Abstract
Ethylmalonic encephalopathy (EE) is a devastating infantile metabolic disorder affecting the brain, gastrointestinal tract, and peripheral vessels. High levels of ethylmalonic acid are detected in the body fluids, and cytochrome c oxidase activity is decreased in skeletal muscle. By use of a combination of homozygosity mapping, integration of physical and functional genomic data sets, and mutational screening, we identified GenBank D83198 as the gene responsible for EE. We also demonstrated that the D83198 protein product is targeted to mitochondria and internalized into the matrix after energy-dependent cleavage of a short leader peptide. The gene had previously been known as “HSCO” (for hepatoma subtracted clone one). However, given its role in EE, the name of the gene has been changed to “ETHE1.” The severe consequences of its malfunctioning indicate an important role of the ETHE1 gene product in mitochondrial homeostasis and energy metabolism.
Introduction
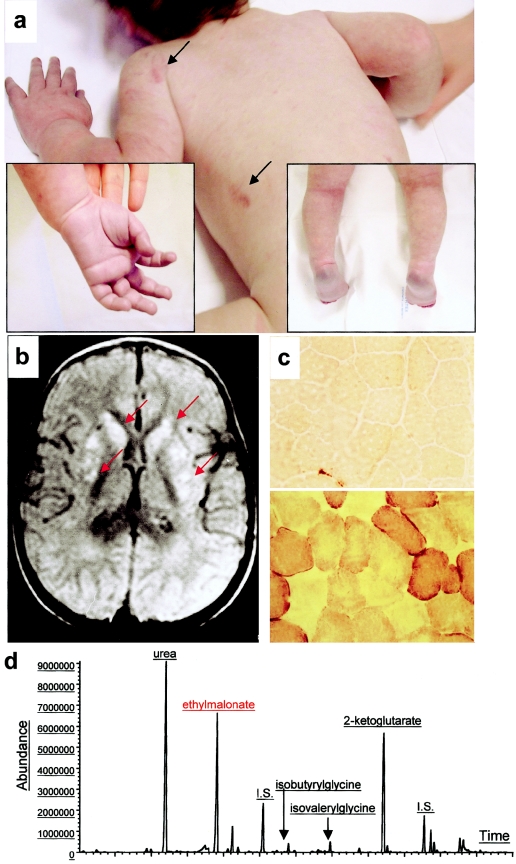
Ethylmalonic encephalopathy (EE [MIM 602473]) is an autosomal recessive disorder that was originally reported in Italian families (Burlina et al. 1991). Most of the patients with EE described thereafter have been, with a few exceptions (Yoon et al. 2001), of Mediterranean (Burlina et al. 1991, 1994; Garavaglia et al. 1994; Garcia-Silva et al. 1997; Grosso et al. 2002) or Arabic (Ozand et al. 1994) descent. EE is characterized by neurodevelopmental delay and regression, prominent pyramidal and extrapyramidal signs, recurrent petechiae, orthostatic acrocyanosis (fig. 1a), and chronic diarrhea, leading to death in the first decade of life. Symmetrical necrotic lesions in the deep gray matter structures are the main neuropathological features of the disease (fig. 1b). These lesions may be related to the toxicity of metabolites that accumulate, or they may be secondary to the vascular changes commonly observed in this disorder. EE is characterized by an unusual combination of biochemical findings, which include persistent lactic acidemia, elevated concentrations of C4 and C5 plasma acylcarnitine species, markedly elevated urinary excretion of ethylmalonic acid (EMA), and elevated C4–6 acylglycines, notably isobutyrylglycine and 2-methylbutyrylglycine. The specific activity of cytochrome c oxidase is reduced in skeletal muscle (fig. 1c) but not in fibroblasts. EMA can be derived from either the carboxylation of butyryl-CoA, as a consequence of disorders of the β-oxidation of short-chain fatty acids, or from 2-ethylmalonic-semialdehyde, the final product of the R-pathway catabolism of isoleucine (Mamer et al. 1976; Nowaczyk et al. 1998). The differential diagnosis of persistent EMA aciduria is short-chain acyl-CoA dehydrogenase deficiency, including the common variants (Corydon et al. 2001), glutaric acidemia type 2 (sometimes described as ethylmalonic adipic aciduria) (Mantagos et al. 1979), Jamaican vomiting sickness (Tanaka et al. 1976), and EE. However, none of the other conditions has been associated with the peculiar clinical features of EE, and extensive studies have consistently failed to document a primary defect, in spite of a unique clinical and biochemical phenotype. To achieve this goal, we have successfully adopted a strategy based on primary linkage analysis, followed by mutation analysis of functionally selected candidate genes.
Figure 1.
Clinical and biochemical features of EE. a, Skin areas with petechiae are indicated by arrows. The boxed pictures show acrocyanosis of hands and feet. b, On T2–fluid-attenuated-inversion-recovery (FLAIR) MRI images of a transverse section of the brain, symmetrical, patchy, high-intensity signals are present in the head of nucleus caudatus and in the putamen (arrows). c, Histochemical reaction to cytochrome c oxidase (COX) in skeletal muscle. The muscle of a patient with EE (top) shows a profound and diffuse reduction in COX reactivity, compared with an age-matched control biopsy (bottom). d, Gas-chromatographic profile of urinary organic acids in EE. The abnormal peak of EMA is indicated in red.
Methods
Patients and DNA Samples
We obtained informed consent from the parents of all probands and siblings, as approved by the institutional review board of the National Neurological Institute “Carlo Besta,” before collecting blood or performing skin biopsies. Several of our probands and families were reported in previous publications describing EE. All patients had clinical and biochemical features typical of the disease. Most of them were deceased before this study began. Besides DNA samples, fibroblast or lymphoblastoid cell lines were established from some patients, but no other tissues are currently available for further biochemical or morphological investigation.
Linkage Analysis
The genomewide search was performed using the ABI PRISM Linkage Mapping Set, version 2.5, on an ABI PRISM 3100 DNA sequencer, and results were processed by GENESCAN software. Alleles were assigned using the GENOTYPER software. Statistical analysis was performed assuming complete penetrance of a recessive disease. Pairwise linkage analysis and multipoint analysis were performed using the LINKAGE computer package and Simwalk2, respectively.
Bioinformatics
Integrative Genomics Analysis
The integrative genomics strategy (Mootha et al. 2003) is based on comparison between the RNA expression profiles of human genes, the products of which are likely to be associated with the mitochondrion, and the RNA expression profiles of the gene set contained in the relevant genomic region—in our case, the 19q13 EE locus. The algorithm reported by Mootha et al. was used to determine the “neighborhood index” for each gene of the locus. The index is a function of the number of mitochondrially related genes found in a series of genes that share an expression profile identical or very similar to that of the proband gene. Identification of the probe set for each of our candidate genes was obtained from the Affymetrix expression profile database specific to the HG-U95A array. The neighborhood index score for each probe set was retrieved from the tables reported by Mootha et al., on an increasingly wider series of genes—namely, 100, 250, and 500.
