Abstract
Recent studies have suggested that a significant fraction of the human genome is contained in blocks of strong linkage disequilibrium, ranging from ∼5 to >100 kb in length, and that within these blocks a few common haplotypes may account for >90% of the observed haplotypes. Furthermore, previous studies have suggested that common haplotypes in candidate genes are generally shared across populations and represent the majority of chromosomes in each population. The conclusions drawn from these preliminary studies, however, are based on an incomplete knowledge of the variation in the regions examined. To bridge this gap in knowledge, we have completely resequenced 100 candidate genes in a population of African descent and one of European descent. Although these genes have been well studied because of their medical importance, we demonstrate that a large amount of sequence variation has not yet been described. We also report that the average number of inferred haplotypes per gene, when complete data is used, is higher than in previous reports and that the number and proportion of all haplotypes represented by common haplotypes per gene is variable. Furthermore, we demonstrate that haplotypes shared between the two populations constitute only a fraction of the total number of haplotypes observed and that these shared haplotypes represent fewer of the African-descent chromosomes than was expected from previous studies. Finally, we show that restricting variation discovery to coding regions does not adequately describe all common haplotypes or the true haplotype block structure observed when all common variation is used to infer haplotypes. These data, derived from complete knowledge of genetic variation in these genes, suggest that the haplotype architecture of candidate genes across the human genome is more complex than previously suggested, with important implications for candidate gene and genomewide association studies.
Introduction
Candidate gene association studies have been suggested as a powerful study design for the identification of common genetic variants involved in common diseases (Risch and Merikangas 1996; Collins et al. 1997). In the simplest of study designs, the frequency of the causal variant(s) is expected to be greater among cases than among controls. For numerous reasons, including insufficient sample size and genetic heterogeneity among cases, studies employing this design have had little success (Ioannidis et al. 2001; Lohmueller et al. 2003). Another possible explanation for the mixed success of these studies is that they typically examine the relationship between a particular phenotype and a single marker within a candidate gene. If the marker surveyed is not the causal variant, this approach relies on the assumption that there is linkage disequilibrium or allelic association between the causal variant and the marker surveyed. Without prior knowledge of the complete genetic variation within the candidate gene or of the structure of linkage disequilibrium in the region, both positive and negative results must be interpreted with caution (reviewed by Cardon and Bell [2001]).
With the recent development of affordable high-throughput sequencing technology, it is now possible to fully catalogue genetic variation in candidate regions for various populations. The most common form of human variation is the SNP: 11 million SNPs with minor allele frequencies of at least 1% are estimated to exist in the human genome, with an average spacing of 1 SNP every 290 bp (Kruglyak and Nickerson 2001). SNPs occurring within regions of functional significance—such as coding regions, splice junctions, and promoter regions—are of particular interest, because changes in these genic regions have been shown to be the most common causes of Mendelian disease and are hypothesized to contribute to common, complex diseases as well (reviewed by Botstein and Risch [2003]). Given the potential applications to genotype-phenotype and disease-gene localization studies, a major goal of the past and current 5-year plans of the Human Genome Project is to catalogue common human variation (Collins et al. 1998, 2003). Both private (Venter et al. 2001) and public (Sachidanandam et al. 2001) resources have been created to facilitate the collection and mining of genetic variation in candidate genes (Coronini et al. 2003), and recent examinations of the public collection suggested that it is adequate for association studies in at least one population (Carlson et al. 2003; Reich et al. 2003).
