Abstract
Carotid intimal medial thickness (IMT) is a heritable quantitative measure of atherosclerosis. A genomewide linkage analysis was conducted to localize a quantitative-trait locus (QTL) influencing carotid IMT. Carotid IMT was measured in 596 men and 629 women from 311 extended families (1,242 sib pairs) in the Framingham Heart Study Offspring cohort. B-mode carotid ultrasonography was used to define mean IMT of the carotid artery segments. Multipoint variance-component linkage analysis was performed. Evidence for significant linkage to internal carotid artery (ICA) IMT (two-point log odds [LOD] score 4.1, multipoint LOD score 3.4) was found 161 cM from the tip of the short arm of chromosome 12; these results were confirmed using the GENEHUNTER package (multipoint LOD score 4.3). No LOD scores >2.0 were observed for common carotid artery (CCA) IMT. Association analysis of a single-nucleotide–polymorphism variant of SCARB1 (minor allele frequency 0.13), a gene in close proximity to the region of peak linkage, revealed a protective association of the missense variant allele in exon 1 of SCARB1, with decreased ICA IMT compared with subjects homozygous for the common allele. Although the exon 1 variant contributed 2% to overall variation in ICA IMT, there was no significant change in the peak LOD score after adjustment in the linkage analyses. These data provide substantial evidence for a QTL on chromosome 12 influencing ICA IMT and for association of a rare variant of SCARB1, or a nearby locus, with ICA IMT. Because this rare SCARB1 variant does not account for our observed linkage, further investigations are warranted to identify additional candidate-gene variants on chromosome 12 predisposing to atherosclerosis phenotypes and clinical vascular disease.
Introduction
Atherosclerotic cardiovascular disease (CVD) is the leading cause of morbidity and mortality in the United States (American Heart Association 2002). Both genetic and environmental factors have been implicated in the pathogenesis of atherosclerosis (Lusis 2000). Carotid artery intimal medial thickness (IMT) is a measure of atherosclerosis that is independently associated with traditional CVD risk factors (Crouse et al. 1996; Espeland et al. 1999) and coronary atherosclerotic burden (O’Leary et al. 1996; Davis et al. 1999) and that is predictive of subsequent myocardial infarction and stroke (Bots et al. 1997; O’Leary et al. 1999). Carotid IMT is associated with a family history of cardiovascular disease, and we have shown that 35%–45% of the variability in multivariable-adjusted carotid IMT is explained by genetic factors (Fox et al. 2003). However, few large population-based analyses of genomewide linkage exist for vascular phenotypes, and there are few significant genomewide linkage data available in the general population for myocardial infarction or the underlying condition of atherosclerosis.
Thus, we conducted a genomewide linkage analysis in a population-based cohort of families, to identify chromosomal regions linked to interindividual variation in carotid IMT. We followed up with association analyses of polymorphisms in a plausible candidate gene in the region of peak linkage.
Material and Methods
Study Population
In the current investigation, subjects from the Framingham Heart Study Offspring cohort underwent B-mode carotid ultrasonography during examination cycle 6 (1996–1998). Carotid IMT was measured in 596 men and 629 women, included in 311 extended families and 1,242 sib pairs.
The Framingham Heart Study began in 1948 with the enrollment of 5,209 men and women, 28–62 years of age at study entry, with subjects undergoing repeat exams every 2 years (Dawber et al. 1951, 1963). In 1971, 5,124 men and women were enrolled in the offspring cohort of the Framingham Heart Study, which included the children or spouses of the children of the original cohort. Offspring subjects underwent examinations approximately every 4 years; the design and methodology of the offspring cohort has been described elsewhere (Feinleib et al. 1975; Kannel et al. 1979).
For the linkage analysis, 3,532 subjects attended cycle 6 of the total 5,124 subjects that attended the initial offspring exam. Of these, 152 did not undergo carotid ultrasound, 2,137 were excluded because they were not a biologic member of a family that was genotyped in the marker set for the linkage analysis, a further 17 were excluded because they were not part of a biologic family whose members had carotid ultrasound data, and 1 was excluded because of inadequate carotid IMT data. Thus, a total of 1,225 subjects in 311 families comprised the study sample. Of these, all had an available common carotid artery (CCA) measurement, and 1,113 had an available internal carotid artery (ICA) measurement.
For the genotyping of the gene encoding scavenger receptor, class B, type I (SCARB1 [MIM 601040]), previously known as SR-BI, in the association portion of this study, DNA samples from 2,342 individuals who attended cycle 6 were used. Of these, 96 subjects were excluded because they did not undergo carotid ultrasound, and 1 was excluded because of inadequate carotid IMT data. In all, 2,245 subjects had at least one carotid IMT measure and were typed for at least one of the SCARB1 polymorphisms; therefore, they were included in these analyses. A total of 736 subjects appear in both the linkage and association analyses.
Assessment of Risk Factors and Cardiovascular Disease
Details regarding the methods of risk factor measurement and laboratory analysis have been described (Cupples and D’Agostino 1987). Each examination included an extensive cardiovascular disease assessment, 12-lead electrocardiogram (ECG), and blood testing. Measured covariates for the current study were assessed at the time of carotid ultrasonography.
