Abstract
This article provides evidence that selection has been a significant force during the evolution of the human mitochondrial genome. Both gene-by-gene and whole-genome approaches were used here to assess selection in the 560 mitochondrial DNA (mtDNA) coding-region sequences that were used previously for reduced-median-network analysis. The results of the present analyses were complex, in that the action of selection was not indicated by all tests, but this is not surprising, in view of the characteristics and limitations of the different analytical methods. Despite these limitations, there is evidence for both gene-specific and lineage-specific variation in selection. Whole-genome sliding-window approaches indicated a lack of selection in large-scale segments of the coding region. In other tests, we analyzed the ratio of nonsynonymous-to-synonymous substitutions in the 13 protein-encoding mtDNA genes. The most straightforward interpretation of those results is that negative selection has acted on the mtDNA during evolution. Single-gene analyses indicated significant departures from neutrality in the CO1, ND4, and ND6 genes, although the data also suggested the possible operation of positive selection on the AT6 gene. Finally, our results and those of other investigators do not support a simple model in which climatic adaptation has been a major force during human mtDNA evolution.
Introduction
The mitochondrial genome has been the most widely used system for the investigation of the evolutionary history of our species. It has been the system of choice because of its high rate of sequence divergence and its uniparental, maternal inheritance. As a result of the former, there is a high level of “signal” for analysis, whereas the latter property allows evolutionary histories to be reconstructed without the complexities imposed by recombination of paternal and maternal genomes.
Previous studies have shown that the evolution of human mtDNA is characterized by the emergence of distinct lineages or haplogroups among the three major ethnic groups (Torroni and Wallace 1994; Macaulay et al. 1999; Finnilä et al. 2001; Herrnstadt et al. 2002). These mtDNA haplogroups have been used to trace the evolutionary roots of modern humans and their early population dispersals. However, those conclusions were based on simple models of evolution, including the assumption that the divergence of human mtDNA obeyed a molecular clock, that selection had been negligible, and that the striking differences in the worldwide distribution of mtDNA haplogroups reflected founder events. In recent years, there has been increasing evidence that these simple models of mtDNA evolution are inadequate and that selection has played a role in shaping the evolution of the mitochondrial genome (reviewed by Gerber et al. [2001]), including human mtDNA (Hasegawa et al. 1998; Nachman 1998; Wise et al. 1998; Torroni et al. 2001b; Moilanen and Majamaa 2003; Moilanen et al. 2003).
If a neutral model of evolution does not apply to human mtDNA sequences, then our picture of human evolution and population dispersal will have to be re-evaluated with a more appropriate nonneutral model. There is broad agreement that modern humans originated in Africa and that they emerged from there to populate the rest of the world. This model is supported not only by mtDNA analyses, but also by studies of Y-chromosome and nuclear microsatellite data (reviewed by Excoffier [2002]). However, if selection has been operating, then the previous estimates of the rate of mtDNA evolution are called into question, as are the dates for various population events (e.g., Hasegawa et al. [1998]). There are ample reasons to be cautious about “one size fits all” mtDNA divergence rates (Howell et al. [2003] and references therein). The issue of selection versus neutral evolution is not a springboard for challenging or overturning previous mtDNA analyses on a wholesale basis. Rather, the point is that, pending the development and application of more realistic models of mtDNA evolution, the uncertainty of dating is increased.
Previous analyses of selection in human mtDNA have often focused on a single protein-encoding mitochondrial gene, and they have relied on methods, such as the MacDonald-Kreitman test, that compare intraspecies and interspecies variation (reviewed by Kreitman [2000] and Gerber et al. [2001]). It has been observed in several such studies—and in several different species—that there is an excess of nonsynonymous mtDNA polymorphisms relative to fixed sequence changes, a result that supports the mildly deleterious model of evolution (e.g., Nachman et al. 1996, 1998; Rand and Kann 1996; Hasegawa et al. 1998). Other analytical approaches have also provided evidence for the action of selection on mtDNA (Templeton 1996; Wise et al. 1998). Mishmar et al. (2003) recently obtained evidence for selection using a gene-by-gene approach, and these investigators concluded that climate adaptations were a major evolutionary driving force.
In addition to gene-by-gene analysis, there are also whole-genome sliding-window approaches, although these are just now being applied to human mtDNA sequences. Ballard (2000a, 2000b) used both types of approaches in his analysis of 22 complete Drosophila mitochondrial genomes. He showed that Drosophila mtDNA evolution was not compatible with the neutral model, and he was able to discern lineage-specific selection in the patterns of synonymous and nonsynonymous polymorphisms. We report here our tests of human mtDNA evolution with a set of 560 coding-region sequences that were used elsewhere for phylogenetic analyses (Herrnstadt et al. 2002). Both gene-by-gene and whole-genome approaches were used to broaden the analysis.
Material and Methods
mtDNA Sequences and Population Sampling
We have assembled and analyzed complete mtDNA coding-region sequences for 560 maternally unrelated individuals of European, African, and Asian descent. In these analyses, the coding region spans nucleotide positions (nts) 577–16023, and it includes a small number of intergenic noncoding nucleotides. The 560 mtDNA coding-region sequences used in this analysis are published on the MitoKor Web site (Herrnstadt et al. 2002), and these sequences have recently been corrected for errors (Herrnstadt et al. 2003).