Antibodies
A polyclonal antibody (anti-ETHE1C17) against an oligopeptide encompassing amino acids 190 to 206 in the C-terminus of the ETHE1 protein was raised in rabbits by Neosystem, according to standard procedures. The following mouse monoclonal antibodies were used in the work: antihemoagglutinin (anti-HA) epitope of the influenza virus (Roche) and anti–succinate-dehydrogenase (anti-SDH) (30-kDa subunit) (Molecular Probes). In all experiments, the anti-ETHE1C17 antiserum was used at a 1:2,000 dilution, the anti-HA antibody at a final concentration of 2 μg/ml, and the anti–SDH antibody at a final concentration of 5 μg/ml.
Cell Cultures and Immunofluorescence Studies
All cell cultures were carried out in Dulbecco's modified Eagle medium, supplemented with 10% fetal calf serum at 37°C in a 5% CO2 atmosphere. Immunofluorescence (IF) was carried out on coverslip-plated cells. In some experiments, cells were preincubated with 100 nM of MitoTracker Red dye (Molecular Probes) for 45 min at 37°C, followed by fixation and incubation with primary- and fluorescent-dye–conjugated secondary antibodies, as described elsewhere (Tiranti et al. 1997). IF was visualized by a confocal microscope (Biorad).
Western Blot Analysis
Approximately 2 × 106 cells were trypsinized, pelleted, sonicated, and solubilized, as described elsewhere (Tiranti et al. 1999). SDS-polyacrylamide gel of 100–200 μg protein/lane and western blot analysis were performed by use of the ECL chemiluminescence kit (Amersham), as described by Tiranti et al. (1999).
Cell Fractionation and Biochemical Assays
Standard methods were used for the preparation of cell lysates, as well as digitonin-treated fibroblasts (Tiranti et al. 1999) and mitochondrial/postmitochondrial fractions in cultured cells (Fernandez-Silva et al. 1997). The specific activities of citrate synthase (Srere 1969) and lactic dehydrogenase (Cornberg 1955) were measured as enzymatic markers for mitochondrial and cytosolic fractions, respectively. Glyoxalase II (Glyo-II)–specific activity was measured spectrophotometrically, after the glutathione-dependent reduction of DTNB (5,5′-di-thio-bis-(2)-nitrobenzoate) to TNB (thio-nitrobenzoate) at 412 nm, by use of S-lactoyl-glutathione (Sigma) as a substrate (Ridderström et al. 1996). The reaction was carried out at 37°C in 100 mM MOPS (3-[N-morpholino]propanesulphonic acid), pH 7.5, 200 μM DTNB, 0.1% Triton X-100, and 900 μM S-lactoyl-glutathione, with a molar extinction coefficient (ɛ) of 13.6 mM−1.
Mitochondrial Targeting
For mitochondrial targeting (Tiranti et al. 1997), cellular proteins were radiolabeled with 35S-methionine for 2 hr in the presence or absence of 20 μM valinomycin. The specific translation products were immunoprecipitated using the anti-HA–specific monoclonal antibody in the presence of the protein A-agarose (Roche) and electrophoresed through a 15% SDS-polyacrylamide gel. After fixation in 10% acetic acid and 25% isopropanol, the gel was washed for 20 min in Amplify reagent (Amersham) and layered onto a phosphorimaging screen (Biorad). After overnight exposure, autoradiography was carried out in a Molecular Imager apparatus (Biorad).
For proteinase K protection, mitochondria from human fibroblasts, freshly isolated as described elsewhere (Fernandez-Silva et al. 1997), were incubated for 20 min at 37°C with 0, 100, and 250 μg/ml proteinase K and with 0.5% Triton X-100, supplemented with 250 μg/ml proteinase K. For suborganellar localization, human fibroblast mitochondria were treated with three cycles of freezing and thawing and six sonication strokes on ice-cold water, and the membrane and soluble fractions were separated by ultracentrifugation at 100,000 × g for 1 hr at 4°C. After SDS-PAGE through a 15% polyacrylamide gel, the ETHE1 protein was visualized by western blotting by use of anti-ETHE1C17.
Miscellaneous RNA and DNA Manipulations
ETHE1 Cloning
A full-length human ETHE1 cDNA was retrotranscribed and PCR-amplified from total RNA extracted from human fibroblasts, by use of standard procedures (Tiranti et al. 1997). The following oligonucleotides were used for RT-PCR: 5′-GCTCGGCCTCCGCTCATTCCTGAC-3′ (forward primer) and 5′-GGCAGTGGGTGTCTGCACCCCACA-3′ (reverse primer). Each of the 30 PCR amplification cycles was 70°C for 2 min and 95°C for 1 min. The cDNA was cloned into the plasmid vector of the TOPO-TA cloning kit (Invitrogen), according to the instruction manual of the kit.
Generation and Cloning of ETHE1HA Recombinant Proteins
The human ETHE1 cDNA was tagged on the 3′ end with the sequence encoding an epitope of the influenza virus HA, essentially as described elsewhere (Tiranti et al. 1997). A second ETHE1 cDNA encoding a shorter HA-tagged variant protein starting at P21 amino acid residue (ETHE121) was PCR-amplified from the HA-tagged ETHE1-encoding construct, by use of suitable oligonucleotides as primers. Both constructs were inserted into the eukaryotic-expression plasmid vector pcDNA3.1 (Invitrogen) by use of suitable restriction sites. The recombinant plasmids were transfected by electroporation in Cos-7 (for transient expression) or HeLa (for stable expression) cells, as described elsewhere (Tiranti et al. 1997).
Northern Blot
A premade multiple-tissue northern blot (MTN), containing 1 μg/lane of purified polyA+ RNA (Clontech), was preincubated in ExpressHyb hybridization solution (Clontech) for 30 min at 65°C and then was hybridized for 2 hr at 65°C with a human ETHE1 cDNA probe (32P), which was radiolabeled using the Ready-To-Go DNA Labelling Beads kit (Amersham). The MTN filter was exposed overnight and visualized by a Molecular Imager apparatus (Biorad).
Genotyping
Nucleotide sequence analysis was carried out on a 3100 ABI Automated Sequencer on samples prepared using the BigDye Termination kit (Applied Biosystems). Data were elaborated using the Secscape software (Applied Biosystems). Oligonucleotides for the PCR amplification of the seven exons of the human ETHE1 gene are listed in table 1.
Table 1.
