Abstract
Stüve-Wiedemann syndrome (SWS) is a severe autosomal recessive condition characterized by bowing of the long bones, with cortical thickening, flared metaphyses with coarsened trabecular pattern, camptodactyly, respiratory distress, feeding difficulties, and hyperthermic episodes responsible for early lethality. Clinical overlap with Schwartz-Jampel type 2 syndrome (SJS2) has suggested that SWS and SJS2 could be allelic disorders. Through studying a series of 19 families with SWS/SJS2, we have mapped the disease gene to chromosome 5p13.1 at locus D5S418 (Zmax=10.66 at θ=0) and have identified null mutations in the leukemia inhibitory factor receptor (LIFR or gp190 chain) gene. A total of 14 distinct mutations were identified in the 19 families. An identical frameshift insertion (653_654insT) was identified in families from the United Arab Emirates, suggesting a founder effect in that region. It is interesting that 12/14 mutations predicted premature termination of translation. Functional studies indicated that these mutations alter the stability of LIFR messenger RNA transcripts, resulting in the absence of the LIFR protein and in the impairment of the JAK/STAT3 signaling pathway in patient cells. We conclude, therefore, that SWS and SJS2 represent a single clinically and genetically homogeneous condition due to null mutations in the LIFR gene on chromosome 5p13.
Introduction
Stüve-Wiedemann syndrome (SWS [MIM 601559]) belongs to the group of the bent-bone dysplasias and is characterized by bowing of the lower limbs, with internal cortical thickening, wide metaphyses with abnormal trabecular pattern, and camptodactyly (Stüve and Wiedemann 1971; Cormier-Daire et al. 1998). Additional features include feeding and swallowing difficulties, as well as respiratory distress and hyperthermic episodes, which cause death in the first months of life. The rare survivors develop progressive scoliosis; spontaneous fractures; bowing of the lower limbs, with prominent joints and dysautonomia symptoms, including temperature instability; absent corneal and patellar reflexes; and smooth tongue (Kozlowski and Tenconi 1996; Superti-Furga et al. 1998; Chen et al. 2001; Al-Gazali et al. 2003; Di Rocco et al. 2003). Clinical and radiological overlap with Schwartz-Jampel type 2 syndrome (SJS2) has suggested that SWS and SJS2 could be a single entity (Al-Gazali et al. 1996; Cormier-Daire et al. 1998; Superti-Furga et al. 1998). Here, we report the mapping of the SWS/SJS2 gene on chromosome 5p13.1 and the identification of null mutations in the leukemia inhibitory factor receptor (LIFR [GenBank accession number NM_002310]) gene in 19 affected families. It appears, therefore, that SWS and SJS2 represent a single clinically and genetically homogeneous condition.
Material and Methods
Patients
A total of 24 children from 19 families (14 consanguineous and 5 nonconsanguineous families) were included in the study. Among them, 10 have been reported elsewhere (Al-Gazali et al. 1996, 2003; Chabrol et al. 1997; Cormier-Daire et al. 1998; Superti-Furga et al. 1998; Di Rocco et al. 2003), and 9 additional families (families 2, 3, 9–11, 13, 14, 18, and 19) fulfilled the diagnostic criteria for SWS, namely bowing of the long bones with flared metaphyses, feeding difficulties, respiratory distress, and hyperthermic episodes (fig. 1). Among the 24 affected children, 16 died in the first years of life, and 8 are still alive (those from families 4, 5, 11, 12, 16, 17, and 19). All survivors presented with a severe bowing of the long bones, with prominent joints, severe spinal deformity, and normal mental development. Neurological symptoms included temperature instability, absent corneal and patellar reflexes, and smooth tongue. Radiological changes included undertubulation of diaphyses and rarefaction and striation of metaphyses.
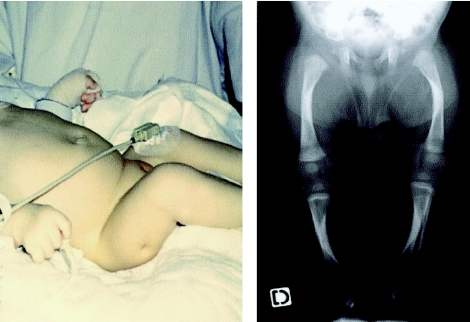
Figure 1.