These 560 mtDNA sequences include 435 that belong to European haplogroups, 69 that belong to Asian/Native American haplogroups, and 56 that belong to African haplogroups. It should be noted that, since the article by Herrnstadt et al. (2002), there have been some recommended changes to haplogroup nomenclature. What was termed “haplogroup L1” in that publication refers to a paraphyletic group of mtDNA sequences. L1a sequences have been suggested to be members of the “root” clade of modern humans, and they are now termed “L0a” (e.g., Mishmar et al. [2003]). The L0+L1 sequences analyzed here include representatives from clade L0a and from haplogroups L1b and L1c. We have “lumped” these clades together here in order to analyze the largest number of sequences. It should also be noted that the haplogroup L3 sequences refer here more narrowly to the sub-Saharan branches (in the broadest definition, haplogroup L3 would include sequences from superclades M and N). What was originally termed “haplogroup K” is now recognized to be a branch of haplogroup U, and haplogroup U—as used here—includes both U and K sequences. Finally, it appears that our haplogroup I mtDNA sequences (Herrnstadt et al. 2002) may be more appropriately assigned to superclade N1.
The 560 mtDNA sequences were obtained from individuals who live or lived in the United States or in the United Kingdom (Herrnstadt et al. 2002), so the question of population sampling arises. All of the mtDNA haplogroups are geographically widespread. The earlier studies concluded that mtDNAs that belonged to European haplogroups showed little, if any, geographical structuring. As more analyses have accumulated, however, it now appears that some structuring does occur (Richards et al. 2002), although these haplogroups are found, with rare exception, in populations throughout Europe and the Near East. In a similar fashion, the African and Asian haplogroups show continental, widespread distributions (e.g., Salas et al. [2002]). The balance between random genetic drift and selection is a function of population size, with the former making a relatively greater contribution in small populations. On the basis of the available information, there is no evidence that our sampling scheme is biased and that therefore we are detecting the effects of drift rather than selection.
Analysis of Synonymous and Nonsynonymous Substitutions
For some tests of neutrality, we compared the numbers of synonymous (S) and nonsynonymous (A) substitutions for one group of sites or sequences with those of a second group. In these tests, we used a “simple counts” approach to derive the numbers of substitutions. To maintain consistency among the different tests and to avoid any weighting of different substitutions, a substitution was counted only once for each continental group of sequences (European, Asian/Native American, and African). That is, even when a substitution has arisen multiple times independently (homoplasy) within, for example, European sequences, it is still counted as a single substitution. For example, a substitution at nt 5046 has arisen in both African and European sequences, and it is counted as a change in both sequence sets. In contrast, a substitution at nt 13393 has arisen in both European haplogroups J and U, but it is counted as a single change in our analyses. This approach will be biased conservatively in the case of negative selection, because it will tend to more severely underestimate the occurrences of synonymous substitutions. For example, in table 1 of Herrnstadt et al. (2002), there are 147 substitutions that map to protein-encoding genes and that occur in multiple haplogroups. Taking these substitutions as an estimator of homoplasy, only 50 (34%) of those sequence changes are nonsynonymous. That is, homoplastic substitutions tend to be synonymous (see also Moilanen and Majamaa [2003]). Conversely, our approach will tend to inflate or overestimate the numbers of nonsynonymous substitutions, a point that will be addressed in the relevant portion of the “Results” section.
Whole-Genome Analysis of mtDNA Sequences
Investigations into whether there is regional variation within the human mtDNA genome—in terms of a global, uniform model of neutral evolution in all regions of the mitochondrial genome—were performed using PLATO (Partial Likelihoods Assessed through Optimisation), version 2.11, as described elsewhere (Grassly and Holmes 1997). PLATO uses a likelihood approach with sliding-window analysis to detect anomalously evolving regions in DNA sequences, where such regions are most likely created by selection.
The PLATO program requires a sequence alignment, as well as a phylogenetic tree. For the analyses reported here, the HKY model of sequence evolution was used. PLATO calculates the independent likelihoods at each site within a window, as well as a measure of the average likelihood with respect to the window. Windows with likelihood values that fall below certain values are correlated with the poorest fit to the global model derived from the whole sequence. The statistical significance of regions of low likelihood are then determined using a Monte Carlo simulation-based method in which the regions are assigned a Z value that indicates how far the likelihood is from the expected value, under the global model. PLATO also performs a Bonferroni correction to adjust the significance level to account for multiple comparisons, because the program tests all possible window sizes from the assigned minimum window size (10 bp) to half the sequence length (Grassly and Holmes 1997). Owing to the computational intensity of maximum likelihood methods, all tests here were run with mtDNA sequences broken down into four overlapping segments that spanned nts 577–4877, 4327–8577, 8077–12327, and 11827–16077. That is, our approach tested for selection in 4-kb-sequence blocks in an effort to reduce the “noise” that is likely to arise from analysis of smaller segments of the mitochondrial genome. As a result, the maximum number of sequences that could be analyzed in a single test was four.
Results
Whole–Mitochondrial Genome Analysis/Sliding-Window Tests
Previous tests of the effects of selection during human mtDNA evolution have generally involved a single gene. Our initial approach was to use a whole-genome sliding-window analysis (as implemented in PLATO), both to detect overall violations of neutrality and then to determine if any such violations were region specific. Sliding-window analysis makes use of windows of varying sizes (regions of an mtDNA alignment) to discern mtDNA regions that do not fit with a global (null) phylogenetic hypothesis that is calculated a priori from the data. Because of computational limits, our sliding-window tests were performed with small sets of human mtDNA sequences drawn from the total database of 560 coding-region sequences.