Oligonucleotides for PCR Amplification of ETHE1 Exons
| Exon | Forward Primer | Reverse Primer |
| 1 | 5′-TCCGTGGCCCCTTTAAGGCGT-3′ | 5′-CCCGGAGTTCCGTCCTTGCT-3′ |
| 2 | 5′-AGCAAGGACGGAACTCCGGG-3′ | 5′-TCCCATTCCACTGACGCTGCA-3′ |
| 3 | 5′-TGCAGCGTCAGTGGAATGGGA-3′ | 5′-AAGAAGTCCAGGCCACCACC-3′ |
| 4 | 5′-GGGCCAGTTTCATCTAGAAGGC-3′ | 5′-GCCCCCTAAAAGTCTAATGTCC-3′ |
| 5 | 5′-GGAAGGGGTTAGAGTCTTCTGT-3′ | 5′-GAGACTGGTCGTCTTCATGCC-3′ |
| 6 | 5′-AGTTCTGAGAGGCCTGAGGCA-3′ | 5′-ACCCAGGAGTCCAAGACCCCA-3′ |
| 7 | 5′-AGTGGGGCCTGGAACTCTAC-3′ | 5′-CGGCCCAGAAACCCAATTGG-3′ |
PCR amplification conditions were 94°C for 1 min, 59°C for 30 s, 72°C for 1 min, for 32 cycles, plus an initial denaturation step at 94°C for 3 min and a final extension at 72°C for 2 min. Exons 2 and 7 were amplified in a buffer containing 3.1 mM MgCl2, 35% deionized formamide, and 10% glycerol (final concentrations). PCR amplification cycles were as above, except that the annealing temperature was 57°C.
Results
Identification of the EE Locus on Chromosome 19q13
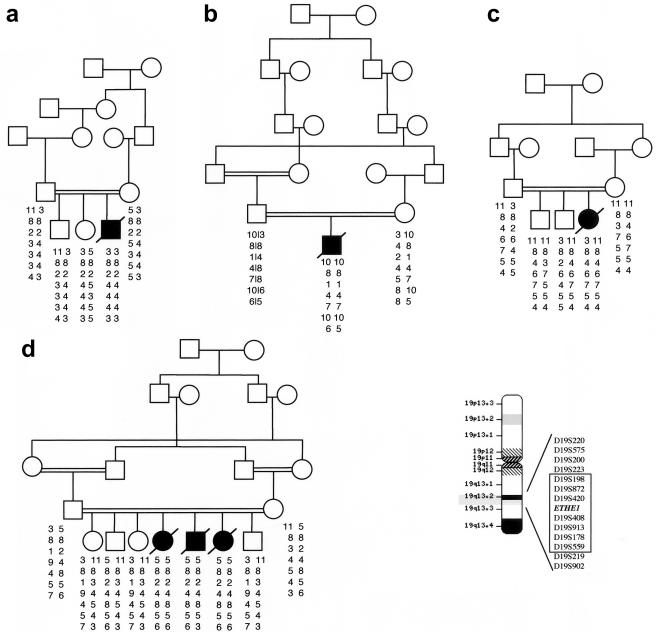
A full genomewide scan of four consanguineous and two nonconsanguineous families (fig. 2) showed a maximum pairwise LOD score of 4.35 at a recombination frequency (θ) of 0 with marker D19S420 on chromosome 19q13. The critical interval was defined within a region of ∼3 cM, between markers D19S223 and D19S559. Multipoint analysis (Lathrop et al. 1984) detected a maximum location score of 7.21 (fig. 3a). Haplotype reconstruction (Sobel and Lange 1996) revealed a homozygous condition caused by identity-by-descent alleles in all affected individuals of the four consanguineous families. The two nonconsanguineous families contributed no further narrowing of the critical region. One of these families, family E, showed an extended homozygous region in the proband, suggesting unrecognized parental consanguinity.
Figure 2.
Pedigrees. The blackened symbols indicate the subjects with EE, and the unblackened symbols indicate the clinically healthy individuals. Beside each symbol are shown the individual haplotypes, covering a 3-cM region on chromosome 19q13. The markers from which the haplotypes were constructed and their genetic distances are shown in the boxed area against a chromosome-19 idiotype. The ETHE1 gene is flanked by markers D19S420 and D19S408.
Figure 3.
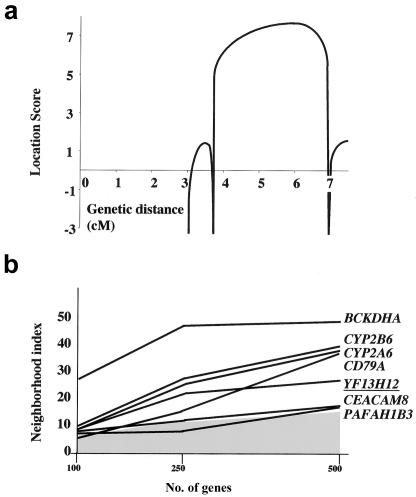
Genetic findings. a, Multipoint analysis of the EE locus for the families shown in figure 2. b, Integrative genomics strategy based on calculation of the mitochondrial neighborhood index. On the X-axis is shown the number of genes with expression profiles that were considered for the calculation of the index. The graph reports the values obtained for genes (UCSC annotation) from the 19q13 EE locus for which scores were equal to or higher than a threshold (gray area) corresponding to the score reported by Mootha et al. for the LRPPRC gene. YF13H12 (underlined) corresponds to a partial-length ETHE1 cDNA.
Search for Candidate Genes
The genomic sequence between markers D19S223 and D19S559 contains ∼130 known or predicted genes, based on concordant data obtained from the NCBI, CELERA, and UCSC genome browser databases. Because of the clinical and biochemical features of EE, we assumed that the responsible protein is likely to be involved in mitochondrial metabolism. We first excluded mutations in BCKDHA, which encodes the α polypeptide of branched-chain ketoacid dehydrogenase E1, as well as mutations in TOM40, which encodes the 40-kDa subunit of the outer mitochondrial membrane transport machinery. We then selected candidate genes that predict proteins of unknown function, which may potentially be related to mitochondria. We followed an integrative genomics approach that was originally developed to identify LRPPRC, the gene responsible for cytochrome c oxidase deficiency, Saguenay–Lac-Saint-Jean type (MIM 220111) (Mootha et al. 2003). According to this strategy, a “neighborhood index” was given to all genes of the critical region; this index reflects the similarity of their RNA-expression profiles to those of known mitochondrial genes.