Carotid Intimal Medial Thickness Assessment
Subjects underwent ultrasonography according to a modification of a standard protocol (O’Leary et al. 1999). Imaging was conducted by a single trained sonographer, using a Toshiba SSH-140A imaging unit and 7.0-MHz transducer for the CCA and a 5.0-MHz transducer for the ICA.
The following views were obtained from the right and left sides: two longitudinal views of the distal common carotid, one at end diastole and one at end systole, and two longitudinal views of the ICA at end diastole. Measurements were made by a single trained sonographer blinded to all clinical information, and they were overread by one of the investigators (J.F.P.).
All studies were read according to a standardized protocol. The high-resolution images of the ICA and CCA were analyzed to calculate near- and far-wall IMT, lumen diameter, and vessel width at each arterial site. All measurements of lumen and wall thickness were calculated with a specially designed computer program (O’Leary et al. 1996, 1999). On the basis of 25 replicate readings by two separate readers, intraclass correlation coefficients for the mean and maximum ICA and CCA IMT were 0.74, 0.74, 0.86, and 0.90, respectively.
A summary measure of the mean wall thickness of the ICA was defined as the mean of the wall thickness for the near and far wall on both the left and right sides. Similar measurements were obtained for the CCA.
Genotyping
Leukocyte DNA was extracted from 5–10 mL of whole-blood or buffy-coat specimens by use of a standard protocol (Gross-Bellard et al. 1973; Miller et al. 1988). Aliquots of DNA from members of the largest Framingham Heart Study families were sent in four batches to the Mammalian Genotyping Service Laboratory at the Marshfield Clinic (Marshfield, WI). A 10-cM density genomewide scan was performed (marker set 8A, heterozygosity 0.77). Genotype-data cleaning, including verification of family relationships and Mendelian inconsistencies, has been described elsewhere (Levy et al. 2000). In brief, familial relationships were checked using the SIBKIN program in ASPEX, which uses maximum-likelihood estimation to test whether the genotyping data are consistent with the specified relationships, and adjustments were made accordingly. In addition, random genotyping errors were checked using Gentest, a precursor to the INFER procedure, in PEDSYS. When we discovered a genotyping error and could not attribute it to a specific person in a nuclear family, all genotypes for that nuclear family were assigned as missing.
Known mutations in SCARB1 (Acton et al. 1999; Osgood-McWeeney et al. 2000) were identified by direct sequencing of PCR products. Three polymorphisms were typed: one in exon 1, one in exon 8, and one in intron 5. SCARB1 genotyping was carried out by allelic discrimination by use of the 5′ nuclease assay with fluorogenic probes on a 7700 Sequence Detection System (PE Applied Biosystems), as described elsewhere (Osgood-McWeeney et al. 2000).
Linkage Analysis
Our statistical analysis focused on mean IMT measures of the ICA and CCA. The heritabilities for CCA and ICA IMT have been published elsewhere (Fox et al. 2003). Before multivariable adjustment, the heritabilities were 0.67 for CCA IMT and 0.43 for ICA IMT. After multivariable adjustment, the heritabilities were 0.43 and 0.35, respectively.
Because the distributions of carotid measures were skewed, normalized deviates were used. Multivariable-adjusted normalized deviates were calculated using the SAS/STAT user’s guide, version 8 (SAS Institute Inc. 2000); all linkage data presented is multivariable adjusted. For these adjustments, we used multiple linear regression separately for men and women, to obtain standardized residuals. These standardized residuals were then ranked, and normalized deviates were calculated. Covariates in the multivariable models included age, systolic blood pressure, number of cigarettes per day, total and HDL cholesterol, log-triglycerides, diabetes status (yes/no), body mass index, antihypertensive treatment (yes/no), menopausal status (yes/no), and hormone-replacement therapy in women (yes/no). All covariates were continuous except where otherwise specified.
For the linkage analysis, there were a total of 311 extended families, with 2–24 individuals each. There were 1,242 sib pairs (sibships range from two to seven individuals), 100 avuncular pairs, 39 half siblings, and 716 first cousins. Subjects were contained in sibships in the following distribution: 47% had two members, 32% had three members, 14% had four members, and 7% had five or more members.
Two-point and multipoint quantitative-trait linkage analyses were conducted with the normalized deviates for the carotid IMT measures by use of the SOLAR package (Almasy and Blangero 1998). This approach uses information on available marker data and the family structure to impute genotypes for untyped individuals. Estimates of the proportion of alleles with shared identity by descent (IBD) are then obtained for all relative pairs. By imputing genotypes for untyped individuals, SOLAR makes use of all individuals with carotid IMT measures in the linkage analysis. To assess linkage, a fitted model with no genetic marker information (polygenic model) is used and compared with a fitted model that incorporates genetic marker information at a specific marker (two-point analysis) or across a chromosome (multipoint analysis). To obtain the log odds (LOD) score, the log (base 10) of the ratio of the likelihoods of the marker-specific and polygenic models is computed. We have verified our results by use of GENEHUNTER (Kruglyak et al. 1996), an alternative statistical linkage program, conducted to address the suggestion by some critics of SOLAR that the multipoint IBD estimates calculated by GENEHUNTER are superior to those calculated by SOLAR (Ekstrom 2001).