Clinical and radiological features of patient 3. The general appearance is characterized by bowing of the lower limbs, with skin dimple and camptodactyly. X-rays of the lower limb show bowing of the long bones, with internal cortical thickening at the concave site and irregular metaphyses.
Blood samples were obtained with the written consent of the patients and unaffected relatives. Genomic DNA was extracted from leukocytes, according to standard procedures.
For homozygosity mapping, we used a panel of 400 markers at an average distance of 10 cM. Oligonucleotide sequences of the OSMR (oncostatin M receptor) CA-repeat were: forward 5′-AAG GGT GAT GCC AGT TCA AG-3′ and reverse 5′-CAC AAA GGC CAT CAC AAC CT-3′. The MLINK program of the Linkage software package was used to calculate two-point LOD scores between the disease phenotype and each of the markers, under the assumption of a recessive disease with a mutant allele frequency of 0.01.
A series of 42 intronic primers was designed to amplify the 20 coding exons of the LIFR gene. Amplification products were purified and sequenced using the fluorescent dideoxy-terminator method on an automatic sequencer, ABI 3100. Total RNAs were extracted from human primary cultured cells (chondrocytes, osteoblasts, and skin fibroblasts) through use of the RNeasy Mini Kit (Qiagen GmbH). Complementary DNA was synthesized by priming with random hexamers in the presence of MuLV reverse transcriptase, according to the manufacturer’s protocol (GeneAmp RNA PCR Core Kit, Roche GmbH). Thirty-five to 40 PCR cycles were performed at an annealing temperature of 56°C to amplify a 397-bp fragment specific for the LIFR gene (forward 5′-CCTCATCTTAGATGTGTCTC-3′; reverse 5′-TTCTCCTCACCTGGCATTAC-3′). Sense and antisense primers used for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification were as follows: sense, 5′-AGACAGCCGCATCTTCTTGT-3′; antisense, 5′-CCACAGTCTTCTGAGTGGCA-3′ (587 bp).
For binding experiments, the leukemia inhibitory factor (LIF) was iodinated using the chloramine T method (Godard et al. 1992) at a specific radioactivity of 10,720 counts per minute/fmol. Binding experiments were performed in PBS-BSA. Cells (0.3×106/well in 96-well round-bottom plates) were incubated with increasing concentrations of labeled LIF in a final volume of 50 μl. Nonspecific binding was evaluated by including a 100-fold excess of unlabeled cytokine. Incubation was performed under agitation for 90 min at 4°C. Cell-bound and unbound fractions were separated by centrifugation through a layer of dibutylphthalate (90%) and paraffin oil (10%). Regression analysis of the binding data was performed using a one-equilibrium–binding equation.
For western blotting, skin fibroblasts were grown in Dulbecco’s modified Eagle medium (with 10% fetal bovine serum). The cells were incubated in a serum-free medium for 5 min or 30 min with 20 ng/ml LIF (CHEMICON International), were washed twice with PBS, and were lysed in radioimmunoprecipitation buffer (50 mM Tris-Cl; pH 7.5; 150 mM NaCl; 1% Nonidet P-40; 0.5% sodium deoxycholate; 1 mM EDTA; 1μg ml−1 leupeptin; 1μg ml−1 aprotinin; 0.2 mM phenylmethylsulfonyl fluoride). Total proteins were quantified by colorimetric dosage; protein extracts were run on NuPAGE 4%–12% Bis-Tris Gels (Invitrogen) under reducing conditions, were transferred onto immobilon membranes (Millipore), and were incubated with anti-STAT 3 and antiphospho-STAT3 antibodies (Cell Signaling Technology).
For immunocytochemistry, fibroblasts seeded in culture chambers (FALCON) were stimulated for 15 min with LIF (20 ng/ml), then were fixed with 4% paraformaldehyde, were blocked with 10% sheep serum, and were incubated with anti-STAT3 antibody (1:200) (Cell Signaling Technology) at room temperature. A fluorescent dye-conjugated antibody (DAKO) was added for 1 h. Cells were then examined with a Zeiss LSM 510 confocal microscope.