ETHE1 Is the Gene Responsible for EE
Among the seven genes (including BCKDHA) that were given the highest scores (fig. 3b), a likely candidate was a gene annotated as UCSC entry YF13H12, generically associated with a protein “present in thyroid.” A BLAST search against human ESTs showed that YF13H12 corresponded to a partial-length cDNA of GenBank D83198. This gene was previously known as “HSCO” (hepatoma subtracted cDNA library clone one), a gene overexpressed in human hepatoma cell cultures (Higashitsuji et al. 2002). However, in light of its role as the gene responsible for ethylmalonic encephalopathy, the name of the gene has been changed to “ETHE1.” We showed that human ETHE1 is ubiquitously expressed as a single, ∼1,000-nucleotide (nt)–long transcript (fig. 4a). The ETHE1 gene encodes a protein that shares similarities with human Glyo-II (fig. 4b). We observed that this protein contains an amino (N)-terminal sequence of 24 amino acids, which was missing in the polypeptide predicted by YF13H12. This N-terminal region is rich in apolar and basic (mostly R) residues, similar to mitochondrial leader peptides (Attardi and Schatz 1988). When analyzed by dedicated software packages available online (Mitoprot-2, PSORT-II, Predotar, and TargetP), the predicted ETHE1 protein sequence showed a very high likelihood of being targeted to mitochondria (not shown). This analysis also suggested two potential cleavage sites for the mitochondrial processing peptidase (MPP), between either A20 and P21 or R11 and Q12.
Figure 4.
ETHE1 transcript and protein. a, Northern blot analysis of ETHE1 transcript on polyA+ mRNA extracted from different human tissues. B, brain; H, heart; M, skeletal muscle; C, colon; T, thymus; S, spleen; K, kidney; L, liver; I, small intestine; P, placenta; Lu, lung; Bl, blood. The 1,000-nt ETHE1 transcript is indicated by an arrow. b, Multiple alignments of the ETHE1 protein sequences in different species. Protein sequences were aligned using CLUSTALW. Amino acid substitutions are shown below the alignment, except for the I replacing the first M. A thick bar indicates the β-lactamase signature.
Mutation Analysis
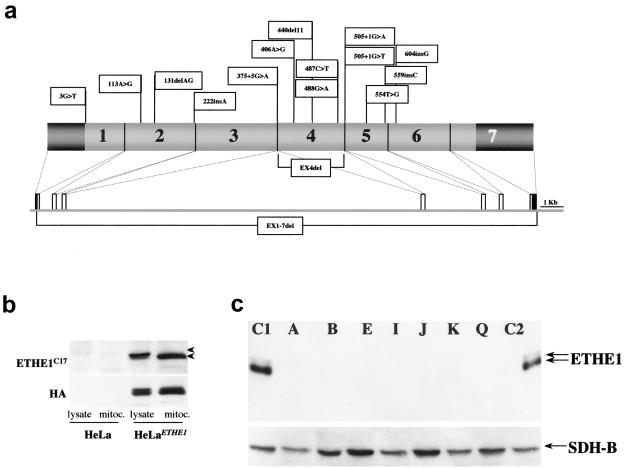
In light of these observations, we sequenced the seven exons (fig. 5a) of the ETHE1 gene from patients with EE (table 2). Homozygous mutations were found in all probands from the four consanguineous families (A–D) originally used for gene mapping. The proband from family E carried a homozygous mutation, whereas the proband from family F was a compound heterozygote. Mutations were also found in eight additional probands belonging to four consanguineous (G–J) and two nonconsanguineous (K, L) families, as well as in four unrelated singleton patients (M–Q). Cosegregation of the mutations with the disease was demonstrated in all families (not shown).
Figure 5.
Characterization of ETHE1 gene and protein (ETHE1) in patients with EE. a, ETHE1 mutations identified in our patients. Mutations are indicated along the schematic representation of the ETHE1 cDNA. Exons are numbered in the cDNA scheme. The dark grey areas represent the 5′ and 3′ UTRs. The genomic organization of human ETHE1 is represented below the cDNA. b, Western blot analysis on cell lysates and isolated mitochondria from HeLa- and HeLaETHE1-derivative cell lines, using the anti-ETHE1C17 and anti-HA antibodies. The filter was treated with anti-ETHE1C17 and then stripped and rehybridized with the anti-HA antibody. The two antibodies show a virtually identical pattern in the HeLaETHE1 cell line, whereas ETHE1 protein CRM is barely detected by anti-ETHE1C17 antibody in naive, untransfected HeLa cells. c, Western blot analysis on fibroblast lysates was performed using anti-ETHE1C17; a monoclonal antibody specific for SDH-B, the 30-kDa subunit of succinate dehydrogenase, was used as a control. The same filter was first treated with anti-ETHE1C17 and then stripped and rehybridized with the anti-SDH antibody. C1 and C2 denote control fibroblasts. The upper and lower arrows indicate the precursor and mature ETHE1 polypeptides, respectively. Lanes A, B, E, I, J, K, and Q show fibroblasts from patients with EE. Each letter refers to the corresponding proband (and mutations) listed in table 2.
Table 2.
Mutation Screening[Note]
| Familiesa | No. of Patients | Mutation | Localization | Predicted Transcript or Protein Abnormality |
| A | 1 | 604_605insG | EX6 | V202fsX220 |
| B | 1 | 3G→T | EX1 | M1I |
| C | 3 | del exon 4 | EX4 | Not translated |
| D | 1 | 487C→T | EX4 | R163W |
| E | 1 | 406A→G | EX4 | T136A |
| F | 1 | 221_222insA/440_450del11 | EX2/4 | Y74X/H147fsX176 |
| G | 1 | 221_222insA | EX2 | Y74X |
| H | 2 | del exons 1–7 | EX1–7 | Not translated |
| I | 1 | del exon 4 | EX4 | Not translated |
| J | 1 | 505+1G→T | EX4/IN4 | Splicing error |
| K | 1 | 375+5G→A | EX3/IN3 | Splicing error |
| L | 2 | 131_132delAG/488G→A | EX2/4 | E44fsX105/R163Q |
| M | 1 | 592_593insC | EX5 | H198fsX220 |
| N | 1 | 487C→T | EX4 | R163W |
| O | 1 | 505+1g→A | EX4/IN4 | Splicing error |
| P | 1 | 487C→T | EX4 | R163W |
| Q | 1 | 113a→G/554T→G | EX2/5 | Y38C/L185R |
Note.— Nucleotides are numbered considering the A of the first ATG codon as +1. Amino acids are numbered considering the initiation M as +1.
A–D, families shown in figure 3; E–L, additional families in which segregation analysis could be performed; M–Q, families composed of singleton patients (no other family member available for analysis).
Most of the 16 different mutations found in our cohort (table 2 and fig. 5a) were loss-of-function mutations producing a stop, a frameshift, or aberrant splicing. In one consanguineous family, the entire gene was missing, and, in two others, exon 4 of the gene was missing in both alleles. Six missense mutations were detected, all predicting amino acid changes in highly conserved positions (fig. 4b).
Haplotype reconstruction in the two families lacking exon 4 showed an identical set of microsatellite polymorphisms, indicating that these families are related. This result is concordant with a common geographic origin of the families. An identical missense point mutation, 487C→T, predicting an R163W amino acid change, was found in three unrelated probands, but in this case the haplotypes differed from each other, suggesting that the mutational event had occurred either independently or in a very ancient common progenitor. None of the missense mutations was detected in 200 DNA samples from unrelated control individuals from Italy.