In addition to the continuing efforts in human variation discovery, the next challenge is harnessing the information garnered from these efforts to understand the relationship between genotypes and phenotypes (Collins et al. 2003). Because it is currently infeasible to genotype every available SNP in a genetic-association study, key questions include which and how many SNPs must be chosen for an association study so that it has sufficient power to detect an association with a disease-causing variant (Kruglyak 1999). To answer these questions, it is important to describe the amount of linkage disequilibrium and the properties of haplotype structure across the human genome. Several studies have recently suggested that the human genome can be organized into haplotype blocks such that blocks are defined as regions of strong linkage disequilibrium (Daly et al. 2001; Patil et al. 2001; Dawson et al. 2002; Gabriel et al. 2002; Phillips et al. 2003). Generally, the few haplotypes within these blocks can be resolved with a few SNPs, greatly reducing the number of SNPs required for an association study. Haplotype blocks are separated by regions exhibiting low levels of linkage disequilibrium, consistent with evidence of historical recombination. Local hotspots of recombination (Daly et al. 2001; Jeffreys et al. 2001; Gabriel et al. 2002), as well as stochastic recombination events and other forces (Subrahmanyan et al. 2001; Wang et al. 2002; Phillips et al. 2003; Zhang et al. 2003), have been proposed as factors that shape block lengths across the genome. Inspired by these results, the International HapMap Project has been launched to produce a genomewide haplotype map in several world populations for association studies (reviewed by Wall and Pritchard [2003]).
Given the heightened interest in haplotype structure across the human genome, we sought to describe haplotype diversity within a European-descent (ED) and an African-descent (AD) population across 100 candidate genes related to inflammation, lipid metabolism, and and blood pressure regulation. The candidate genes chosen for analysis were completely resequenced for discovery of variation in 47 individuals; thus, these data represent one of the most comprehensive catalogues of genetic variation for these genes, many of which have been targets of previous SNP discovery efforts (Cambien et al. 1999; Halushka et al. 1999; Stephens et al. 2001a; Nakajima et al. 2002; Taylor et al. 2002; Tiret et al. 2002). Given this comprehensive collection of genetic variation, we show that the average number of SNPs per gene is higher for these candidate genes in both populations than previously documented, as is the number of common SNPs. Furthermore, using the variation identified through our resequencing efforts, we demonstrate that the number of inferred haplotypes per gene varies greatly across genes as well as between the two populations within genes. Also, the proportion of chromosomes represented by haplotypes shared between the two populations is lower than previous estimates. Finally, we show that reducing the number of SNPs from all common SNPs to a subset representing common coding variation reduces the number of common haplotypes and increases the apparent size of haplotype blocks observed. These data have important implications for the use of haplotypes in candidate gene association studies and genomewide studies in populations with different histories.
Material and Methods
Variation Discovery
As part of an ongoing project known as the “SeattleSNPs Program for Genomic Applications” (PGA), we resequenced >100 genes involved in the inflammation process for the purpose of cataloguing common DNA variation. A list of completed genes, as well as a list of genes in progress and under consideration for resequencing, can be found on the University of Washington–Fred Hutchinson Cancer Research Center (UW-FHCRC) Variation Discovery Resource Web site. For each gene, we resequenced the genomic region spanning the longest reference transcript in LocusLink, including exons and introns, ∼2,500 base pairs (bp) upstream of the gene and ∼1,500 bp downstream of the gene. All DNA variation data identified through these resequencing efforts and the corresponding individual genotypes and allele frequencies were deposited into GenBank and dbSNP and are available on our Web site. A small proportion of the genes described here (ACE, AGT, AGTR1, APOB, HMGCR, LDLR, and REN) were resequenced for other projects related to the investigation of lipid metabolism and the renin-angiotensinogen system (Rieder et al. 1999) as part of a Pharmacogenetics Research Network. The chromosomal locations of the 100 genes presented here are available in table A1 (online only).
Each gene was resequenced for variation discovery in two populations: an ED population (n=23) and an AD population (of African Americans; n=24). The expected detection rate for common (>5% minor allele frequency) SNPs with these sample sizes has been estimated at 99%, whereas the detection rate for all SNPs (>1% minor allele frequency) has been estimated at 87% (Kruglyak and Nickerson 2001). On the basis of these sample sizes, the allele and haplotype frequencies from this survey are expected to be similar to frequencies estimated in larger populations. We genotyped 43 sites across 6 genes in 408 control individuals and found that allele frequencies from this larger sample are strongly correlated with allele frequencies estimated from this resequencing survey (R2=0.83; data not shown).