Results
For linkage analysis, 11 families with SWS/SJS2 (10 consanguineous families and 1 nonconsanguineous family) were included in this study. Using a homozygosity-mapping strategy, we found linkage of the disease gene to chromosome 5p13.1 (Zmax=10.66 at θ=0 at locus D5S418) (table 1). A recombination event between locus D5S1964 and the OSMR gene in family 1 defined the proximal boundary of the genetic interval, and another recombination event between locus D5S418 and locus D5S1457 in family 13 defined the distal boundary of the candidate region (3.2 Mb) (fig. 2A).
Table 1.
Two-Point LOD Scores for Linkage of an SWS Locus to Chromosome 5p13.1
|
LOD at θ = |
||||||||
| Loci | Distancea | .00 | .05 | .10 | .20 | .30 | Zmax | θ |
| D5S1964 | −∞ | 5.4 | 4.87 | 3.35 | 1.89 | 5.4 | .05 | |
| .986 | ||||||||
| OSMR-CA | 8.99 | 7.59 | 6.24 | 3.77 | 1.86 | 8.99 | .00 | |
| 1.04 | ||||||||
| D5S2022 | 9.86 | 8.44 | 7.07 | 4.56 | 2.56 | 9.86 | .00 | |
| .097 | ||||||||
| D5S418 | 10.66b | 9.21 | 7.78 | 5.06 | 2.76 | 10.66 | .00 | |
| 1.07 | ||||||||
| D5S1457 | 1.05 | 3.09 | 2.81 | 1.85 | .19 | 3.09 | .05 | |
The distance between loci is given in megabases, according to the human genome working draft (UCSC Genome Bioinformatics).
Maximum LOD score.
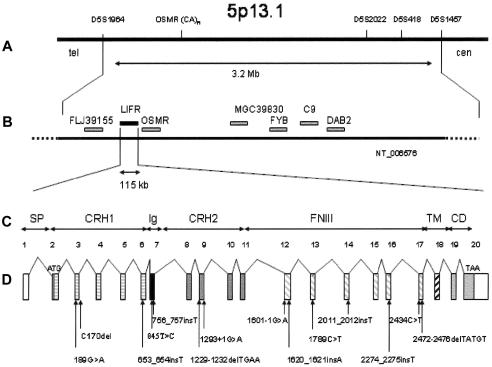
Figure 2.
Map of the region encompassing the candidate genes and genomic organization of LIFR. A, The common region of homozygosity is located between markers D5S1964 and D5S1457. B, The seven candidate genes identified in that region. C and D, Exon-intron structure of the 115-kb LIFR. Mutations were identified in four extracellular domains.
A search of the human genome resources of the National Center for Biotechnology Information (NCBI) resulted in several putative disease genes within the candidate region, including FLJ39155 (similar to perlecan), disabled-2 (DAB2), complement 9 (C9), Fyn-binding protein (FYB), oncostatin M receptor (OSMR), and LIFR (Gearing et al. 1993; fig. 2B). The LIF receptor is a heterodimer complex composed of the gp190 subunit referred to as “LIFR” and the gp130 subunit (Gearing et al. 1991). This receptor is able to bind multiple cytokines, including LIF, OSM, ciliary neurotrophic factor (CNTF), cardiotrophin 1, and neurotrophin-1/B-cell stimulating factor 3 (Gearing et al. 1991; Metcalf 2003). LIFR (gp190) was considered to be a good candidate because LIFR−/− mice present with reduction of fetal bone volume, with an increased number of osteoclasts, reduction of astrocyte numbers in spinal cord and brain, and perinatal death (Ware et al. 1995).
LIFR is composed of 20 exons and encodes a 1,097–amino acid transmembrane protein, which is composed of six different domains: two cytokine receptor homology domains (CRH1 and CRH2), one Ig-like domain (Ig), one type III fibronectin domain with three modules (FNIII), one transmembrane domain (TM), and one cytoplasmic domain (CD) (fig. 2C) (Gearing et al. 1991; Tomida and Gotoh 1996; Bitard et al. 2003). A total of 14 distinct mutations was identified in the 11 initial families and in eight additional cases (in families 5, 7–10, 12, 14, and 19) (table 2). An identical frameshift insertion (653_654insT) was identified in families from the United Arab Emirates, suggesting a founder effect in that region. Similarly, the same mutation (R597X) was identified in one family from Portugal (family 6), in two Gypsy families (families 7 and 8), in one French family (family 9), and in one Swiss family (family 12) (in a compound heterozygote). It is interesting that 12/14 mutations predicted premature termination of translation. The mutations were located in CRH1 (3/14), in CRH2 (2/14), in the Ig-like domain (2/14), or in the FNIII domains of the protein (7/14) (fig. 2D). The mutations were not identified in 210 control chromosomes.