Western Blot Analysis
A polyclonal antibody was raised in rabbit against a synthetic peptide corresponding to the last 17 amino acids on the carboxy (C)-terminal end of the human ETHE1 protein product (anti-ETHE1C17). The specificity of the anti-ETHE1C17 antibody was tested by immunostaining HeLa cell lysates that stably express a recombinant, influenza-virus-HA–tagged ETHE1 (ETHEHA) protein product. The western blot pattern obtained using the anti-ETHE1C17 antibody was virtually identical to that produced by an anti-HA monoclonal antibody, indicating the specificity of anti-ETHE1C17 for the ETHE1 protein (fig. 5b). The anti-ETHE1C17 antibody was used to immunostain ETHE1 protein–specific crossreacting material (CRM) for western blot analysis on fibroblast cell lines from patients with EE and controls (fig. 5c). A band corresponding to the mature ETHE1 polypeptide was clearly detected in fibroblast lysates from two control individuals. A much fainter band of slightly slower mobility was also detected in the same samples and was attributed to the precursor ETHE1 protein species retaining the mitochondrial N-terminal leader peptide (see below). No ETHE1 protein CRM was detected in any EE fibroblast cell line, including missense, splicing, and deletion mutations.
The ETHE1 Gene Product Is a Mitochondrial Matrix Protein
Import of the ETHE1HA Protein In Vivo
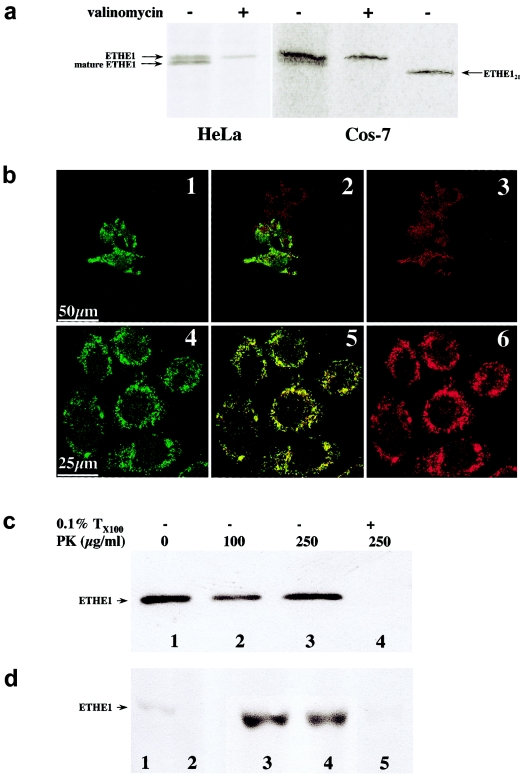
Using an anti-HA–specific monoclonal antibody, two 35S-radiolabeled protein species, of ∼30 kDa and ∼29 kDa, were immunoprecipitated from COS-7ETHE1 and HeLaETHE1 cells. The 30-kDa polypeptide is the radiolabeled ETHE1HA protein. The second, slightly shorter product is the mature form of the ETHE1HA protein, resulting from the intramitochondrial cleavage of a leader peptide (Attardi and Schatz 1988). This is cleaved from the N-terminal end of the polypeptide, since the mature protein still contains the HA epitope, which is fused with the protein C-terminus. The import process is energy dependent (Attardi and Schatz 1988), since it was inhibited by treatment with valinomycin, a mitochondrion-specific ionophore that abolishes the mitochondrial proton-motive force (ΔΨ).
We estimated that the leader peptide must be <20 amino acid residues, because the size of the mature ETHE1HA protein is bigger than that of ETHE121HA, a recombinant ETHE1HA protein variant missing the first 20 amino acids on the N-terminus.
Immunofluorescence Studies
In COS-7 cells transiently expressing the ETHE1HA protein (COS-7ETHE1), as well as in stably expressing HeLa cells (HeLaETHE1) (fig. 6b), the immunofluorescence pattern specific to the ETHE1HA protein was virtually identical to that obtained by using MitoTracker, a mitochondrion-specific immunofluorescent dye.
Figure 6.
Mitochondrial localization of the ETHE1 protein. a, Mitochondrial import in vivo. Autoradiographs of 35S-radiolabeled HA-tagged ETHE1 protein, immunoprecipitated from HeLaETHE1 and Cos-7ETHE1 cell lysates. Arrows indicate the HA-tagged ETHE1 precursor protein, mature ETHE1 protein, and ETHE121, a shortened HA-tagged ETHE1 protein species lacking the first 20 amino acid residues on the N-terminus. Experiments were carried out in the presence (+) or absence (−) of valinomycin. b, Confocal immunofluorescence on Cos-7ETHE1 (panels 1–3 depict transient transfection) and HeLaETHE1 (panels 4–6 depict stable transfection). The green fluorescence in panels 1 and 4 corresponds to ETHE1HA protein–specific immunoreactivity. The red fluorescence in panels 3 and 6 corresponds to mitochondrial staining by MitoTracker, a mitochondrion-specific dye. The two immunofluorescence patterns largely overlapped, as shown by confocal merge images in panels 2 and 5. Magnification 40×. c, Proteinase K (PK)-protection assay by western blot analysis on freshly isolated mitochondria from human fibroblasts. Lanes 1–3 show intact isolated mitochondria treated with 0, 100, 250 μg/ml PK; lane 4 shows mitochondria treated with 0.5% Triton X-100 before digestion with 250 μg/ml PK. Anti-ETHE1C17 was used as the primary antibody. d, Suborganellar localization of the ETHE1 protein by western blot analysis on mitochondria isolated from human fibroblasts. Lane 1, cell lysate; lane 2, postmitochondrial supernatant (10,000 × g); lane 3, intact mitochondria; lane 4, mitochondrial membrane fraction; lane 5, mitochondrial soluble fraction. Anti-ETHE1C17 was used as the primary antibody.
Protease-Protection Test
To determine which mitochondrial compartment contains the mature ETHE1 protein product, a protease-protection test (McQuibban et al. 2003) was performed on mitochondria isolated from human fibroblasts. As shown in figure 6c, the ETHE1 protein–specific CRM was protected from proteolytic digestion with increasing concentrations of proteinase K in intact mitochondria, whereas it was completely digested by proteinase K in detergent-treated mitochondria. These results indicate that the mature ETHE1 protein product is located in the inner compartment of mitochondria.