We used SOLAR to estimate power and type 1 error. We used the function “simqtl” in SOLAR (Almasy and Blangero 1998) to simulate both a linked and an unlinked quantitative trait for power and type 1 error analysis, respectively. Given the family structure and the approximate number of individuals with trait values used in these analyses, we have 63%–95% power to detect a QTL that accounts for 25%–35% of the variation in the carotid IMT traits if we use a LOD score >2 as our critical value, and we have 37%–84% power for a LOD score >3. In addition to power, we conducted 100 simulations of our genomewide scan by use of unlinked phenotypes, to assess the risk of false-positive results. The genomewide chance is only 4% of a LOD score in our data being >3 when there is no genomewide linkage. This indicates that the chance of our results being a false positive is <4%, controlling for genomewide multiple testing.
Association Analysis
To evaluate the association between the SCARB1 gene and carotid IMT, a total of 2,245 individuals were included in the analysis. These individuals had at least one carotid IMT measure and were genotyped for at least one of the three SCARB1 polymorphisms considered. The majority of individuals (1,435) included in these analyses were members of families, with 2–14 individuals each. Association analyses for polymorphisms in the SCARB1 gene were analyzed using the SAS procedure PROC MIXED (SAS Institute Inc. 2000). This methodology uses a maximum-likelihood procedure, assuming a normal distribution of the phenotypes, to estimate the regression coefficients in a mixed-effect model that incorporates fixed effects for known covariates and specific genotypes and random effects to account for the correlation among members in the same families. Normalized deviates of crude IMT measures were used in these analyses to meet the normality assumption.
To evaluate individual effects of the SCARB1 polymorphisms on carotid IMT, we created two dichotomous variables. The significance of covariates and genotype effects was assessed by comparing a fitted model in which fixed effects for known covariates, genotypes, and random effects were included, with a model in which the two dummy variable-regression coefficients were set to zero. We calculated crude and multivariable-adjusted models, including the following covariates: age, sex, systolic blood pressure, number of cigarettes per day, total and HDL cholesterol, log-triglycerides, diabetes status (yes/no), body mass index, antihypertensive treatment (yes/no), menopausal status (yes/no), and hormone-replacement therapy in women (yes/no). To parallel the linkage analysis, the association analysis was repeated using sex-specific multivariable-adjusted residuals.
Results
Study Sample
The characteristics of the study samples used in the linkage and association analyses are shown in table 1. The mean age was 56 years in the linkage sample and 59 years in the association sample. Mean cholesterol, triglycerides, and HDL cholesterol were similar in both groups; the prevalence of hypertension and hypertension treatment was higher in the association sample.
Table 1.
Baseline Characteristics of Study Subjects[Note]
|
Finding in Sample |
||
| Characteristic | Linkage(n=1,225) | Association(n=2,245) |
| Age (years)a | 56 ± 10 | 59 ± 10 |
| Sex (% male) | 49 | 48 |
| Systolic blood pressure (mm Hg)a | 125 ± 18 | 129 ± 19 |
| Total cholesterol (mg/dL)a | 206 ± 43 | 205 ± 40 |
| Triglycerides (mg/dL)a | 144 ± 178 | 142 ± 151 |
| High-density lipoprotein cholesterol (mg/dL)a | 51 ± 15 | 51 ± 16 |
| Body mass index (kg/m2) a | 28.2 ± 5.5 | 28.0 ± 5.1 |
| Current smoking (%) | 17 | 15 |
| Postmenopause (%) | 70 | 79 |
| Hormone replacement therapy (%) | 22 | 28 |
| Prevalent hypertension (%) | 35 | 43 |
| Hypertension treatment (%) | 24 | 30 |
| Diabetes (%) | 5 | 6 |
| Cardiovascular disease (%) | 9 | 11 |
| Carotid artery IMT (mm)a: | ||
| Common | .59 ± .15 | .61 ± .16 |
| Internal | .56 ± .36 | .59 ± .39 |
Note.— To convert total and high-density lipoprotein from mg/dL to mmol/L, multiply by .0259. To convert triglycerides from mg/dL to mmol/L, multiply by .0113.
Mean ± SD.
Mean CCA and ICA IMT values are presented in table 1. The CCA values ranged from 0.33–3.25 mm; the ICA values ranged from 0.20–3.94 mm. The correlation between ICA and CCA IMT is 0.48 (P<.001).