The first such tests were performed with African mtDNA coding-region sequences, because they are the most divergent and should thus have the highest signal. One test involved sequences from “root” clade L0a (149), haplogroup L1c (173 and 207), and haplogroup L1b (514). (The numbers in parentheses refer to the individual sequence numbers reported elsewhere [Herrnstadt et al. 2002]). This test failed to indicate any significant departures from neutrality within any of the four mitochondrial coding-region segments. We next tested haplogroup L2 sequences, because of the previous report of departure from a molecular clock in the haplogroup (Torroni et al. 2001b). A sliding-window analysis of haplogroup L2 sequences was performed only with haplogroups L2d (153), L2b (175), and L2a (401 and 142). Haplogroup L3 sequences were also tested on a group basis, because this haplogroup gave rise to Asian and European mtDNAs. The sequences used in this test included haplogroups L3b (164), L3d (140), and L3e (216 and 180). Another test included coding-region sequences from haplogroups L3b (164 and 309), L3d (381), and L3e (216). Finally, we tested for neutrality in a more diverged set of African mtDNAs that comprised sequences from clade L0a (149) and from haplogroups L1b (158), L2a (142), and L3e (180). In all of these tests, there were no coding-region segments in which departure from neutrality could be detected.
Additional sliding-window tests were then performed to compare mtDNA coding-region sequences from different ethnic groups. Thus, we analyzed sequences from clade L0a (149) and haplogroups L3e (216) and H (100 and 526) to investigate haplogroup H, which is the most prevalent European mtDNA haplogroup and which samples a population that has undergone a relatively recent population expansion. Another test involved sequences from haplogroups L1b (514), B (205), and K (416 and 223). Haplogroup K is a subclade of European haplogroup U. A test of sequences from haplogroups L1c (194), J (243 and 370), and K (474) was performed to investigate evolution in other major European haplogroups. The final test involved sequences from clade L0a (149), B (179 and 205), and H (100), which involves sequences from all three major ethnic groups (African, Asian, and European, respectively). Again, none of these tests showed any regions of the mitochondrial genome that departed from a neutral model of evolution.
Gene-by-Gene Analysis of Synonymous and Nonsynonymous Substitutions in Protein-Encoding mtDNA Genes
To investigate the substitution rates in each protein encoded by the human mtDNA and to assess the role of selection, the numbers of synonymous (S) and nonsynonymous (A) substitutions in the 13 protein-encoding genes were investigated. In the first analyses, the substitutions were stratified into two classes on the basis of our reduced-median-network analyses (Herrnstadt et al. [2002]; other examples of phylogenetically stratified contingency analyses can be found in articles by Templeton [1996] and Wise et al. [1998]). The first class includes those polymorphisms that we designate as “haplogroup associated” and that are defined operationally as “those that occur at an internal node within a network.” In practical terms, each of these substitutions defines a subclade of at least two mtDNA sequences, although some of these substitutions occurred in the ancestor of the entire haplogroup (see further discussion in the article by Herrnstadt et al. [2002]). It should also be noted that this group includes haplogroup-associated substitutions that are homoplastic and that have also arisen independently in other mtDNA haplogroups. Among the haplogroup-associated substitutions, there was a total of 89 nonsynonymous and 330 synonymous polymorphisms, thus yielding an A/S ratio of 0.27 (table 1).
Table 1.
Analysis of Nonsynonymous (A) and Synonymous (S) Haplogroup-Associated and Private Polymorphisms[Note]
|
No. of Haplogroup-Associated Substitutions |
No. of PrivateSubstitutions |
|||||||
| mtDNAGene | A | S | A/S | A | S | A/S | Pa | NIb |
| ND1 | 9 | 26 | .35 | 23 | 40 | .58 | NS | 1.66 |
| ND2 | 12 | 31 | .39 | 15 | 44 | .34 | NS | .88 |
| CO1 | 3 | 54 | .06 | 28 | 49 | .57 | <.0001 | 10.29 |
| CO2 | 4 | 13 | .31 | 10 | 29 | .35 | NS | 1.12 |
| AT8 | 3 | 5 | .60 | 13 | 12 | 1.08 | NS | 1.81 |
| AT6 | 14 | 17 | .82 | 34 | 21 | 1.62 | NS | 1.97 |
| CO3 | 3 | 17 | .18 | 18 | 33 | .55 | ∼.05 | 3.09 |
| ND3 | 3 | 9 | .33 | 3 | 20 | .15 | NS | 0.45 |
| ND4L | 1 | 8 | .13 | 5 | 8 | .63 | NS | 5.00 |
| ND4 | 2 | 41 | .05 | 25 | 53 | .47 | <.0004 | 9.67 |
| ND5 | 18 | 51 | .35 | 33 | 88 | .38 | NS | 1.06 |
| ND6 | 3 | 22 | .14 | 17 | 21 | .81 | <.0116 | 5.94 |
| CYb | 14 |
36 |
.39 | 33 |
55 |
.60 | NS | 1.54 |
| Total | 89 | 330 | .27 | 257 | 473 | .54 | ≪.0001 | 2.02 |
Note.— Both types of polymorphisms are described in greater detail in the “Methods” section (see also Herrnstadt et al. [2002]).