Suborganellar Localization
To establish whether the mature ETHE1 polypeptide is a matrix- or a membrane-bound protein, western blot analysis was carried out using anti-ETHE1C17 against different suborganellar fractions of human mitochondria. As shown in figure 6d, marked enrichment of ETHE1 protein CRM was obtained in the total mitochondrial fraction compared with the cell lysate, whereas no CRM was observed in the supernatant obtained after isolation of mitochondria. When the suborganellar fractions were analyzed, virtually all the ETHE1 protein–specific CRM was found in the soluble fraction, which largely corresponds to the mitochondrial matrix, whereas no ETHE1 protein–specific CRM was observed in the lane corresponding to the membrane fraction. Identical results were consistently obtained on mitochondria isolated from rat and mouse liver (not shown).
Glyo-II Activity Assay
To test whether the mature ETHE1 protein is a mitochondrial Glyo-II, we measured the Glyo-II–specific activity in isolated mitochondria from naive HeLa and HeLaETHE1 cells. As shown in figure 2d, the ETHE1 protein CRM is virtually absent in naïve HeLa cells, whereas it is robustly expressed in HeLaETHE1 cells. Nevertheless, very low, comparable Glyo-II activities were measured in isolated mitochondria from both cell lines, whereas abundant activities were detected in the postmitochondrial supernatants, due to the presence of cytosolic Glyo-II (table 3). Likewise, Glyo-II activity was present in the cytosolic fraction of both control and EE fibroblasts, whereas it was almost undetectable after the elimination of the cytosolic fraction from the same cells by treatment with digitonin (table 3). In any case, no difference was measured between control and EE cells in the mitochondrial-enriched fraction.
Table 3.
Glyo-II–Specific Activities
|
Mean ± SD Glyo-II Activity(nM/min/mg protein) |
||
| Cell Type (No. of Samples) | Cytosol | Mitochondria |
| HeLa (n=4) | 77.2 ± 13.4 | 5.2 ± 2.5a |
| HeLaETHE1(n=4) | 69.5 ± 6.3 | 7.2 ± 2.3a |
| Control fibroblasts (n=5) | 45.4 ± 9.1 | 3.0 ± 0.96b |
| EE fibroblasts (n=4) | 54.3 ± 9.4 | 3.2 ± 1.25b |
Isolated mitochondria.
Mitochondrion-enriched fraction from digitonin-treated cells.
Discussion
Using a combination of homozygosity mapping, integration of physical and functional genomic data sets, and mutational screening, we identified ETHE1 as the gene responsible for EE. An integrative genomic approach was adopted to select a list of a few candidate-for-function genes out of >130 sequences present in the critical interval defined by linkage analysis. The success of this strategy in our project, as well as in the original work on the identification of the gene responsible for Leigh syndrome, French-Canadian type, confirms it as a powerful method for the discovery of genes responsible for mitochondrial, and possibly nonmitochondrial, inherited disorders.
Localization of ETHE1 Mature Protein in the Mitochondrial Matrix
We identified ETHE1 as a candidate gene on the basis of the observation that its expression is concordant with that of human genes known to encode proteins targeted to and/or functionally associated with mitochondria. Previous observations by Higashitsuji et al. (2002) suggested that ETHE1 (formerly HSCO) is diffusely present in the cytoplasm of cultured cells and is occasionally found in the nucleus, possibly acting as a shuttle protein controlling the nucleus-cytoplasm trafficking of the transcription factor NF-κB. In immunofluorescence experiments, we consistently failed to observe a nuclear localization of the ETHE1 protein in human cell cultures, in both native conditions and after treatment with leptomycin B, an inhibitor of nuclear export (not shown). By contrast, we have clearly demonstrated that the main, if not exclusive, localization of this protein is confined to the mitochondrial matrix. In particular, we showed that, for most proteins targeted to mitochondria, a canonical mitochondrial leader peptide, which is present at the N-terminus of the full-length ETHE1 protein, addresses the protein to the organelle and is cleaved off after internalization in the inner mitochondrial compartment through an energy-dependent process, presumably carried out by MPP. This conclusion is the result of several experimental approaches: in vivo mitochondrial import assays; immunofluorescence-based colocalization of the ETHE1 protein–specific CRM with mitochondrion-specific dyes; protease-based protection assays on intact and disrupted organelles; and ETHE1 protein immunotitration on human cell fractions, isolated mitochondria, and mitochondrial subfractions.
Structure and Function of the ETHE1 Protein
The function of the ETHE1 mature protein is presently unknown. The ETHE1 protein is a phylogenetically conserved protein, sharing high homology with Glyo-II. Besides Glyo-II, a BLAST-P search, with the human ETHE1 predicted protein sequence as a probe, resulted in the identification of highly similar proteins in all metazoan species, in plants, such as Arabidopsis thaliana, and in fungi, such as Saccharomyces cerevisiae (fig. 3b). In contrast with the remaining portion of these protein sequences, the first 20–30 amino acid residues on the N-terminus appear to be poorly conserved. However, when the sequences shown in figure 4b were analyzed by organellar target–prediction softwares, a high score for mitochondrial targeting was obtained for all of them, with the exception of Glyo-II, which is known to be a cytosolic enzyme. These results suggest the existence of a distinct family of mitochondrial proteins that is present in all eukaryotes and is structurally—and possibly functionally—similar to the human ETHE1 protein.
The glyoxalase system catalyzes the conversion of toxic 2-oxoaldehydes into the corresponding 2-hydroxy acids (Thornalley 1990). The main substrate seems to be methylglyoxal, which is formed as a by-product of glycolysis from triose phosphates through the action of triose-phosphate isomerase but also via other metabolic routes. As the first step of the glyoxalase system, methylglyoxal reacts spontaneously with reduced glutathione to form a hemithioacetal. Glyoxalase I converts hemithioacetal into S-D-lactoylglutathione, which is further metabolized to D-lactate and glutathione by Glyo-II. Both glyoxalase I and II are cytosolic enzymes. However, in the yeast S. cerevisiae, in addition to the cytosolic Glyo-II, encoded by the gene GLO2, a second Glyo-II, encoded by the GLO4 gene, is targeted to mitochondria (Bito et al. 1997). The GLO4 protein contains an N-terminal leader peptide with a Mitoprot-2–predicted MPP cleavage site at +21, similar to the prediction for the ETHE1 protein. However, the leader peptide of GLO4 has been experimentally demonstrated to be much shorter—only 10 amino acids, including the initiation M residue (Bito et al. 1997). These results are similar to those obtained by us for the ETHE1 precursor protein, which also appears to have a very short leader peptide, possibly between R11 and Q12, as predicted by the TargetP program. It is interesting that only the mature form of GLO4, lacking the 10-amino-acid–long mitochondrial leader peptide, shows Glyo-II activity, whereas the uncleaved precursor form is completely inactive (Bito et al. 1999). Glyo-II, GLO4, and the ETHE1 protein all contain a metallo-β-lactamase region that, in the ETHE1 protein sequence, encompasses amino acid residues 35–196. A consensus signature sequence, HXHXD(X)H, is conserved throughout the β-lactamase protein family and is believed to be the Zn(II) ligand (Aravind 1999). In addition, the ETHE1 protein sequences in humans, mice, and other organisms contain the Y residue, which is present in human Glyo-II (at position 175) and in GLO4 (at position 200) and is believed to be part of the binding site for glutathione (Ridderström et al. 2000). The similarities of the ETHE1 protein with both Glyo-II and GLO4, the conservation of the β-lactamase signature and the glutathione binding site, as well as the abnormalities in mitochondrial metabolic pathways associated with EE, all suggest that the main role of the mature ETHE1 protein is that of an enzyme, possibly a metallo-β-lactamase. Nevertheless, our experiments, performed on mammalian cell cultures, failed to demonstrate a significant Glyo-II activity in isolated mitochondria, using D-lactoylglutathione as a substrate. A likely possibility is that the ETHE1 protein could still be a mitochondrial metallo-β-lactamase involved in the metabolism of an unknown substrate. Alternatively, its activity as a mitochondrial Glyo-II could vary in different tissues and growth conditions; for instance, robust Glyo-II activity has been reported in isolated mitochondria from rat (Talesa et al. 1989) and bovine liver (Talesa et al. 1990), and our preliminary data (not shown) have confirmed these findings.