Table 2.
LIFR Mutations Identified in the 19 Families with SWS/SJS2
| Family | Ethnic Origin | ConsanguineousParents | No. ofAffectedChildren | Nucleotide Change | Amino Acid Change | MutatedExon(s) | PredictedConsequenceon theProtein | Domain |
| 1a | Yugoslavian | Yes | 2 | 756_757insT | K253X | 7 | Truncated | Ig-like |
| 2 | Senegalese | Yes | 1 | 2434C→T | R812X | 17 | Truncated | FNIII |
| 3 | Turkish | Yes | 1 | 2274_2275insT | Y559L and frameshift causing a stop, 13 codons downstream | 16 | Truncated | FNIII |
| 4a | Italian | Yes | 1 | 1620_1621insA | T541N and frameshift causing a stop, 7 codons downstream | 12 | Truncated | FNIII |
| 5a | Italian | No | 1 | C170del | T57I and frameshift causing a stop, 54 codons downstream | 3 | Truncated | CRH1 |
| 6a | Gypsy | Yes | 1 | 1789C→T | R597X | 13 | Truncated | FNIII |
| 7a | Gypsy | No | 1 | 1789C→T | R597X | 13 | Truncated | FNIII |
| 8a | Portuguese | Yes | 1 | 1789C→T | R597X | 13 | Truncated | FNIII |
| 9 | French | Yes | 1 | 1789C→T | R597X | 13 | Truncated | FNIII |
| 10 | French | No | 1 | 189G→A/(1293+1G→A+1122_1291del) | W63X/splicing mutation and frameshift causing a stop, 13 codons downstream | 3, 9 | Truncated | CRH1/CRH2 |
| 11 | Canadian | No | 1 | 845T→C/1229-1232 delTGAA | S279P/frameshift causing a stop, 10 codons downstream | 7, 9 | Missense/truncated | Ig-like/CRH2 |
| 12a | Swiss | No | 2 | 1789C→T/2011_2012insT | R597X/frameshift causing a stop, 12 codons downstream | 13, 14 | Truncated | FNIII |
| 13 | Israeli Arab Muslim | Yes | 2 | 1601-1G→A | Splicing mutation | 12 | FNIII | |
| 14 | Israeli Arab Muslim | Yes | 1 | 2472-2476delTATGT | S824R and frameshift causing a stop, 39 codons downstream | 17 | Truncated | FNIII |
| 15a | Omanib | Yes | 2 | 653_654insT | Frameshift causing a stop, 2 codons downstream | 6 | Truncated | CRH1 |
| 16a | Yemenib | Yes | 1 | 653_654insT | Frameshift causing a stop, 2 codons downstream | 6 | Truncated | CRH1 |
| 17a | Yemenib | Yes | 2 | 653_654insT | Frameshift causing a stop, 2 codons downstream | 6 | Truncated | CRH1 |
| 18 | Omanib | Yes | 1 | 653_654insT | Frameshift causing a stop, 2 codons downstream | 6 | Truncated | CRH1 |
| 19 | Omanib | Yes | 1 | 653_654insT | Frameshift causing a stop, 2 codons downstream | 6 | Truncated | CRH1 |
Families 1 and 8 (Cormier-Daire et al. 1998), 4 and 5 (Di Rocco et al. 2003), 6 and 7 (Chabrol et al. 1997), 12 (Superti-Furga et al. 1998), and 15–17 (Al-Gazali et al. 1996, 2003) have been reported elsewhere. Mutations were present at the homozygous state in affected children from families 1–9 and families 13–19.
From United Arab Emirates.
