ, Donahue LR (1999) Quantitative trait loci for bone density in C57BL/6J and CAST/EiJ inbred mice. Mamm Genome 10:1043–1049 10.1007/s003359901159 [DOI] [PubMed] [Google Scholar]
- Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ (2001) Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res 16:1195–1206 [DOI] [PubMed] [Google Scholar]
- Bell NH (1997) Bone and mineral metabolism in African-Americans. Trends Endocrinol Metab 8:240–245 10.1016/S1043-2760(97)00065-9 [DOI] [PubMed] [Google Scholar]
- Bell NH, Shary J, Stevens J, Garza M, Gordon L, Edwards J (1991) Demonstration that bone mass is greater in black than in white children. J Bone Miner Res 6:719–723 [DOI] [PubMed] [Google Scholar]
- Boehnke M, Cox NJ (1997) Accurate inference of relationships on sib-pair linkage studies. Am J Hum Genet 61:423–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannon AD, Hanlon JT, Landerman R, Gold DT (1999) Association of race and other potential risk factors with nonvertebral fractures in community-dwelling elderly women. Am J Epidemiol 149:1002–1009 [DOI] [PubMed] [Google Scholar]
- Christian JC, Yu P-L, Slemanda CW, Johnston CC Jr (1989) Heritability of bone mass: a longitudinal study in aging male twins. Am J Hum Genet 44:429–433 [PMC free article] [PubMed] [Google Scholar]
- Cox NJ (2001) Challenges in identifying genetic variation affecting susceptibility to type 2 diabetes: examples from studies of the calpain-10 gene. Hum Mol Genet 10:2301–2305 10.1093/hmg/10.20.2301 [DOI] [PubMed] [Google Scholar]
- Devoto M, Shimoya K, Caminis J, Ott J, Tenenhouse A, Whyte M, Sereda L, Hall S, Considine E, Williams CJ, Tromp G, Kuivaniemi H, Ala-Kokko L, Prockop DJ, Spotila L (1998) First-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density on chromosomes 1p, 2p, and 4p. Eur J Hum Genet 6:151–157 10.1038/sj.ejhg.5200169 [DOI] [PubMed] [Google Scholar]
- Duncan EL, Cardon LR, Sinsheimer JS, Wass JA, Brown MA (2003) Site and gender specificity of inheritance of bone mineral density. J Bone Miner Res 18:1531–1538 [DOI] [PubMed] [Google Scholar]
- Fullerton SM, Bartoszewicz A, Ybazeta G, Horikawa Y, Bell GI, Kidd KK, Cox NJ, Hudson RR, Di Rienzo A (2002) Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. Am J Hum Genet 70:1096–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Ott J (1997) Relationship estimation in affected sib pair analysis of late-onset disease. Eur J Hum Genet 5:69–77 [PubMed] [Google Scholar]
- Jones G, Nguyen TV (2000) Associations between maternal peak bone mass and bone mass in prepubertal male and female children. J Bone Miner Res 15:1998–2004 [DOI] [PubMed] [Google Scholar]
- Karasik D, Myers RH, Cupples LA, Hannan MT, Gagnon DR, Herbert A, Kiel DP (2002) Genome screen for quantitative trait loci contributing to normal variation in bone mineral density: the Framingham study. J Bone Miner Res 17:1718–1727 [DOI] [PubMed] [Google Scholar]
- Kelly PJ, Hopper JL, Macaskill GT, Pocock NA, Sambrook PN, Eisman JA (1993) Changes in axial bone density with age: a twin study. J Bone Miner Res 8:11–17 [DOI] [PubMed] [Google Scholar]
- Klein RF, Calos AS, Vartanian A, Chambers VK, Turner RJ, Phillips T, Belknap JK, Orwoll ES (2001) Confirmation and fine mapping of chromosomal regions influencing peak bone mass in mice. J Bone Miner Res 16:1953–1961 [DOI] [PubMed] [Google Scholar]