All DNAs representing healthy individuals were obtained from Coriell Cell Repository. Samples representing the AD population were selected from the African-American Human Variation Panel (HD50AA: individuals NA17101-17116 and NA17133-NA17140), and samples representing the ED population were selected from available Centre d’Etude du Polymorphisme Humain (CEPH) reference panel DNAs. The Coriell Cell Repository numbers for each of the CEPH samples resequenced for SNP discovery described here are NA06990, NA07019, NA07348, NA07349, NA10830, NA10831, NA10842, NA10843, NA10842-NA10845, NA10848, NA10850-NA10854, NA10857, NA10858, NA10860, NA10861, NA12547, NA12548, and NA12560.
A complete description of our resequencing protocol can be found on the UW-FHCRC Variation Discovery Resource Web site. In brief, overlapping primers for PCR were designed to span the gene, with an average amplicon size of 980 bp and an average overlap between amplicons of 197 bp. The PCR products were sequenced using standard dye primer and termination chemistry on an ABI 3700. Data analysts assembled the sequence data for each gene onto a reference genomic sequence, using Phred and Phrap, and edited the resulting alignments for accuracy, using Consed. Polymorphisms were then identified, using PolyPhred 4.0, from pairwise comparisons of individual sequence chromatograms within regions with an average quality score >40 using Phred. Analysts reviewed all polymorphisms identified by PolyPhred for false positives associated with features of the surrounding sequence or biochemical artifacts. If the polymorphism identified was an insertion/deletion polymorphism, analysts manually genotyped each sample and designed primers from the other strand to sequence past the polymorphism. Using this protocol for variation discovery, we recently determined that the error rate is well below 1% for genotype calls from the sequence data (Carlson et al. 2003).
Statistical Methods
For all analyses described here, 100 genes in which >85% of the gene was successfully resequenced for SNP discovery were considered (see table A1 [online only] for a list of individual genes). Haplotypes were inferred by the statistical software package PHASE, version 1.0.1 (Stephens et al. 2001b), which allows for missing genotype data and a large number of sites per gene. Haplotypes were inferred using the default settings of PHASE from genotype data using either all biallelic polymorphisms available or biallelic polymorphisms with a minor allele frequency >5% present in each population (African-descent and European-descent), as well as for combined genotype data from both populations (hereafter referred to as the “combined” sample). To mimic a variation discovery strategy targeting coding regions, we simulated the resequencing strategy reported by Stephens et al. (2001a) by selecting common SNPs (MAF >5%) from exons, 100 bp of introns on each side of each exon, 1 kb upstream of the gene, and 100 bp downstream of the gene. Haplotypes were then inferred using PHASE.
We calculated descriptive summary statistics for each gene (total number of haplotypes, mean number of haplotypes, median number of haplotypes, standard deviation, and variance). A linear regression was performed on the number of haplotypes per gene in the ED population versus the number of haplotypes per gene in the AD population to describe differences in haplotype diversity within a gene between populations. We also calculated descriptive summary statistics on the effective number of haplotypes for all 100 genes. The effective number of haplotypes, analogous to the effective number of alleles (Ewens 1964; Hartl and Clark 1997), was calculated as ne=1/Σp2i, where pi is the frequency of the ith haplotype and ne is the effective number of haplotypes. Expected haplotype heterozygosity (H) was calculated using the equation 1-(1/ne), where ne is the effective number of haplotypes. The maximum possible number of haplotypes in the absence of intragenic recombination, gene conversion, and recurrent mutation was calculated as min(s+1,2x), where s is the number of biallelic polymorphisms per gene and x is the sample size.
To compare sharing of similar (but not identical) haplotypes between populations, divergence between haplotypes was measured as the number of alleles that differed between a pair of haplotypes given the size of the gene. For example, for a 15-kb gene, 0.02% haplotype divergence would describe a pair of haplotypes which differed at three sites.
The focus of most analyses presented here is to describe the distribution of haplotypes in terms of number of haplotypes and frequency of haplotypes for each candidate gene. Previous studies of PHASE have demonstrated that eliminating rare sites from the data set improves the accuracy of haplotype inference (Lin et al. 2002) and that the software estimates haplotype frequencies well (Xu 2002), compared with molecularly determined haplotypes. To test the accuracy of the number of haplotypes per gene inferred by PHASE, we inferred haplotypes in a data set of four X-linked genes containing 24 males. We duplicated the haplotypes and then randomly paired the X-chromosomes to create a diploid, “mixed male” data set. These four genes (ACE2, AGTR2, PFC, and F9) had a total of 124 sites at MAF >5% for an average of 24.8 sites per gene. We then compared the number of haplotypes inferred from this mixed male data set at MAF >5% with known haplotypes from the 24 males. The number of haplotypes per gene observed in the inferred mixed male data set correlated well with the “true” number of haplotypes observed (R2=0.97). Thus, the relative distribution of haplotypes described here is expected to be accurate.