Multivariable-Adjusted Two-Point and Multipoint Linkage
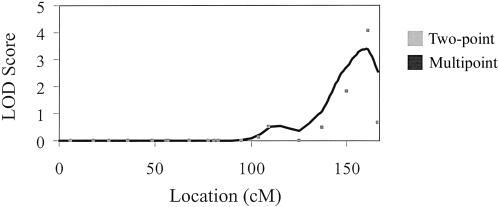
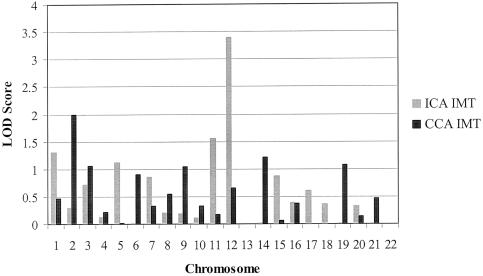
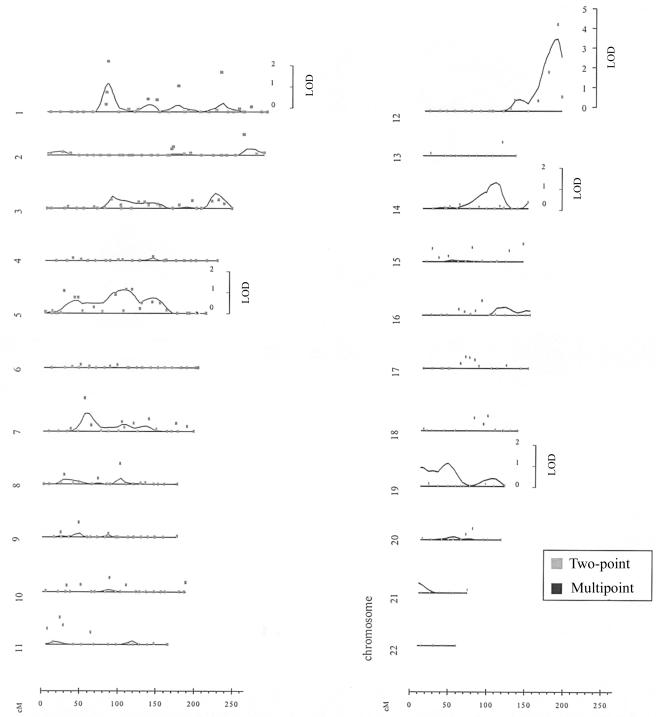
For the ICA, we observed a maximum two-point LOD score of 4.1 at 161 cM on chromosome 12, with a small secondary peak on chromosome 1 (LOD score 2.4 at 76 cM) and a maximum multipoint LOD score of 3.4 at 160 cM on chromosome 12 (fig. 1). The marker at the peak LOD score on chromosome 12 was D12S1045. The highest multipoint LOD scores for ICA and CCA IMT for each of the 22 autosomes are presented in figure 2. For CCA IMT, we observed a peak multipoint LOD score of 1.99 on chromosome 2 at 105 cM. The marker nearest to the peak LOD score on chromosome 2 was D2S1790. Using GENEHUNTER, we confirmed our results on chromosome 12, with a multipoint LOD score of 4.3 in the same region identified by SOLAR. Table 2 presents all LOD scores >1.0 for ICA and CCA IMT. The two-point and multipoint ICA IMT linkage results for all chromosomes appear as an online-only supplement.
Figure 1.
Two-point and multipoint LOD scores for ICA IMT, chromosome 12
Figure 2.
Peak multipoint LOD scores for ICA and CCA IMT by chromosome
Table 2.
All Markers and Locations with Two-Point LOD Scores >1.0 for ICA and CCA IMT[Note]
| Type and Marker | Chromosome | Location (cM) | LOD Score |
| ICA: | |||
| D1S2134 | 1 | 76 | 2.37 |
| D1S1653 | 1 | 164 | 1.22 |
| D1S1678 | 1 | 218 | 1.86 |
| D5S817 | 5 | 23 | 1.06 |
| D5S1725 | 5 | 98 | 1.12 |
| D5S1462 | 5 | 105 | 1.14 |
| D7S817 | 7 | 50 | 1.58 |
| D11S199 | 11 | 17 | 1.28 |
| D12S2078 | 12 | 150 | 1.82 |
| D12S1045 | 12 | 161 | 4.06 |
| CCA: | |||
| D1S3721 | 1 | 74 | 1.08 |
| D2S1790 | 2 | 103 | 1.57 |
| D2S2972 | 2 | 114 | 1.03 |
| D3S1768 | 3 | 62 | 1.16 |
| D3S2459 | 3 | 119 | 1.44 |
| D9S319 | 9 | 54 | 1.53 |
| D12S1301 | 12 | 56 | 1.70 |
| D12S1294 | 12 | 78 | 1.18 |
| D14S606 | 14 | 92 | 1.75 |
| D19S586 | 19 | 33 | 1.47 |
Note.— Genetic distance provided by Marshfield map location.
Figure 4.
Two-point and multipoint linkage results for ICA IMT
Association Analysis of Common Variants in SCARB1
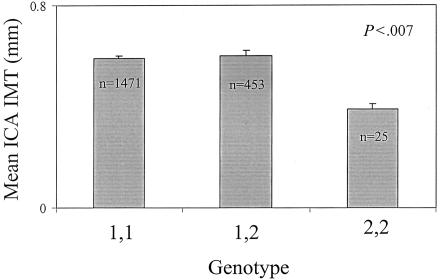
SCARB1 is an atherosclerosis candidate gene that resides at 124.9 Mb, ∼5 million base pairs away from the microsatellite marker D12S1045. Allele frequencies for the rare alleles in exons 1 and 8 and intron 5 were 0.13, 0.49, and 0.09, respectively. All polymorphisms were in Hardy-Weinberg equilibrium. In crude analyses, the exon 1 variant was significantly associated with ICA IMT (P<.007); crude (i.e., prior to the calculation of normalized deviates) ICA IMT values, arranged by exon 1 variant genotype, are presented in figure 3. In multivariable-adjusted analyses, subjects who are homozygous for the rare allele had a mean ICA IMT that was 0.5 normalized SDs lower than the rest of the population (P<.02 for differences among all three groups). The use of sex-specific multivariable-adjusted residuals resulted in similar differences in the magnitude of mean ICA IMT, as compared with the rest of the population, but the P value was attenuated (P=.12). Including the exon 1 variant as a covariate in the variance-component analysis in SOLAR revealed that it accounted for 2% of the overall variability in ICA IMT, whereas the additional covariates accounted for 36% of the total variation in CCA IMT, and 21% of the total variation for ICA IMT. In further multivariable analyses with ICA IMT, there were no significant associations with variants in either exon 8 or intron 5.