RT-PCR analysis of LIFR RNA transcripts detected a single amplification product in osteoblasts, chondrocytes, and fibroblasts from controls. By contrast, RT-PCR analysis of the fibroblast RNA transcripts detected either no amplification product (patient 4) or a weak signal (patients 7 and 10) (fig. 3A).
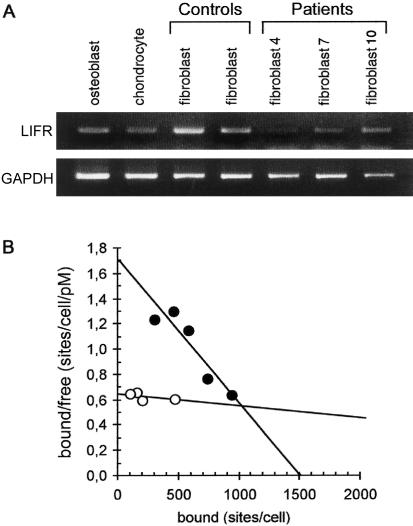
Figure 3.
RT-PCR detection of LIFR transcript and Scatchard analysis of high-affinity receptor. A, RT-PCR analyses were done using RNA isolated from control osteoblasts, chondrocytes, and fibroblasts and from patient fibroblasts (patients 4, 7, and 10). The specific transcript was detected in control osteoblasts, chondrocytes, and fibroblasts but was almost absent in SWS fibroblasts. B, Scatchard analysis showing specific binding of the iodinated LIF in control fibroblasts (blackened circles) and the absence of binding in patient 10 fibroblasts (unblackened circles).
Binding experiments on patient and control fibroblasts through use of iodinated LIF revealed that fibroblasts from patients 1, 4, 7, and 10 failed to express high-affinity LIF receptors, whereas control fibroblasts bound LIF with an apparent Kd of 0.9 nM and a maximum capacity of 1,500 sites/cell (fig. 3B). Binding of LIF to the LIFR complex is known to induce signaling through the JAK/STAT and MAPK pathways. After phosphorylation by JAK, STAT3 molecules dimerize and translocate to the nucleus, where they bind to cytokine-response elements and ultimately activate gene transcription (Schiemann et al. 1997; Starr et al. 1997; Dziennis and Habecker 2003). The activation of STAT3 in response to LIF was tested in patient and control fibroblasts. Western blot analysis of fibroblast lysates from patients 1, 4, and 7 with an antiphospho STAT3 antibody showed that STAT3 failed to be phosphorylated in response to LIF (5-min stimulation), whereas LIF normally triggered STAT3 phosphorylation in control cells (fig. 4A). Similar results were obtained after 30 min (data not shown). Consistently, immunofluorescence analysis showed that incubation with LIF (15 min) triggered nuclear localization of STAT3 in control fibroblasts, whereas fibroblasts from patient 10 exhibited an exclusive cytoplasmic staining (fig. 4B).
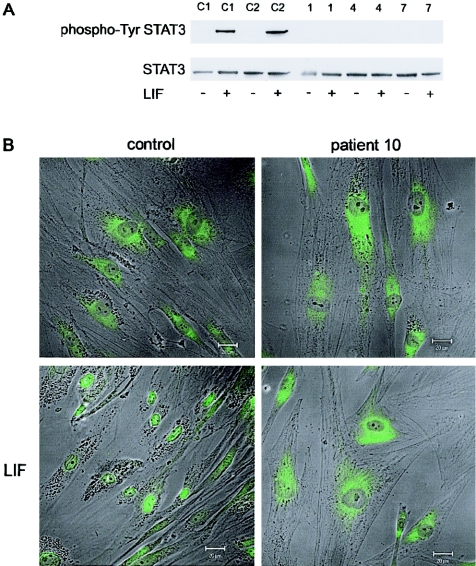
Figure 4.
Analysis of the STAT pathway in control and patient fibroblasts. A, Patient (patients 1, 4, and 7) and control cell lysates (C1 and C2) analyzed by western blot by use of antiphospho-Tyr STAT3 or STAT3 antibodies. Fibroblasts were stimulated with LIF (20 ng/ml) for 5 min. Only control cells gave a Phospho-STAT3 signal after LIF stimulation. B, Immunofluorescence of control and patient 10 fibroblasts with an anti-STAT3 antibody in the absence (upper panel) or presence (lower panel) of LIF (20 ng/ml) for 15 min. Only LIF-stimulated control cells showed nuclear localization of the STAT3 protein. Bar = 20μm.