Sites that “tag” or resolve common haplotypes (>5% frequency) and haplotypes inferred from coding variation (MAF >5%) were identified for each gene either manually or by implementing SNPtagger (Ke and Cardon 2003) at default settings. Haplotype blocks were defined using the four-gamete test implemented in the program HaploBlockFinder (Zhang and Jin 2003) at default settings. The four-gamete test performs a pairwise comparison of all SNPs to determine if all four possible haplotypes are present in the population surveyed. If all four haplotypes are present in the population surveyed, D′<1, indicating evidence of historical recombination. Block boundaries, therefore, are determined by SNPs whose pairwise comparisons show evidence of historical recombination (Wang et al. 2002). To apply a less stringent definition of blocks, we set the minimal D′ range to 0.80 in HaploBlockFinder at default settings.
Results
Variation Discovery
One hundred genes, with an average size of 16.5 kb and spanning 18 autosomes, were completely resequenced in two populations with different demographic histories: AD and ED. Both populations were ascertained through the Coriell Cell Repository (the African-American Human Variation Panel HD50AA and unrelated individuals drawn from the parental generation of the CEPH pedigrees). Resequencing 1,616 kb of reference sequence across the human genome identified a total of 8,877 biallelic polymorphisms in the combined sample, with 7,793 and 4,620 in the AD and ED populations, respectively (table 1). These sites should represent nearly complete ascertainment of variable sites in the discovery samples and are expected to describe ∼99% of frequent variants in the broader population (Kruglyak and Nickerson 2001).
Table 1.
Total Number of SNPs in 100 Candidate Genes Related to Inflammation, Lipid Metabolism, and Blood Pressure Regulation, by Population
| Population (No.of Chromosomes) | No. ofSNPs | No. of SNPsper kb | No. of SNPswith MAF >5% | No. of SNPswith MAF >5%,per kb |
| AD (48) | 7,793 | 4.71 | 4,360 | 2.64 |
| ED (46) | 4,620 | 2.79 | 3,053 | 1.85 |
| Combined (94) | 8,877 | 5.37 | 3,992 | 2.41 |
For every gene, the number of biallelic polymorphic sites was greater in the AD population than in the ED population. Overall, the majority of the biallelic polymorphisms identified were SNPs (n=8,397), with the remainder representing insertion/deletions (n=480). All analyses included both SNPs and biallelic indels (hereafter collectively referred to as SNPs). On average, the density of SNPs identified in the combined sample and the AD and ED populations, across all 100 genes, was 5.4, 4.7, and 2.8 SNPs per kb, respectively (table 1), which is comparable to reports from other large surveys (Halushka et al. 1999; Stephens et al. 2001a; Schneider et al. 2003). However, the mean number of SNPs per gene (89, 78, and 46 in the combined sample and the AD and ED populations, respectively) was higher compared with previous reports. Also, we identified a greater number of SNPs in genes that were previously included in other large surveys. As an example, we identified a total of 195 SNPs in ApoB compared with 24 sites identified for the same gene in a separate discovery effort (Cambien et al. 1999). This difference reflects the fact that the present survey resequenced the entire gene in two distinct populations rather than performing SNP discovery on selected portions of each gene (Cambien et al. 1999; Cargill et al. 1999; Stephens et al. 2001a; Tiret et al. 2002) or in a single population (Haga et al. 2002).