Figure 3.
Mean (SE) ICA IMT by genotype of SCARB1; data presented is unadjusted
In a subanalysis conducted on subjects included in the genomewide linkage analysis and typed for SCARB1 (n=736), multipoint LOD scores were calculated, after adjustment for the exon 1 genotype of SCARB1. The multivariable-adjusted LOD score was 2.33 prior to including the SCARB1 exon variant and 2.27 after including the SCARB1 exon 1 variant. The lower peak LOD score is due to the loss of sample size incurred when limiting the analysis to those subjects included in both the linkage and association samples.
Discussion
We found evidence for genomewide linkage to ICA IMT 161 cM from the tip of the short arm of chromosome 12, with a maximum two-point LOD score of 4.1 and multipoint LOD score of 3.4. To our knowledge, this is the first population-based study that has provided strong evidence for linkage of a QTL to a measure of subclinical atherosclerosis on a genomewide scale. We followed up our finding of genomewide linkage with an association study of variants in a highly plausible candidate gene, SCARB1, in the region of linkage. Evidence for association of the exon 1 variant of SCARB1 with ICA IMT was found. However, although this variant is associated with ICA IMT, the homozygotes are quite rare, and it does not account for the linkage observed, suggesting that other variants in this gene or in other genes in the region are responsible for the linkage with carotid IMT.
To date, the available evidence from genomewide linkage analyses of atherosclerosis phenotypes derives from selected populations. A recent report in families with two or more affected relatives identified a region of linkage to premature myocardial infarction on chromosome 14 (LOD score 3.9) (Broeckel et al. 2002). An additional genomewide linkage analysis performed in a genetically isolated population in Finland that was enriched for subjects with premature coronary artery disease demonstrated evidence for linkage on chromosome 2 (LOD score 3.0), ∼45 million base pairs from our peak LOD score for CCA IMT (Pajukanta et al. 2000). Linkage to chromosome 16 (LOD score 3.1) was found in another population with premature coronary artery disease in Mauritius (Francke et al. 2001). A recent study performed in 29 families enriched for hypertension showed evidence for linkage to coronary artery calcification, assessed by electron-beam computed tomography (LOD score 3.2 on chromosome 10) (Lange et al. 2002). Although these studies provide suggestive results, each was drawn from a sample selected for probands with a high burden of cardiac risk factors or premature atherosclerosis, or from a genetically isolated population. Whereas the findings of these prior studies may represent valid evidence for linkage of genetic loci to subclinical and clinical vascular disease phenotypes in rare families, the selected nature of the cohorts in these studies may limit the generalizability of the results.
It is interesting that QTLs for ICA IMT and CCA IMT map to different chromosomes and that our linkage peak for ICA IMT is stronger than that for CCA IMT. It has been suggested that thickening of the CCA intima may be a better marker for total body atherosclerosis, whereas thickening of the ICA intima might represent focal atherosclerotic plaques (O’Leary et al. 1996). Indeed, thickening of the ICA has been shown to be more strongly associated with incident CVD, compared with the CCA (O’Leary et al. 1996). Differential blood flow in regions of arterial bifurcation, such as the ICA, may increase thrombosis, lipid deposition, and atherosclerosis (Grabowski and Lam 1995), whereas laminar blood flow, as seen in the CCA, is inversely related to wall shear stress (Gnasso et al. 1996). Low wall shear stress, via an increase in the duration of time that blood comes into contact with the endothelial wall, may enhance the delivery of atherogenic particles (Gnasso et al. 1996). Thus, different pathophysiologic mechanisms may underlie observed differences in ICA IMT and CCA IMT, suggesting the role of unique sets of genes in their variation.
The region on chromosome 12 near our peak LOD score for ICA IMT contains a number of atherosclerosis candidate genes located very close to the region of linkage. Among them, SCARB1 is implicated in the atherogenic process by a considerable body of prior research. The identification of SCARB1 (Acton et al. 1996) represented a significant advance in understanding the manner in which high-density lipoproteins (HDL) are believed to exert their anti-atherogenic role in cholesterol metabolism through the process of delivering cholesterol from peripheral tissues back to the liver for removal from the body, called “reverse cholesterol transport” (Stein and Stein 1999; Silver et al. 2000). SCARB1, a cell-surface glycoprotein expressed in the liver, adrenal glands, and ovaries, was the first HDL receptor to be well defined and characterized in vitro and in animal studies. In murine atherosclerosis models, the absence of SCARB1 dramatically accelerates the onset of atherosclerosis (Braun et al. 2002), whereas atherosclerosis is suppressed by hepatic overexpression of this gene (Arai et al. 1999; Kozarsky et al. 2000).