- Klein RF, Mitchell SR, Phillips TJ, Belknap JK, Orwoll ES (1998) Quantitative trait loci affecting peak bone mineral density in mice. J Bone Miner Res 13:1648–1656 [DOI] [PubMed] [Google Scholar]
- Koller DL, Econs MJ, Morin P, Christian JC, Hui SL, Parry P, Curran ME, Rodriguez LA, Conneally PM, Joslyn G, Peacock M, Johnston CCJ, Foroud T (2000) Genome screen for QTLs contributing to normal variation in bone mineral density and osteoporosis. J Clin Endocrinol Metab 85:3116–3120 [DOI] [PubMed] [Google Scholar]
- Koller DL, Schriefer J, Sun Q, Shultz KL, Donahue LR, Rose CJ, Foroud, T, Beamer, WG, Turner CH (2003) Genetic effects for femoral biomechanics, structure and density in C57BL/6J and C3H/HeJ inbred mouse strains. J Bone Miner Res 18:1758–1765 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Lander ES (1995) Complete multipoint sib-pair linkage studies. Am J Hum Genet 57:439–454 [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- Looker AC, Orwoll ES, Johnston CCJ, Lindsay RL, Wahner HW, Dunn WL, Calvo M, Harris TB, Heyse SP (1997) Prevalence of low femoral bone density in older US adults from NHANES III. J Bone Miner Res 12:1761–1768 [DOI] [PubMed] [Google Scholar]
- Melton L, Chrischilles E, Cooper C, Lane AW, Riggs BL (1992) How many women have osteoporosis? J Bone Miner Res 7:1005–1010 [DOI] [PubMed] [Google Scholar]
- Melton L Jr (1995) How many women have osteoporosis now? J Bone Miner Res 10:175–177 [DOI] [PubMed] [Google Scholar]
- Niu T, Chen C, Cordell H, Yang J, Wang B, Wang Z, Fang Z, Schork NJ, Rosen CJ, Xu X (1999) A genome-wide scan for loci linked to forearm bone mineral density. Hum Genet 104:226–233 10.1007/s004390050940 [DOI] [PubMed] [Google Scholar]
- Orwoll ES, Belknap JK, Klein RF (2001) Gender specificity in the genetic determinants of peak bone mass. J Bone Miner Res 16:1962–1971 [DOI] [PubMed] [Google Scholar]
- Peacock M, Turner CH, Econs MJ, Foroud T (2002) Genetics of osteoporosis. Endocr Rev 23:303–326 [DOI] [PubMed] [Google Scholar]
- Pietschmann F, Breslau NA, Pak CY (1992) Reduced vertebral bone density in hypercalciuric nephrolithiasis. J Bone Miner Res 7:1383–1388 [DOI] [PubMed] [Google Scholar]
- Reed BY, Gitomer WL, Heller HJ, Hsu MC, Lemke M, Padalino P, Pak CY (2002) Identification and characterization of a gene with base substitutions associated with the absorptive hypercalciuria phenotype and low spinal bone density. J Clin Endocrinol Metab 87:1476–1485 [DOI] [PubMed] [Google Scholar]
- Reed BY, Heller HJ, Gitomer WL, Pak CY (1999) Mapping a gene defect in absorptive hypercalciuria to chromosome 1q23.3-q24. J Clin Endocrinol Metab 84:3907–3913 [DOI] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Pottelberg I, Goemaere S, Zmierczak H, DeBacquer D, Kaufman JM (2003) Deficient acquisition of bone during maturation underlies idiopathic osteoporosis in men: evidence from a three-generation family study. J Bone Miner Res 18:303–311 [DOI] [PubMed] [Google Scholar]
- Wilson SGR, Bansal A, Chiano M, Lindersson M, Langdown M, Prince RL, Thompason D, Thompson E, Bailey M, Kleyn PW, Sambrook MM, Spector TD (2003) Comparison of genome screens for two independent cohorts provides replication of suggestive linkage of bone mineral density to 3p21 and 1p36. Am J Hum Genet 72:144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Consensus Development Conference (1991) Prophylaxis and treatment of osteoporosis. Am J Med 90:107–110 [DOI] [PubMed] [Google Scholar]