P values were determined with Fisher’s exact test. “NS” indicates that the difference between the Ka/Ks ratios was not statistically significant (P>.05).
We calculated an intraspecies neutrality index (NI) that is based on the interspecies index developed and used by Rand and Kann (1996). NI is a ratio of ratios and is calculated as (A-private/A-haplo)/(S-private/S-haplo). The value will be 1.0 under strict neutrality.
The second class comprises “private polymorphisms,” or those changes that occur at the tips of individual branches within a network or phylogenetic tree. Under the operational definition that is the basis of our conservative approach, the vast majority of these substitutions occur in only one of the 560 mtDNA sequences, although some were homoplastic private polymorphisms that occurred independently in two or more haplogroups. There was a total of 257 nonsynonymous private polymorphisms and 473 synonymous ones (table 1), giving an A/S ratio of 0.54. The difference in A/S ratios between the two classes is highly significant (P≪.0001 with Fisher’s exact test) (table 1). In addition, the calculated neutrality index (NI) (see footnote “b” of table 1; also see Rand and Kann [1996]) was 2.02. The difference between the two groups of substitutions is time depth, or evolutionary age. The private substitutions are younger, in terms of evolutionary age, and they will have been exposed to selection for a relatively short period of evolutionary time. The haplogroup-associated substitutions will vary in their ages, with some being very old, in terms of human evolution, and others being much more recent. We also note that the private polymorphisms are defined operationally and that, with a larger set of sequences, some of these substitutions might be found to be haplogroup associated. Nevertheless, the point is that “older” sequence changes in the human mtDNA-coding region have relatively fewer nonsynonymous substitutions than a set of “younger” sequence changes, even when adjusted for the numbers of synonymous substitutions. This relative “loss” of nonsynonymous substitutions during evolution suggests that the human mtDNA has been subjected to negative selection or to a relatively recent relaxation of selective constraints.
Evidence for negative selection was also obtained when the sequences were stratified in a different way. There is a broad consensus that modern humans originated in Africa and then spread to Asia and Europe. Under this model, therefore, Asian and European mtDNAs will be younger, in evolutionary terms, than African mtDNAs. The NI for the African superclade sequences is 4.75, whereas the value for the European mtDNAs is 1.39. The numbers of substitutions in the Asian mtDNAs are much smaller, and the NI of 1.04 has a greater degree of uncertainty than the other NI values. Again, the relatively lower proportion of nonsynonymous substitutions in the evolutionarily older African mtDNAs indicates that negative selection has acted on the human mitochondrial genome during evolution.
In these analyses, a “most conservative counts” approach has been used, in which a substitution is scored as “private” once for each haplogroup, even if network analyses indicate that the substitution has arisen multiple times within a haplogroup. We did estimate an A/S ratio and found that it was not substantially changed when we included such homoplastic private polymorphisms (data not shown). Therefore, there is no evidence that our approach is grossly biased, although this is an area that would benefit from further analysis with much larger sequence sets.
We next considered the action of selection on individual mitochondrial genes, and A/S ratios and NI values were calculated on an individual basis for the 13 protein-encoding mitochondrial genes (table 1). For 11 of the 13 genes, the A/S ratio was lower for the haplogroup-associated polymorphisms than for the private polymorphisms, although only in the case of the CO1, ND4, and ND6 genes was this result statistically significant (see also the results of Rand and Kann [1996] and Hasegawa et al. [1998]). The ratio for the CO3 gene was of borderline significance. We note that the NI values for the first two genes were close to 10, results that indicate that negative selection has been relatively strong. The most marked exception to NI values >1.0 (and therefore suggestive of negative selection) was the ND3 gene (table 1), but the numbers of sequence changes were relatively low. Previous studies of the ND3 gene in sets of more highly diverged mtDNA sequences have indicated the operation of negative selection during evolution (Nachman et al. 1994; Hasegawa et al. 1998). The other noteworthy results were obtained for the AT6 gene, which had the largest A/S ratio (table 1). As will be discussed in greater detail below, the cumulative results suggest the action of positive selection on this gene, despite the fact that the NI for this gene is >1.0, a result that indicates negative selection. Overall, the results shown in table 1 indicate that, in the coding region of the mitochondrial genome, selection acts to reduce the numbers of nonsynonymous changes, although there appears to be variation in the strength of selection on different genes.