Pathogenic Mechanism of ETHE1 Mutations in EE
We have identified 16 different ETHE1 mutations in several families or singleton patients with EE. The following evidence supports the idea that mutations are of pathogenic significance in ETHE1, thus specifying ETHE1 as the EE gene: (1) the absence of mutations in ethnically matched controls; (2) the occurrence of mutations that clearly predict a loss of function of the resulting protein; (3) the occurrence of a few missense mutations that all affect phylogenetically conserved regions in eukaryotes, and, with the exception of the T136A mutation, are largely nonconservative in nature; (4) their strict cosegregation with disease in all families analyzed; and (5) the absence of the mutant protein in several fibroblast cell lines from patients with EE examined by western blot analysis. Taken together, the above observations indicate that EE is a genetically homogeneous disorder caused by the loss of function of mutant ETHE1. This conclusion is fully compatible with the recessive behavior of the disease trait and is again suggestive of a defect in a still-unknown enzymatic activity related to mitochondrial energy metabolism.
Clinical Implications
Even though EE has been described mainly in families from the Mediterranean basin and the Arabian peninsula, we obtained no evidence of the existence of an ancestral haplotype or a cluster of common mutations in our cohort of patients with EE. The identification of the gene responsible for EE offers a tool for early diagnosis and rational genetic counseling. Since the initial report, no more than 30 cases of EE have been described worldwide, leading to the assumption that EE is a very rare autosomal recessive disorder. However, the actual incidence of this condition could have been significantly underestimated because the biochemical phenotype may be incorrectly attributed to other metabolic disorders, particularly defects of the mitochondrial electron-transfer flavoprotein pathway. In the experience of the authors, several cases of EMA aciduria are initially given a biochemical diagnosis of glutaric acidemia type II, but this diagnosis is not confirmed by in vitro enzyme assays and/or molecular studies. We believe that some of these cases could be EE.
Although the function of the ETHE1 protein remains to be elucidated, the evidence that its loss causes EE indicates that the activity of the protein is required for maintaining important metabolic homeostasis in the mitochondria and in the whole organism. This is also supported by the ubiquitous expression of the ETHE1 transcript in humans and by the diffusion of the ETHE1 protein family among eukaryotic species.
With appropriate model systems, in which we systematically manipulate ETHE1 expression, it should be possible to establish the role of this gene in mitochondrial homeostasis, leading to an understanding of EE pathogenesis.
Acknowledgments
We thank the patients and their families, whose collaboration and understanding have made this work possible. We are grateful to Norman Pavelka, Paola Tonin, Graziella Uziel, and Ernst Christensen, for clinical evaluation or technical help; to Giovanni Principato, for critical discussion; and to Barbara Geehan, for revising the manuscript. This work was supported by Fondazione Telethon-Italy (grant 1180), Fondazione Pierfranco & Luisa Mariani (Ricerca 2000), and Ricerca Finalizzata Ministero della Salute (RF-2002/158), as well as a MitEuro network grant from the European Union Framework Program 5. This paper is dedicated to the memory of Paolo Durand.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Affymetrix, http://www.affymetrix.com/analysis/index.affx
- CELERA, http://www.celera.com
- Center for Genome Research, http://www.genome.wi.mit.edu/mpg/isfc/ (for neighborhood index tables)
- CLUSTALW, http://www.ebi.ac.uk/clustalw/
- Mitoprot-2, http://mips.gsf.de/cgi-bin/proj/medgen/mitofilter
- National Center for Biotechnology Information (NCBI) Home Page, http://www.ncbi.nlm.nih.gov/ (for Homo sapiens ETHE1 [formerly HSCO] [GI: 25165940], Mus musculus ETHE1 [formerly HSCO] [GI number: 12963539], Caenorhabditis elegans hypothetical protein C33A12.7 [GI number: 3874677] Arabidopsis thaliana glyoxalase II [gb|AAB17995.1] [GI number: 1644427], Saccharomyces cerevisiae ORF YOR040w [GI number: 1420164], Synechocystis sp. PCC 6803 ORF_ID:slr1259∼hypothetical protein [GI number: 1652935], Drosophila melanogaster CG30022-PA [gb|AAF58646.2] [GI number: 21627488], Xenopus laevis MGC53036 protein [similar to RIKEN cDNA 0610025L15 gene] [GI number: 27371293], Homo sapiens glyoxalase II [GI number: 1237213], Homo sapiens ETHE1 [formerly HSCO] mRNA [accession number: D83198])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for EE and Saguenay–Lac-Saint-Jean type)
- Predotar, http://www.inra.fr/predotar/french.html
- PSORT, http://psort.nibb.ac.jp/
- TargetP, http://www.cbs.dtu.dk/services/TargetP/
- UCSC, http://www.genome.ucsc.edu/ (for human ETHE1 [formerly HSCO] genomic sequence [YF13H12 >hg_15 D83198 chr19:48685921-48708445])
References
- Aravind L (1999) An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol 1:69–91 [PubMed] [Google Scholar]
- Attardi G, Schatz G (1988) Biogenesis of mitochondria. Annu Rev Cell Biol 4:289–333 [DOI] [PubMed] [Google Scholar]
- Bito A, Haider M, Briza P, Strasser P, Breitenbach M (1999) Heterologous expression, purification, and kinetic comparison of the cytoplasmic and mitochondrial glyoxalase II enzymes, Glo2 and Glo4, from Saccharomyces cerevisiae. Protein Expr Purif 17:456–464 10.1006/prep.1999.1151 [DOI] [PubMed] [Google Scholar]
- Bito A, Haider M, Hadler I, Breitenbach M (1997) Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J Biol Chem 272:21509–21519 10.1074/jbc.272.34.21509 [DOI] [PubMed] [Google Scholar]