Discussion
We report here the identification of null mutations in 19 families with SWS/SJS2. These results first confirm that SWS and SJS2 represent a single entity. The same mutations were identified in families originating from the United Arab Emirates, suggesting a founder effect within that ethnic group. All but two mutations predicted the generation of a markedly truncated protein. The absence of any detectable transcript or the marked reduction of the signal through use of RT-PCR analysis of patient fibroblasts suggests that null mutations induced instability of LIFR transcripts. Moreover, Scatchard and western blot/immunofluorecence analysis clearly demonstrate that null LIFR mutations abrogate the LIF-mediated JAK-STAT3 signaling in SWS.
Cytokines play a major role in bone development and have a complex and multiphasic effect on osteoblast proliferation and differentiation and on bone formation (Allan et al. 1990; Liu et al. 2002; Metcalf 2003). It is interesting to note, especially since osteoblasts secrete LIF and express LIFR, that femoral metaphyses of patients with SWS display irregularities of the chondro-osseous junction, with rare, thick, and irregular bone trabeculae and the presence of many osteoclasts (Cormier-Daire et al. 1998). Consistently, the LIFR−/− mice show decreased bone density, especially in the primary spongiosa, with a reduced number of bone spicules and few well-formed trabeculae. A sixfold increase in osteoclast numbers was also observed. By contrast, the developing cartilage bone appeared normal both in LIFR−/− mice (Ware et al. 1995) and in patients with SWS (Cormier-Daire et al. 1998), even though detectable amounts of LIFR were identified in control human chondrocytes (fig. 3). These observations in both humans and mice suggest that the alteration of LIFR has a direct effect on osteoblast differentiation and/or function and, presumably, also on osteoclast activity, leading to a decrease in bone formation and mineralization.
Unlike LIFR−/− mice, which died perinatally, some patients with SWS survive and present with dysautonomia symptoms. Some features are reminiscent of the Schwartz-Jampel syndrome type I, a chondrodystrophic myotonia, which is caused by mutations in the gene encoding perlecan, a component of the neuromuscular junction (Nicole et al. 2000). More recently, cold-induced sweating syndrome has been ascribed to mutations in the CRLF1 (cytokine receptor like factor 1) gene also called “CNTF II” (Lesser and Lo 2000; Knappskog et al. 2003). Both CNTF and LIF are neurotrophic cytokines, involved in the development of the mammalian nervous system, with profound effects on the survival and maintenance of motor neurons in brain stem and spinal cord (Li et al. 1995; Schweizer et al. 2002). CNTF and LIF stimulate cholinergic differentiation in sympathetic neurons, induce choline acetyltransferase gene expression, and promote the survival of cholinergic neurons in vitro and in vivo (Dziennis and Habeker 2003). One can hypothesize, therefore, that the manifestations of dysautonomia in SWS can be ascribed to impairment of STAT3 signaling, which is responsible for induction of the cholinergic and peptidic properties of sympathetic neurons.
Conversely, the IL-6 type cytokines also play important roles in the immune system during hematopoiesis and inflammation, heart development, adipocyte lipid transport, and reproduction, which were not altered in SWS. This feature can be explained by function redundancies—that is, the shared use of the receptor subunit gp130. Whereas IL-6 and IL-11 induce homodimerization of gp130, the other IL-6 type cytokines lead to heterodimerization of gp130 with the LIF receptor (Starr et al. 1997; Metcalf 2003). Apart from exerting these redundant effects, each cytokine is additionally endowed with specific functions. The restricted pattern of cytokine expression and the differential signaling abilities in a cell-type specific manner may explain cytokine-specific effects.
In conclusion, our study shows that SWS and SJS2 are a single genetic entity and are due to null mutations in LIFR. The study emphasizes the specific function of LIFR in bone remodeling and in the autonomous nervous system. We hope that identifying the involvement of LIFR in a disorder of the autonomous nervous system will shed light on the hitherto poorly understood mechanisms of dysautonomia in humans, particularly in constitutional, autoimmune, and neurodegenerative disorders.