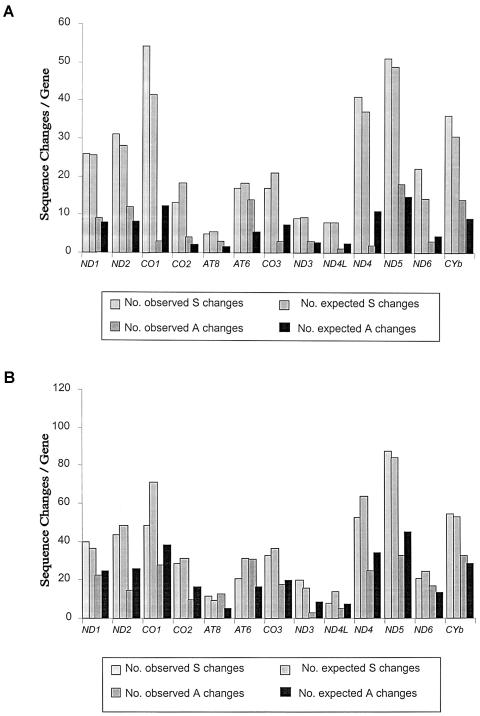
As another test of selection, we compared the numbers of observed substitutions relative to the number expected (see fig. 1 and the details in the legend). Neutral evolution predicts that all genes should evolve at a relatively constant rate (the molecular clock). Specifically, there should not be any significant discrepancies between the numbers of observed and predicted substitutions, but this is not the result that we obtained. For both haplogroup-associated and private polymorphisms, the results for synonymous substitutions are broadly compatible with a molecular clock, the null hypothesis. That is, the number of synonymous substitutions was proportional to the number of sites in the gene that could undergo such sequence changes. At the same time, we note that the fit to the neutral model was of borderline statistical significance, especially for the private polymorphism class (see the similar results of Hasegawa et al. [1998]).
Figure 1.
Plots of sequence changes/gene for haplogroup-associated (A) and private (B) polymorphisms. The plots contain the observed number of substitutions within each of the two major classes (the exact numbers are presented in table 1). The expected numbers of substitutions in each class are based on the total number of changes in that class and the predicted number of sites in each gene (determined using the DnaSP program [Rozas and Rozas 1999]). For the haplogroup-associated synonymous polymorphisms, a contingency test of the neutral expectation yielded a P value >.2 (12 df); thus, the data provide no evidence to reject the neutral model. These results indicate that there are no mutational hotspots for synonymous substitutions in the coding region. In contrast, when the nonsynonymous changes were analyzed, there was a clear deviation from the neutral expectation, and the contingency test yielded a P value <.001. For the private synonymous polymorphisms (B), a contingency test against the neutral expectation yielded a P value >.05 (12 df); thus, the neutral model holds. In contrast, when the data for the nonsynonymous changes were analyzed, there was a clear deviation from the neutral expectation, and the contingency test yielded a P value <.001.
In contrast, the nonsynonymous changes show marked deviations from the neutral model for both classes of polymorphisms (P<.001) (fig. 1). For the haplogroup-associated polymorphisms, the mtDNA genes that accounted for the greatest portion of this deviation were CO1 and ND4, which had the largest deficiency of nonsynonymous changes (expected vs. observed), and the AT6 gene, which had the largest excess. For the private polymorphisms group, this deviation was due to both anomalously large (ND2 and ND5) and small (AT6) expected-versus-observed ratios. We thus observed an “excess” of nonsynonymous substitutions for the AT6 gene (fig. 1) for both haplogroup-associated and private polymorphisms. These results are similar to those of Mishmar et al. (2003). This gene is one of the most conserved when interspecies comparison are made, but they observed that it had the highest level of nonsynonymous substitutions on an intraspecies basis (see their fig. 2). These results suggest the occurrence of balancing or positive selection during evolution of this gene, or—alternatively—the relaxation of negative selection (see below).
Tests for Climatic Adaptation
An obvious question that we asked, in view of the recent report of Mishmar et al. (2003), was whether there was any evidence in our coding-region sequences for climatic adaptation. Those investigators analyzed a total of 104 complete mtDNA sequences that were distributed among the major haplogroups. They assumed that African populations represent peoples that evolved in a tropical or subtropical climate, whereas the climates were arctic and subarctic for the Asian (Siberian)/Native American populations and were temperate for the European populations. They calculated Ka/Ks ratios in two different ways: as Ka/Ks+constant and as Ka/Ks+Ka with the Nei-Gojobori method (Rozas and Rozas 1999), which uses pairwise comparison of sequences; they then compared the ratios for the different “climate populations” with a Wilcoxon rank-sum test (see p. 172 and fig. 3 in Mishmar et al. [2003] for further details). They concluded that there were gene-specific differences in Ka/Ks ratios and that these differences reflected climatic adaptation.
The role of climatic adaptation in human evolution—and its possible effects on the mitochondrial genome—is an important issue, and we investigated it in the following way. We used our “conservative counts” data for each of the protein-encoding genes (table 1) and further stratified them into African/tropical, European/temperate, and Asian–Native American/arctic climate groups. We then used Fisher’s exact test to determine if the A/S ratios significantly differed for each gene and for each of the pairwise comparisons of major “climate” groups.
The results of the A/S ratio analyses for the African/tropic–European/temperate comparison are shown in table 2. It was found that only two genes, ND1 and CYb, showed a statistically significant difference between these two climate groups. For comparison, Mishmar et al. (2003) reported significant differences in the Ka/Ks+constant ratios of the ND2, CO1, CO2, AT6, ND4, ND5, ND6, and CYb genes (see their fig. 3). When Ka/Ks+Ka ratios were used, they reported significant differences in the ND1, ND2, CO1, CO2, AT6, AT8, ND5, ND6, and CYb genes (the ratio for the ND4 gene appears to be of borderline statistical significance; see their table 2). It is difficult to believe that their tests, which involve fewer sequences, are more sensitive than ours, so an explanation for the disparate results is not at hand, although it is a concern that the numbers of substitutions for many of the genes are relatively small.
Table 2.