- Burlina AB, Dionisi-Vici C, Bennet MJ, Gibson KM, Servidei S, Bertini E, Hale DE, Schimidt-Sommerfeld E, Sabetta G, Zacchello F, Rinaldo P (1994) A new syndrome with ethylmalonic aciduria and normal fatty acid oxidation in fibroblasts. J Pediatr 124:79–86 [DOI] [PubMed] [Google Scholar]
- Burlina AB, Zacchello F, Dionisi-Vici C, Bertini E, Sabetta G, Bennet MJ, Hale DE, Schimidt-Sommerfeld E, Rinaldo P (1991) New clinical phenotype of branched-chain acyl-CoA oxidation defect. Lancet 338:1522–1523 10.1016/0140-6736(91)92338-3 [DOI] [PubMed] [Google Scholar]
- Cornberg A (1955) Lactic dehydrogenase. Methods Enzymol 1:441–443 [Google Scholar]
- Corydon MJ, Vockley J, Rinaldo P, Rhead WJ, Kjeldsen M, Winter V, Riggs C, Babovic-Vuksanovic D, Smeitink J, De Jong J, Levy H, Sewell AC, Roe C, Matern D, Dasouki M, Gregersen N (2001) Role of common variant alleles in the molecular basis of short-chain acyl-CoA dehydrogenase deficiency. Pediatr Res 49:18–23 [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G (1997) The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds the DNA as a monomer, with evidence pointing to intramolecular leucine-zipper interactions. EMBO J 16:1066–1079 10.1093/emboj/16.5.1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia B, Colamaria V, Carrara F, Tonin P, Rimoldi M, Uziel G (1994) Muscle cytochrome c oxidase deficiency in two Italian patients with ethylmalonic aciduria and peculiar clinical phenotype. J Inher Metab Dis 17: 301–303 [DOI] [PubMed] [Google Scholar]
- Garcia-Silva MT, Ribes A, Campos Y, Garavaglia B, Arenas J (1997) Syndrome of encephalopathy, petechiae, and ethylmalonic aciduria. Pediatr Neurol 17:165–170 10.1016/S0887-8994(97)00048-9 [DOI] [PubMed] [Google Scholar]
- Grosso, S, Mostardini R, Farnetani MA, Molinelli M, Berardi R, Dionisi-Vici C, Rizzo C, Morgese G, Balestri P (2002) Ethylmalonic encephalopathy, further clinical and neuroradiological characterization. J Neurol 249:1446–1450 10.1007/s00415-002-0880-4 [DOI] [PubMed] [Google Scholar]
- Higashitsuji H, Higashitsuji H, Nagao T, Nonoguchi K, Fujii S, Itoh K, Fujita J (2002) A novel protein overexpressed in hepatoma accelerates export of NF-κB from the nucleus and inhibits p53-dependent apoptosis. Cancer Cell 2:335–346 10.1016/S1535-6108(02)00152-6 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel J-M, Julier C, Ott J (1984) Strategies for multilocus analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamer OA, Tjoa SS, Scriver CR, Klassen GA (1976) Demonstration of a new mammalian isoleucine catabolic pathway yielding an R series of metabolites. Biochem J 160:417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantagos S, Genel M, Tanaka K (1979) Ethylmalonic-adipic aciduria: in vivo and in vitro studies indicating deficiency of activities of multiple acyl-CoA dehydrogenases. J Clin Invest 64:1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban GA, Saurya S, Freeman M (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423:537–641 10.1038/nature01633 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, Mitchell GA, Morin C, Mann M, Hudson TJ, Robinson B, Rioux JD, Lander ES (2003) Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci USA 100:605–610 10.1073/pnas.242716699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowaczyk MJ, Lehotay DC, Platt BA, Fisher L, Tan R, Phillips H, Clarke JT (1998) Ethylmalonic and methylsuccinic aciduria in ethylmalonic encephalopathy arise from abnormal isoleucine metabolism. Metabolism 47:836–839 [DOI] [PubMed] [Google Scholar]
- Ozand PT, Rashed M, Millington DS, Sakati N, Hazzaa S, Rahbeeni Z, Al Odaib A, Youssef N, Mazrou A, Gascon GG, Brismar (1994) Ethylmalonic aciduria: an organic acidemia with CNS involvement and vasculopathy. Brain Dev 16:12–22 10.1016/0387-7604(94)90092-2 [DOI] [PubMed] [Google Scholar]
- Ridderström M, Jemth P, Cameron AD, Mannervik B (2000) The active-site residue Tyr-175 in human glyoxalase II contributes to binding of glutathione derivatives. Biochim Biophys Acta 1481:344–348 [DOI] [PubMed] [Google Scholar]
- Ridderström M, Saccucci F, Hellman U, Bergman T, Principato G, Mannervik B (1996) Molecular cloning, heterologous expression, and characterization of human glyoxalase II. J Biol Chem 271:319–323 10.1074/jbc.271.1.319 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores and marker sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Srere PA (1969) Citrate synthase. Methods Enzymol 13: 3–11 [Google Scholar]
- Talesa V, Principato GB, Norton SJ, Contenti S, Mangiabene C, Rosi G (1990) Isolation of glyoxalase II from bovine liver mitochondria. Biochem Int 20:53–58 [PubMed] [Google Scholar]
- Talesa V, Uotila L, Koivusalo M, Principato G, Giovannini E, Rosi G (1989) Isolation of glyoxalase II from two different compartments of rat liver mitochondria: kinetic and immunochemical characterization of the enzymes. Biochim Biophys Acta 993:7–11 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Ramsdell HS, Baretz BH, Keefe MB, Kean EA, Johnson B (1976) Identification of ethylmalonic acid in urine of two patients with the vomiting sickness of Jamaica. Clin Chim Acta 69:105–112 10.1016/0009-8981(76)90478-2 [DOI] [PubMed] [Google Scholar]
- Thornalley PJ (1990) The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem J 269:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini MP, Zeviani M (1999) Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum Mol Genet 8:2533–2540 10.1093/hmg/8.13.2533 [DOI] [PubMed] [Google Scholar]
- Tiranti V, Savoia A, Forti F, D’Apolito MF, Centra M, Rocchi M, Zeviani M (1997) Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening of the Expressed Sequence Tags database. Hum Mol Genet 6:615–625 10.1093/hmg/6.4.615 [DOI] [PubMed] [Google Scholar]
- Yoon HR, Hahn SH, Ahn YM, Jang SH, Shin YJ, Lee EH, Ryu KH, Eun BL, Rinaldo P, Yamaguchi S (2001) Therapeutic trial in the first three Asian cases of ethylmalonic encephalopathy: response to riboflavin. J Inherit Metab Dis 24:870–873 10.1023/A:1013948409790 [DOI] [PubMed] [Google Scholar]