Acknowledgments
We thank Noman Kadhom and Catherine Benoit-Lasselin, for cell cultures, and Sylvie Fournier, for binding experiments. Deborah Scheffer is a recipient from the Association Française de Recherche en Génétique. Part of this work was supported by European Skeletal Dysplasia Network grant number QLGI-CT2001-02188 and Swiss National Foundation grant number 3100A0-100485.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for the human genome working draft)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for LIFR [accession number NM_002310])
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SWS) [PubMed]
References
- Al-Gazali LI, Ravenscroft A, Feng A, Shubbar A, Al-Saggaf A, Haas D (2003) Stüve-Wiedemann syndrome in children surviving infancy: clinical and radiological features. Clin Dysmorphol 12:1–8 10.1097/00019605-200301000-00001 [DOI] [PubMed] [Google Scholar]
- Al-Gazali LI, Varghese M, Varady E, Talabani J, Scorer J, Bakalinova D (1996) Neonatal Schwartz-Jampel syndrome: a common autosomal recessive syndrome in the United Arab Emirates. J Med Genet 33:203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan EH, Hilton DJ, Brown MA, Evely RS, Yumita S, Metcalf D, Gough NM, Nicola NA, Martin TJ (1990) Osteoblasts display receptors for and responses to leukemia-inhibitory factor. J Cell Physiol 145:110–119 [DOI] [PubMed] [Google Scholar]
- Bitard J, Daburon S, Duplomb L, Blanchard F, Vuisio P, Jacques Y, Godard A, Health JK, Moreau JF, Taupin JL (2003) Mutations in the immunoglobulin-like domain of gp190, the leukemia inhibitory factor (LIF) receptor, increase or decrease its affinity for LIF. J Biol Chem 278:16253–16261 10.1074/jbc.M207193200 [DOI] [PubMed] [Google Scholar]
- Chabrol B, Sigaudy S, Paquis V, Montfort MF, Giudicelli H, Pelissier JF, Millet V, Mancini J, Philip N (1997) Stüve-Wiedemann syndrome and defects of the respiratory chain. Am J Med Genet 72:222–226 [DOI] [PubMed] [Google Scholar]
- Chen E, Potter PD, Cohen RA, Lachman RS (2001) Characterization of a long-term survivor with Stüve-Wiedemann syndrome and mosaicism of a supernumerary marker chromosome. Am J Med Genet 101:240–245 10.1002/ajmg.1382 [DOI] [PubMed] [Google Scholar]
- Cormier-Daire V, Superti-Furga A, Munnich A, Lyonnet S, Rustin P, Delezoide AL, De Lonlay P, Giedion A, Maroteaux P, Le Merrer M (1998) Clinical homogeneity of the Stüve-Wiedemann syndrome and overlap with the Schwartz-Jampel type 2. Am J Med Genet 78:146–149 [DOI] [PubMed] [Google Scholar]
- Di Rocco M, Stella G, Bruno C, Doria Lamba L, Bado M, Superti-Furga A (2003) Long-term survival in Stüve-Wiedemann syndrome: a neuro-myo-skeletal disorder with manifestations of dysautonomia. Am J Med Genet 118A:362–368 12687669 [DOI] [PubMed] [Google Scholar]
- Dziennis S, Habecker BA (2003) Cytokine suppression of dopamine β hydroxylase by extracellular signal-regulated kinase-dependent and independent pathways. J Biol Chem 278:15897–15904 10.1074/jbc.M212480200 [DOI] [PubMed] [Google Scholar]
- Gearing DP, Thut CJ, Vandenbos T, Gimpel SD, Delaney PB, King J, Price V, Cosman D, Beckmann MP (1991) Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. Embo J 10:2839–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing DP, Truck T, Huebner K, Overhauser J, Gilbert DJ, Copeland NG, Jenkins NA (1993) The leukemia inhibitory factor receptor (LIFR) gene is located within a cluster of cytokine receptor loci on mouse chromosome 15 and human chromosome 5p12-p13. Genomics 18:148–150 10.1006/geno.1993.1441 [DOI] [PubMed] [Google Scholar]