Comparisons of A/S Ratios among European, Asian, and African Sequence Sets
|
No. of Substitutions(A, S)a |
No. of Substitutions(A, S)a |
No. of Substitutions(A, S)a |
|||||||
| Gene | AFR | EUR | Pb for AFR vs. EUR | AFR | ASIA | Pb for AFR vs. ASIA | ASIA | EUR | Pb for ASIA vs. EUR |
| ND1 | 4, 21 | 20, 34 | .040 | 4, 21 | 8, 10 | .083 | 8, 10 | 20, 34 | .589 |
| ND2 | 5, 24 | 23, 47 | .145 | 5, 24 | 3, 14 | 1.000 | 3, 14 | 23, 47 | .255 |
| CO1 | 7, 30 | 25, 64 | .370 | 7, 30 | 5, 18 | 1.000 | 5, 18 | 25, 64 | .608 |
| CO2 | 3, 11 | 8, 24 | 1.000 | 3, 11 | 8, 6 | .120 | 8, 6 | 8, 24 | .048 |
| AT8 | 2, 6 | 10, 11 | .408 | 2, 6 | 6, 2 | .132 | 6, 2 | 10, 11 | .238 |
| AT6 | 7, 14 | 32, 22 | .071 | 7, 14 | 12, 4 | .005 | 12, 4 | 32, 22 | .378 |
| CO3 | 4, 18 | 23, 30 | .063 | 4, 18 | 0, 12 | .273 | 0, 12 | 23, 30 | .005 |
| ND3 | 2, 6 | 5, 19 | 1.000 | 2, 6 | 1, 4 | 1.000 | 1, 4 | 5, 19 | 1.000 |
| ND4L | 0, 6 | 5, 9 | .260 | 0, 6 | 1, 2 | .333 | 1, 2 | 5, 9 | 1.000 |
| ND4 | 3, 31 | 19, 61 | .074 | 3, 31 | 10, 15 | .009 | 10, 15 | 19, 61 | .129 |
| ND5 | 8, 41 | 36, 95 | .172 | 8, 41 | 10, 16 | .047 | 10, 16 | 36, 95 | .345 |
| ND6 | 2, 15 | 13, 24 | .106 | 2, 15 | 7, 4 | .010 | 7, 4 | 13, 24 | .162 |
| CYb | 7, 33 | 31, 51 | .024 | 7, 33 | 12, 15 | .026 | 12, 15 | 31, 51 | .651 |
mtDNA sequences are stratified by continent of origin (AFR = Africa; EUR = Europe; ASIA = Asia/Native American). A = nonsynonymous substitutions; S = synonymous substitutions. Note that, in these analyses, haplogroup-associated and private substitutions are pooled into a single number.
P values were determined with Fisher’s exact test.
One potential concern is that our set of European coding-region sequences was predominantly composed of haplogroup H sequences (226 of 435). Although this proportion is representative of the population, the results obtained might largely reflect evolution in haplogroup H rather than in all of the European mtDNA haplogroups. Therefore, we reran the European-African analyses with a smaller set of sequences in which only 50 randomly chosen haplogroup H mtDNAs were included (thereby better “balancing” the number of sequences per haplogroup). Under these conditions, significant differences were obtained for the CO3 and CYb genes (data not shown). Using the smaller set of haplogroup H mtDNAs decreases the CO3 P value (from .063 to .023), whereas that for the ND1 gene increases (from .040 to .072).
The results for the African/tropical versus Asian–Native American/arctic comparisons are also shown in table 2. With our sequences, we found significant differences in the AT6, ND4, ND5, ND6, and CYb genes. For comparison, Mishmar et al. (2003) found differences in the ND1, ND2, ND5, ND6, CO1, CO3, AT6, and CYb genes through analysis of the Ka/Ks+constant ratios (their fig. 3). With the Ka/Ks+Ka ratio approach, significant differences were reported for the ND1, ND3, ND5, ND6, CO1, CO3, AT6, AT8, and CYb genes (table 2 of Mishmar et al. [2003]). For one gene and one comparison—the AT6 gene for the African/Asian–Native American comparison—Mishmar et al. (2003) used an approach similar to ours, and the results of both approaches agree (see their fig. 3). Finally, we performed a European/temperate versus Asian/arctic comparison and found significant differences in A/S ratios for the CO2 and CO3 genes.
Our results, therefore, do not agree with those of Mishmar et al. (2003), although we do obtain a number of significant differences between continental sequence sets. The most striking results pertain to the AT6 gene. For both the Asian and European sequence sets, but not for the African sequences, the number of nonsynonymous substitutions is greater than the number of synonymous ones. Again, these results suggest that this gene has been subject to balancing or positive selection, at least in the Asian and European mtDNA gene pools. Furthermore, these results show why analysis of the total or combined sequence sets (table 1) yielded an NI value >1.0. A value <1.0 would be expected for positive selection, but the relatively larger numbers of substitutions contributed by the African sequences obscure the disparate trend in the European and Asian sequences. Although highly suggestive, it is not yet clear if these continental (or climatological) differences reflect “real” biological effects, or if we are still working with an insufficient number of sequences.