The human SCARB1 gene was characterized and investigated in a southern European population by Acton et al. (1999). Three common SNP variants were described (with allele frequency >0.1) at exon 1 (G→A), exon 8 (C→T), and intron 5 (C→T). The polymorphisms at intron 5 and exon 8 are in linkage disequilibrium with each other and are both probably nonfunctional, whereas the SNP in exon 1 is not in linkage disequilibrium with either of the other two SNPs and encodes an amino acid change from glycine to serine that modifies one of the potential acylation sites in the SCARB1 molecule. In this study by Acton et al. (1999), associations were found between SCARB1 and both HDL and LDL (low-density lipoproteins), suggesting that SCARB1 may play a role in the metabolism of both lipoprotein classes in humans. In addition, associations were detected with triglycerides and body mass index (Acton et al. 1999). In a study of the Framingham Heart Study Offspring cohort, it has been shown that statistically significant interactions exist between the SCARB1 exon 1 variant and type 2 diabetes. Diabetic subjects with the less-common allele have lower lipid concentrations, including lower LDL cholesterol and HDL cholesterol (Osgood et al. 2003). Taken together, these data allow us to hypothesize that the SCARB1 gene may also be involved in several features of the metabolic syndrome.
Although our findings support a role for SCARB1 as an atherosclerosis candidate gene, we acknowledge the limitation that we have not demonstrated that associations of SCARB1 variants with carotid IMT explain the linkage of chromosome 12 to carotid IMT. Our findings illustrate the challenges inherent in attempts to implicate a specific gene and mutation in the etiology of complex diseases by utilizing the results of genomewide linkage analyses (Altshuler et al. 2000). In this regard, we note that several other candidate genes of interest reside near the region of linkage. For example, the neuronal nitric oxide synthase 1 (NOS1 [MIM 163731]) gene lies near the region of peak linkage that was found in the current study, and upregulation of juxtaglomerular NOS1 has been shown to precede glomerulosclerosis in fawn-hooded hypertensive rats (Weichert et al. 2001). In human studies of asthma, polymorphisms in the NOS1 gene were associated with eosinophil cell counts and IgE levels (Immervoll et al. 2001). Thus, NOS1 may play a role in inflammation and human atherosclerosis. Mutations in the TCF1 (MIM 142410) gene, mapped to chromosome 12q24, have been shown to cause biliary-acid–transport defects, with impaired liver-cholesterol synthesis and HDL metabolism (Shih et al. 2001). Maturity-onset diabetes of the young (MODY3 [MIM 600496]) is a rare, dominantly inherited form of early-onset diabetes that is caused by mutations in the hepatocyte nuclear factor-1 alpha gene (HNF-1 alpha, encoded by the TCF1 gene) (Yamagata et al. 1996). The ATP-binding cassette, subfamily B, member 9 (ABCB9) (Zhang et al. 2000) [MIM 605453], a member of the ABC family of genes, also resides near the region of peak linkage that was found in the current study. Several genes in the ABC gene family have been shown to impact lipid metabolism (Borst and Elferink 2002). The insulin-like growth factor 1 gene (IGF1 [MIM 147440]) also resides near the peak linkage region and has been shown to be associated with type 2 diabetes and glucose tolerance (Frayling et al. 2002).
Some additional limitations of our study deserve mention. First, because the phenotype information for the study was not collected until 1996, we may have incurred a survival bias, since subjects with a greater genetic predisposition inducing more severe disease may have died prematurely or been lost to follow-up. However, this loss would likely attenuate our results. Second, most of the subjects in our study were white, potentially limiting the generalizability of the findings to other ethnicities. Last, the sample size and the marker spacing of 10 cM in the genomewide linkage analysis may have limited the power of our study to localize a disease-influencing QTL that is in linkage with carotid IMT. However, we do not believe the low power affects the validity of our primary findings, since we obtained a LOD score of 3.4, which is typically regarded as of sufficient magnitude to be of nearly genomewide significance. Regardless, there are likely to be QTLs that were not detected in our study but that contribute significantly to variability in carotid IMT, and there may be additional genes in other regions of the genome that we have missed, given the low power of our study.
These data suggest substantial evidence for a QTL on chromosome 12 that influences the IMT of the ICA. SCARB1 represents an attractive candidate gene, and a common missense variant in exon 1 of SCARB1 or a nearby locus is associated with lower IMT. However, we have not demonstrated that allelic variants in this candidate gene explain our observed linkage. Although our findings suggest that further investigation is warranted to determine the role of SCARB1 in atherosclerosis, additional variants on chromosome 12, including those in nearby candidate genes, should be examined for evidence of association to atherosclerosis traits. These findings support the continued use of genomewide approaches to discover genetic variants predisposing to quantitative measures of subclinical atherosclerosis.