- Godard A, Heymann D, Raher S, Anegon I, Peyrat MA, Le Mauff B, Mouray E, Gregoire M, Virdee K, Soulilou JP, Moreau JF, Jacques Y (1992) High and low affinity receptors for human interleukin for DA cells/leukemia inhibitory factor on human cells: molecular characterization and cellular distribution. J Biol Chem 267:3214–3222 [PubMed] [Google Scholar]
- Knappskog PM, Majewski J, Livneh A, Nilsen PTE, Brinsgli JS, Ott J, Boman H (2003) Cold-induced sweating syndrome is caused by mutations in the CRLF1 gene. Am J Hum Genet 72:375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski K, Tenconi R (1996) Stüve-Wiedemann dysplasia in a 3 1/2 year old boy. Am J Med Genet 63:17–19 [DOI] [PubMed] [Google Scholar]
- Lesser SS, Lo DC (2000) CNTF II, I presume. Nat Neurosci 3:851–852 10.1038/78738 [DOI] [PubMed] [Google Scholar]
- Li M, Sendtner M, Smith A (1995) Essential function of LIF receptor in motor neurons. Nature 378:724–727 10.1038/378724a0 [DOI] [PubMed] [Google Scholar]
- Liu F, Aubin JE, Malaval L (2002) Expression of leukemia inhibitory factor (LIF)/interleukin-6 family cytokines and receptors during in vitro osteogenesis: differential regulation by dexamethasone and LIF. Bone 31:212–219 10.1016/S8756-3282(02)00806-2 [DOI] [PubMed] [Google Scholar]
- Metcalf D (2003) The unsolved enigmas of leukemia inhibitory factor (LIFR). Stem Cells 21:5–14 [DOI] [PubMed] [Google Scholar]
- Nicole S, Davoine CS, Topaloglu H, Cattolico L, Barral D, Beighton P, Ben Hamida C, Hammouda H, Cruaud C, White PS, Samson D, Urizberea JA, Lehmann-Horn F, Weissenbach J, Hentati F, Fontaine B (2000) Perlecan, the major proteoglycan of basement membranes is altered in patients with Schwartz-Jampel syndrome (chondrodystrophic myotonia). Nat Genet 26:480–483 10.1038/82638 [DOI] [PubMed] [Google Scholar]
- Schiemann WP, Bartoe JL, Nathanson NM (1997) Box 3-independent signaling mechanisms are involved in leukemia inhibitory factor receptor α- and gp130-mediated stimulation of mitogen-activated protein kinase. J Biol Chem 272:16631–16636 10.1074/jbc.272.26.16631 [DOI] [PubMed] [Google Scholar]
- Schweizer U, Gunnersen J, Karch C, Wiese S, Holtmann B, Takeda K, Akira S, Sendtner M (2002) Conditional gene ablation of Stat3 reveals differential signaling requirements for survival of motoneurons during development and after nerve injury in the adult. J Cell Biol 156:287–297 10.1083/jcb.200107009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr R, Novak U, Wilson TA, Ingleses M, Murphy V, Alexander WS, Metcalf D, Nicola NA, Hilton DJ, Ernst M (1997) Distinct roles for leukemia inhibitory factor receptor α-chain and gp130 in cell type-specific signal transduction. J Biol Chem 272:19982–19986 10.1074/jbc.272.32.19982 [DOI] [PubMed] [Google Scholar]
- Stüve A, Wiedemann MR (1971) Congenital bowing of the long bones in two sisters. Lancet 2:495 10.1016/S0140-6736(71)92666-3 [DOI] [PubMed] [Google Scholar]
- Superti-Furga A, Tenconi R, Clementi M, Eich G, Steinmann B, Bolthsauser E, Giedion A (1998) Schwartz-Jampel type 2 and Stüve-Wiedemann syndrome: a case for lumping. Am J Med Genet 78:150–154 [DOI] [PubMed] [Google Scholar]
- Tomida M, Gotoh O (1996) Structure of the gene encoding the human differentiation-stimulating factor/leukemia inhibitory factor receptor. J Biochem 120:201–205 [DOI] [PubMed] [Google Scholar]
- Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, Cheng L, Donovan PJ, Peschon JJ, Bartlett PF, Willis CR, Wright BD, Carpenter MK, Davidson BL, Gearing DP (1995) Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development 121:1283–1299 [DOI] [PubMed] [Google Scholar]