Discussion
We have applied a number of tests of selection, both gene by gene and whole genome, to the coding regions of 560 human mtDNAs that encompass all three major ethnic groups (African, Asian, and European). Evidence for selection was found in some tests but not in others. Furthermore, it is often difficult to “isolate” the effects of selection from other processes, such as recent population expansions or bottlenecks. Nevertheless, the preponderance of evidence indicates that negative selection operated on the coding region of the human mitochondrial genome during evolution. Selection appears to have influenced the pattern of sequence divergence for most, and perhaps all, of the 13 protein-encoding genes, although there is evidence that some genes have been more vulnerable to the effects of selection than others (table 1) (see also Rand and Kann [1996] and Hasegawa et al. [1998]). These conclusions are supported by other recent analyses, such as those of Moilanen and Majamaa (2003) and Moilanen et al. (2003). At the same time, evolution is a hugely complicated process, and we caution against drawing conclusions at this time that are firm or simple. For example, we have interpreted the results of A/S analyses as indicating the operation of negative selection, but a relaxation of selective constraints is also possible. Similarly, the excess nonsynonymous substitutions in the AT6 gene may reflect either positive selection or the relaxation of negative selection in European and Asian populations.
When the PLATO sliding-window whole-genome method was used in our analysis, there was no evidence for the action of selection when large spans of the coding region were analyzed. We used the most sensitive of the whole-genome methods that were employed by Ballard (2000a, 2000b) in his analysis of selection in Drosophila, but the Drosophila mtDNAs are clearly much more highly diverged than are human mtDNAs. In contrast, Moilanen et al. (2003) have recently performed a finer-grained sliding-window analysis. They found that the departure from neutrality increases as one slides the window through the coding region; for example, the F* value is more negative in the region of nts 13000–16000 than it is in the region of nts 1000–3000 (see their fig. 3). Furthermore, they found that there was a region of the ND5 gene that showed less diversity in the European haplogroup JT sequences. Our sliding-window analyses were coarser grained, and we were focused on detecting selection in large regions of the mtDNA.
The most striking results were obtained from our phylogenetically stratified contingency analyses of synonymous and nonsynonymous substitutions. There was a highly significant “loss” of nonsynonymous substitutions among relatively old site changes, results that suggest the operation of negative selection. These results thus agree with and extend previous single-gene tests for the operation of selection (see also Moilanen and Majamaa [2003] and Moilanen et al. [2003]). Those previous studies involved comparisons of intraspecific and interspecific mtDNA variation, whereas we relied strictly on analysis of intraspecific sequence changes. We avoided interspecies analyses here because there are long-standing concerns about possible anomalies caused by the use of nonhuman primate mtDNA as an outgroup sequence in phylogenetic analyses of human mtDNA sequences (see especially the broader discussion in Gerber et al. [2001] on the limitations of the MacDonald Kreitman test). Weiss and von Haeseler (2003) have recently analyzed complete mtDNA sequences for humans, chimpanzees, bonobos, gorillas, and orangutans; they found that sequence evolution has not been homogeneous within primates. It is the availability of large mtDNA sequence sets that allows the intraspecies analyses to be performed, but we raise the concern that the sequence sets may still not be large enough.
It should also be noted that, whereas the results here indicate that a substantial proportion of nonsynonymous substitutions are mildly deleterious, it is also likely that some sites have evolved under conditions either of neutrality or of positive selection (see especially Yang and Swanson [2002]). Further analysis with methods that involve site partitioning will be necessary for finer-grained analyses. The application of such methods to primate mtDNA sequences indicates that there is a small proportion of sites that have evolved under positive selection (Yang et al. 2000). Those methods should be very useful for determining which sites in the AT6 gene have been subject to positive selection and which to negative selection. Furthermore, we have not addressed the possibility that synonymous substitutions in the human mtDNA have been subject to selection, although the available evidence suggests that there is little codon-usage bias in mammalian nuclear genes (see the discussion in Zeng et al. [1998]). Finally, the present analyses have been limited to the mtDNA coding region, and the rapidly diverging control region has not been investigated here. It is widely assumed that selection has not influenced the evolution of the control region. However, negative selection has been offered as one explanation for the disparity between pedigree and phylogenetic (or population) rates of mtDNA-sequence divergence in both the coding and control regions (see the discussion in Howell et al. [2003] and references therein).
We were not able to reproduce the results of Mishmar et al. (2003), which does not in itself mean that their conclusion is wrong with regard to the role of climatic adaptation as a major force during human mtDNA evolution. However, there are a number of potential limitations to their study. (1) The Nei-Gojobori method for estimation of Ks/Ka ratios, which is based on a pairwise comparison of sequences, will overweight some sites. (2) Their assumptions about climate are simplistic. For example, European populations (and their mtDNA sequences) have evolved under conditions of marked climatic change (Torroni et al. 2001a). (3) They are unable to separate the effects of climate from other effects, including time of evolution. Thus, the results presented here indicate that selection has had a relatively greater impact on the evolutionarily “older” African mtDNAs, which is in accord with the “Out of Africa” model. In addition, Moilanen et al. (2003) have used the same methodology as have Mishmar et al. (2003) on a larger sequence set, and they showed that there were significant differences in Ka/Ks ratios when European superhaplogroups were compared. That is, there were significant differences within a single, large “climate group,” which is a result not in accord with those of Mishmar et al. (2003). If all of these issues are taken into consideration, the approach of Mishmar et al. (2003) may not be an appropriate methodology with which to study climatic adaptation, and the development of alternative methods appears essential if we are to address this important issue.