Acknowledgments
We would like to acknowledge Drs. Daniel Levy, Alan Herbert, and David Altshuler for their helpful comments on this manuscript. This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute’s Framingham Heart Study (NIH-NHLBI N01-HC-25195) and by the National Institute of Neurological Disorders and Stroke (NIH-NINDS 5R01-NS17950-20), as well as by NIH-NHLBI grant HL54776 and USDA Research Service contracts 53-K06-5-10 and 58-1950-9-001.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCARB1, NOS1, TCF1, MODY3, ABCB9, and IGF1)
References
- Acton S, Osgood D, Donoghue M, Corella D, Pocovi M, Cenarro A, Mozas P, Keilty J, Squazzo S, Woolf EA, Ordovas JM (1999) Association of polymorphisms at the SR-BI gene locus with plasma lipid levels and body mass index in a white population. Arterioscler Thromb Vasc Biol 19:1734–1743 [DOI] [PubMed] [Google Scholar]
- Acton S, Rigotti A, Landschulz KT, Xu S, Hobbs HH, Krieger M (1996) Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271:518–520 [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Daly M, Kruglyak L (2000) Guilt by association. Nat Genet 26:135–137 10.1038/79839 [DOI] [PubMed] [Google Scholar]
- American Heart Association (2002) 2003 heart and stroke statistical update. American Heart Association, Dallas [Google Scholar]
- Arai T, Wang N, Bezouevski M, Welch C, Tall AR (1999) Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor BI transgene. J Biol Chem 274:2366–2371 10.1074/jbc.274.4.2366 [DOI] [PubMed] [Google Scholar]
- Borst P, Elferink RO (2002) Mammalian ABC transporters in health and disease. Annu Rev Biochem 71:537–592 10.1146/annurev.biochem.71.102301.093055 [DOI] [PubMed] [Google Scholar]
- Bots ML, Hoes AW, Koudstaal PJ, Hofman A, Grobbee DE (1997) Common carotid intima-media thickness and risk of stroke and myocardial infarction: the Rotterdam Study. Circulation 96:1432–1437 [DOI] [PubMed] [Google Scholar]
- Braun A, Trigatti BL, Post MJ, Sato K, Simons M, Edelberg JM, Rosenberg RD, Schrenzel M, Krieger M (2002) Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ Res 90:270–276 10.1161/hh0302.104462 [DOI] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, Jacob HJ, Schunkert H (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30:210–214 10.1038/ng827 [DOI] [PubMed] [Google Scholar]
- Crouse JR, Goldbourt U, Evans G, Pinsky J, Sharrett AR, Sorlie P, Riley W, Heiss G (1996) Risk factors and segment-specific carotid arterial enlargement in the Atherosclerosis Risk in Communities (ARIC) cohort. Stroke 27:69–75 [DOI] [PubMed] [Google Scholar]
- Cupples LA, D'Agostino RB (1987) Some risk factors related to the annual incidence of cardiovascular disease and death using pooled repeated biennial measurements: Framingham Study, 30-year follow-up. In: Kannel WB, Polf PA, Garrison RJ (eds) The Framingham Heart Study: an epidemiological investigation of cardiovascular disease. NIH Publication 87–203, section 34. Government Printing Office, Washington, DC [Google Scholar]
- Davis PH, Dawson JD, Mahoney LT, Lauer RM (1999) Increased carotid intimal-medial thickness and coronary calcification are related in young and middle-aged adults: the Muscatine study. Circulation 100:838–842 [DOI] [PubMed] [Google Scholar]
- Dawber TR, Kannel WB, Lyell LP (1963) An approach to longitudinal studies in a community: the Framingham Heart Study. Ann NY Acad Sci 107:539–556 [DOI] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE (1951) Epidemiologic approaches to heart disease: the Framingham study. Am J Public Health 41:279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom CT (2001) Power of multipoint identity-by-descent methods to detect linkage using variance component models. Genet Epidemiol 21:285–298 10.1002/gepi.1035 [DOI] [PubMed] [Google Scholar]
- Espeland MA, Tang R, Terry JG, Davis DH, Mercuri M, Crouse JR III (1999) Associations of risk factors with segment-specific intimal-medial thickness of the extracranial carotid artery. Stroke 30:1047–1055 [DOI] [PubMed] [Google Scholar]
- Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP (1975) The Framingham Offspring Study. Design and preliminary data. Prev Med 4:518–525 [DOI] [PubMed] [Google Scholar]
- Fox CS, Polak JF, Chazaro I, Cupples A, Wolf PA, D’Agostino RA, O’Donnell CJ (2003) Genetic and environmental contributions to atherosclerosis phenotypes in men and women: heritability of carotid intima-media thickness in the Framingham Heart Study. Stroke 34:397–401 10.1161/01.STR.0000048214.56981.6F [DOI] [PubMed] [Google Scholar]
- Francke S, Manraj M, Lacquemant C, Lecoeur C, Lepretre F, Passa P, Hebe A, Corset L, Yan SL, Lahmidi S, Jankee S, Gunness TK, Ramjuttun US, Balgobin V, Dina C, Froguel P (2001) A genome-wide scan for coronary heart disease suggests in Indo- Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 10:2751–2765 10.1093/hmg/10.24.2751 [DOI] [PubMed] [Google Scholar]