On the Number of Haplotypes
Haplotypes were statistically inferred from genotypes by use of the software PHASE (Stephens et al. 2001b), a method that has been shown to accurately infer haplotypes (Lin et al. 2002) and to accurately estimate overall haplotype frequencies from diploid sequence data (Xu et al. 2002). Totals of 4,278, 2,826, and 1,844 haplotypes, corresponding to an average of 43, 28, and 18 haplotypes per gene, were identified in the combined sample and the AD and ED populations, respectively (table 2). None of the genes in either the AD or ED population exceeded the theoretical maximum number of haplotypes under the assumption of no recombination, gene conversion, or recurrent mutation (see the “Statistical Methods” section). Overall, as might be expected (Jorde et al. 2000), the mean (28) and median (27) number of haplotypes per gene was higher in the AD population than in the ED population (18 and 18). In all genes but one (IL10RA), the number of unique haplotypes in the AD population was greater than the number of haplotypes in the ED population.
Table 2.
Total Number of Inferred Haplotypes and Average Expected Haplotype Heterozygosity in 100 Candidate Genes Related to Inflammation, Lipid Metabolism, and Blood Pressure Regulation, by Population
| Population (No.of Chromosomes) | No. ofHaplotypes(All SNPs) | H | No. ofHaplotypesfor SNPswith MAF >5% | H for SNPswith MAF >5% |
| AD (48) | 2,826 | .92 | 2,263 | .90 |
| ED (46) | 1,844 | .83 | 1,323 | .78 |
| Combined (94) | 4,278 | .91 | 3,007 | .87 |
Because all available SNPs regardless of minor allele frequency were used to infer haplotypes, a large number of unique haplotypes observed in each population included haplotypes with sites observed only once (singletons) or twice (doubletons) in the population. Correctly assigning phase at rare sites using statistical methods is challenging, particularly for singletons (Stephens et al. 2001b; Lin et al. 2002). Because of the uncertainty associated with rare sites and because it is expected that SNPs with a minor allele frequency of at least 5% will be more useful for genetic association studies than rare sites (Risch 2000), we reinferred haplotypes using only sites with a minor allele frequency of >5% (MAF >5%). By use of only common sites (MAF >5%) from this data set, a total of 3,007, 2,263, and 1,323 haplotypes—corresponding to an average of 30, 23, and 13 haplotypes per gene—were inferred in the combined sample and the AD and ED populations, respectively (table 2; fig. 1). The range of the number of haplotypes per gene was similar for the AD and ED populations: the AD population varied between 4 and 46 haplotypes, whereas the ED population varied from 2 to 41 haplotypes. As with haplotypes inferred using all sites, for most genes, the number of distinct haplotypes was greater in the AD population than in the ED population: only 4 of 100 genes (HMGCR, IL1A, IL10RA, and IL2) had a greater number of unique haplotypes in the ED population than in the AD population.
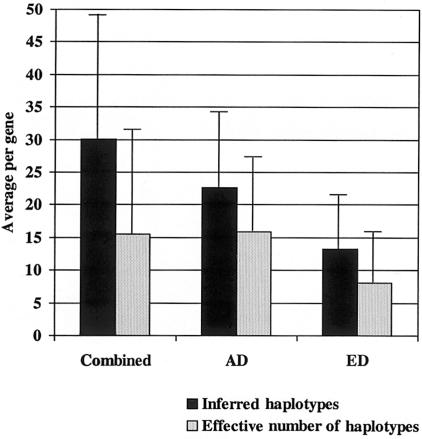
Figure 1.
The average number of haplotypes (blackened bars) and effective haplotypes (gray bars) per gene inferred from common SNPs (MAF >5%), by population. X-axis: population; Y-axis: the average number per gene. The thin bars represent the standard deviation.
Each gene has a distribution of haplotype frequencies that includes rare and common haplotypes. To quantify the number of common haplotypes in our data set, we determined the number of haplotypes with a frequency of >5% in each gene for both populations. The mean number of common haplotypes per gene (frequency >5%) was 4.7, 5.0, and 4.5 in the combined sample and the AD and ED populations, respectively. On average, common haplotypes represented only 56% of chromosomes in the AD population and 75% of chromosomes in the ED population, reflecting the fact that a large proportion of chromosomes in the AD population were represented by rare haplotypes.