Selection, one may say with some confidence, has shaped the evolution of the human mitochondrial genome. This miniscule piece of genetic information has already been a key part of the study of human evolution and population dispersal, and it seems likely to continue playing an important role as we tease out the role of selection.
Acknowledgments
This research was supported in part by National Science Foundation grant BSC-9910871 (to N.H.) and by a Wellcome Trust Collaboration grant (to D.M.T. and N.H.). J.L.E. is a Medical Research Council Bioinformatics training fellow. We thank Barbara Howell for her help with preparation of the manuscript. The expert assistance of Dr. H.-J. Bandelt (University of Hamburg) in navigating the intricacies of haplogroup nomenclature is also gratefully acknowledged.
Electronic-Database Information
The URL for data presented herein is as follows:
- Mitokor, http://www.mitokor.com/science/560mtdnasrevision.php (for the revised 560 mtDNA coding-region sequences; “zip” and “sit” files also available)
References
- Ballard JWO (2000a) Comparative genomics of mitochondrial DNA in Drosophila simulans. J Mol Evol 51:64–75 [DOI] [PubMed] [Google Scholar]
- ——— (2000b) Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J Mol Evol 51:48–63 [DOI] [PubMed] [Google Scholar]
- Excoffier L (2002) Human demographic history: refining the recent African origin model. Curr Opin Genet Dev 12:675–682 10.1016/S0959-437X(02)00350-7 [DOI] [PubMed] [Google Scholar]
- Finnilä A, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AS, Loggins R, Kumar S, Dowling TE (2001) Does nonneutral evolution shape observed patterns of DNA variation in animal mitochondrial genomes? Ann Rev Genet 35:539–566 10.1146/annurev.genet.35.102401.091106 [DOI] [PubMed] [Google Scholar]
- Grassly NC, Holmes EC (1997) A likelihood method for the detection of selection and recombination using nucleotide sequences. Mol Biol Evol 14:239–247 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Cao Y, Yang Z (1998) Preponderance of slightly deleterious polymorphism in mitochondrial DNA: nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol Biol Evol 15:1499–1505 [DOI] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Preston G, Howell N (2003) Errors, phantom and otherwise, in human mtDNA sequences. Am J Hum Genet 72:1585–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Smejkal CB, Mackey DA, Chinnery PF, Turnbull DM, Herrnstadt C (2003) The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates. Am J Hum Genet 72:659–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M (2000) Methods to detect selection in populations with applications to the human. Annu Rev Genomics Hum Genet 1:539–559 10.1146/annurev.genom.1.1.539 [DOI] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macalay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC (2003) Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA 100:171–176 10.1073/pnas.0136972100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moilanen JS, Finnilä S, Majamaa K (2003) Lineage-specific selection in human mtDNA: lack of polymorphisms in a segment of MTND5 gene in haplogroup J. Mol Biol Evol 20:2132–2142 12949126 [DOI] [PubMed] [Google Scholar]
- Moilanen JS, Majamaa K (2003) Phylogenetic network and physicochemical properties of nonsynonymous mutations in the protein-coding genes of human mitochondrial DNA. Mol Biol Evol 20:1195–1210 10.1093/molbev/msg121 [DOI] [PubMed] [Google Scholar]
- Nachman MW (1998) Deleterious mutations in animal mitochondrial DNA. Genetica 102–103:61–69 [PubMed] [Google Scholar]
- Nachman MW, Boyer SN, Aquadro CF (1994) Nonneutral evolution at the mitochondrial NADH dehydrogenase subunit 3 gene in mice. Proc Natl Acad Sci USA 91:6364–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Brown WM, Stoneking M, Aquadro CF (1996) Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142:953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand DM, Kann LM (1996) Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol 13:735–748 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Torroni A, Bandelt H-J (2002) In search of geographical patterns in European mitochondrial DNA. Am J Hum Genet 71:1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, Lareu M-V, Sobrino B, Sánchez-Diz P, Macaulay V, Carracedo A (2002) The making of the African mtDNA landscape. Am J Hum Genet 71:1082–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR (1996) Contingency tests of neutrality using intraspecific/interspecific gene trees: the rejection of neutrality for the evolution of the mitochondrial cytochrome oxidase II gene in the hominoid primates. Genetics 144:1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, et al (2001a) A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo R, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R (2001b) Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69:1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Wallace DC (1994) Mitochondrial DNA variation in human populations and implications for detection of mitochondrial DNA mutations of pathological significance. J Bioenerg Biomembr 26:261–271 [DOI] [PubMed] [Google Scholar]
- Weiss G, von Haeseler A (2003) Testing substitution models within a phylogenetic tree. Mol Biol Evol 20:572–578 10.1093/molbev/msg073 [DOI] [PubMed] [Google Scholar]
- Wise CA, Sraml M, Eastseal S (1998) Departure from neutrality at the mitochondrial NADH dehydrogenase subunit 2 gene in humans, but not in chimpanzees. Genetics 148:409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Krabbe Pedersen A-M (2000) Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Swanson WJ (2002) Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol Biol Evol 19:49–57 [DOI] [PubMed] [Google Scholar]
- Zeng L-W, Comeron JM, Chen B, Kreitman M (1998) The molecular clock revisited: the rate of synonymous vs. replacement change in Drosophila. Genetica 102–103:369–382 9720289 [DOI] [PubMed] [Google Scholar]