- Frayling TM, Hattersley AT, McCarthy A, Holly J, Mitchell SM, Gloyn AL, Owen K, Davies D, Smith GD, Ben Shlomo Y (2002) A putative functional polymorphism in the IGF-I gene: association studies with type 2 diabetes, adult height, glucose tolerance, and fetal growth in U.K. populations. Diabetes 51:2313–2316 [DOI] [PubMed] [Google Scholar]
- Gnasso A, Carallo C, Irace C, Spagnuolo V, De Novara G, Mattioli PL, Pujia A (1996) Association between intima-media thickness and wall shear stress in common carotid arteries in healthy male subjects. Circulation 94:3257–3262 [DOI] [PubMed] [Google Scholar]
- Grabowski EF and Lam FP (1995) Endothelial cell function, including tissue factor expression, under flow conditions. Thromb Haemost 74:123–128 [PubMed] [Google Scholar]
- Gross-Bellard M, Oudet P, Chambon P (1973) Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem 36:32–38 [DOI] [PubMed] [Google Scholar]
- Immervoll T, Loesgen S, Dutsch G, Gohlke H, Herbon N, Klugbauer S, Dempfle A, Bickeboller H, Becker-Follmann J, Ruschendorf F, Saar K, Reis A, Wichmann HE, Wjst M (2001) Fine mapping and single nucleotide polymorphism association results of candidate genes for asthma and related phenotypes. Hum Mutat 18:327–336 10.1002/humu.1194 [DOI] [PubMed] [Google Scholar]
- Kannel WB, Feinleib M, McNamara PM, Garrison RJ, Castelli WP (1979) An investigation of coronary heart disease in families. The Framingham offspring study. Am J Epidemiol 110:281–290 [DOI] [PubMed] [Google Scholar]
- Kozarsky KF, Donahee MH, Glick JM, Krieger M, Rader DJ (2000) Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler Thromb Vasc Biol 20:721–727 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lange LA, Lange EM, Bielak LF, Langefeld CD, Kardia SL, Royston P, Turner ST, Sheedy PF, Boerwinkle E, Peyser PA (2002) Autosomal genome-wide scan for coronary artery calcification loci in sibships at high risk for hypertension. Arterioscler Thromb Vasc Biol 22:418–423 10.1161/hq0302.105721 [DOI] [PubMed] [Google Scholar]
- Levy D, DeStefano AL, Larson MG, O’Donnell CJ, Lifton RP, Gavras H, Cupples LA, Myers RH (2000) Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the Framingham Heart Study. Hypertension 36:477–483 [DOI] [PubMed] [Google Scholar]
- Lusis AJ (2000) Atherosclerosis. Nature 407:233–241 10.1038/35025203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DH, Polak JF, Kronmal RA, Manolio TA, Burke GL, Wolfson SK Jr (1999) Carotid-artery intima and media thickness as a risk factor for myocardial infarction and stroke in older adults. Cardiovascular Health Study Collaborative Research Group. N Engl J Med 340:14–22 10.1056/NEJM199901073400103 [DOI] [PubMed] [Google Scholar]
- O’Leary DH, Polak JF, Kronmal RA, Savage PJ, Borhani NO, Kittner SJ, Tracy R, Gardin JM, Price TR, Furberg CD (1996) Thickening of the carotid wall: a marker for atherosclerosis in the elderly? Cardiovascular Health Study Collaborative Research Group. Stroke 27:224–231 [DOI] [PubMed] [Google Scholar]
- Osgood D, Corella D, Demissie S, Cupples LA, Wilson PW, Meigs JB, Schaefer EJ, Coltell O, Ordovas JM (2003) Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the Framingham Study. J Clin Endocrinol Metab 88:2869–2879 10.1210/jc.2002-021664 [DOI] [PubMed] [Google Scholar]
- Osgood-McWeeney D, Galluzzi JR, Ordovas JM (2000) Allelic discrimination for single nucleotide polymorphisms in the human scavenger receptor class B type 1 gene locus using fluorescent probes. Clin Chem 46:118–119 [PubMed] [Google Scholar]
- Pajukanta P, Cargill M, Viitanen L, Nuotio I, Kareinen A, Perola M, Terwilliger JD, Kempas E, Daly M, Lilja H, Rioux JD, Brettin T, Viikari JS, Ronnemaa T, Laakso M, Lander ES, Peltonen L (2000) Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet 67:1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc. (2000) SAS/STAT user’s guide, version 8, SAS Institute, Cary, NC [Google Scholar]
- Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M (2001) Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 27:375–382 10.1038/86871 [DOI] [PubMed] [Google Scholar]
- Silver DL, Jiang XC, Arai T, Bruce C, Tall AR (2000) Receptors and lipid transfer proteins in HDL metabolism. Ann NY Acad Sci 902:103–111 [DOI] [PubMed] [Google Scholar]
- Stein O, Stein Y (1999) Atheroprotective mechanisms of HDL. Atherosclerosis 144:285–301 10.1016/S0021-9150(99)00065-9 [DOI] [PubMed] [Google Scholar]
- Weichert W, Paliege A, Provoost AP, Bachmann S (2001) Upregulation of juxtaglomerular NOS1 and COX-2 precedes glomerulosclerosis in fawn-hooded hypertensive rats. Am J Physiol Renal Physiol 280:F706–F714 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox RD, Lathrop GM, Boriraj VV, Chen X, Cox NJ, Oda Y, Yano H, Le Beau MM, Yamada S, Nishigori H, Takeda J, Fajans SS, Hattersley AT, Iwasaki N, Hansen T, Pedersen O, Polonsky KS, Bell GI (1996) Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 384:455–458 10.1038/384455a0 [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhang W, Liu L, Fisher CL, Hui D, Childs S, Dorovini-Zis K, Ling V (2000) Characterization of ABCB9, an ATP binding cassette protein associated with lysosomes. J Biol Chem 275:23287–23294 10.1074/jbc.M001819200 [DOI] [PubMed] [Google Scholar]