Although both populations averaged approximately the same number of common haplotypes per gene (>5% frequency), the number and proportion of common haplotypes varied greatly across genes, as well as between the AD and ED populations. For example, at MAF >5%, five genes did not have a single common haplotype in the AD population (AGTR1, IL1R1, LDLR, PON1, and SELP), whereas only one gene in the ED population (LDLR) did not have a single common haplotype. The lack of common haplotypes in these genes probably reflects elevated rates of recombination, as all five genes have weak linkage disequilibrium across the gene (Wall and Pritchard 2003; Nickerson Group Web site). Up to 11 common haplotypes were identified in the AD population (F2), whereas the maximum number of common haplotypes in the ED population was 8 (SCYA2 and SFTPB). The proportion of chromosomes represented by common haplotypes ranged from a low of ∼6% to a high of 100% in both the AD and ED populations.
Because common haplotypes represent only approximately half of the chromosomes in the AD population, we sought to better describe the number of more frequent haplotypes in each population by calculating the effective number of haplotypes per gene (ne) in each population (see the “Statistical Methods” section). The effective number of haplotypes is the number of equally frequent haplotypes that would produce the same observed homozygosity (Hartl and Clark 1997). By use of sites with MAF >5% to infer haplotypes, the mean number of effective haplotypes per gene was 15, 16, and 8 in the combined sample and the AD and ED populations, respectively (fig. 1).
The effective number of haplotypes is directly related to haplotype heterozygosity (the probability that an individual carries two different haplotypes; see the “Statistical Methods” section). On average, the expected haplotype heterozygosity was higher in the AD population (0.92 and 0.90) than in the ED population (0.83 and 0.78), when either all SNPs or SNPs with MAF >5% were used (table 2). At MAF >5%, haplotype heterozygosity ranged from 0.67 (TNF) to 0.98 (AGTR1) in the AD population and from 0.36 (IL5) to 0.97 (SELP) in the ED population. For five genes (IL1A, IL10RA, IL21R, IL10, and LTB), the expected haplotype heterozygosity was higher in the ED population than in the AD population. The higher average expected haplotype heterozygosity in the AD population is consistent with previous reports of haplotypes constructed from biallelic polymorphisms in other candidate genes (Bonnen et al. 2002; Kauppi et al. 2003; Schneider et al. 2003).
Haplotype Diversity within and between Populations
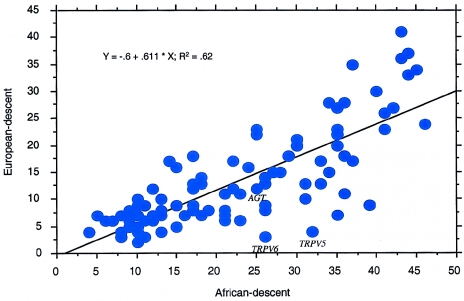
As mentioned previously, the range of the number of haplotypes per gene was similar between the AD and ED populations. However, although the range was similar, the number of haplotypes for specific genes varied between populations (fig. 2 and table A1 [online only]). These gene-to-gene differences between populations at MAF >5% were reflected in the higher mean (fig. 1) and median (22 vs. 11) number of haplotypes per gene, as well as a larger standard deviation (11 vs. 9) in the AD population than in the ED population. Also, a linear regression of the number of haplotypes per gene in the ED population versus the number of haplotypes per gene in the AD population revealed dramatic differences between populations for some genes (R2=0.62) (fig. 2). For example, the genes TRPV5 and TRPV6 have many more inferred haplotypes in the AD population (32 and 26, respectively) as compared with the ED population (4 and 3, respectively). The dramatic differences in the number of haplotypes for these two genes can be attributed to the dramatic differences in SNP density for these genes between the AD and ED populations (table A1 [online only]). For other genes, the difference could not be explained by the difference in SNP density between populations. For example, for AGT, the AD and ED populations have approximately the same number of SNPs per gene (56 and 58, respectively), yet the AD population has twice as many haplotypes (25) and effective haplotypes (15.6) as the ED population (12 and 6.2, respectively), presumably because of greater historical recombination.
Figure 2.
The number of haplotypes per gene in the ED population is positively correlated with the number of haplotypes per gene in the AD population. However, the correlation is not perfect (R2=0.62), and it identifies genes in which the number of haplotypes per gene differs dramatically between the two populations. X-axis: number of haplotypes inferred by PHASE per gene in the AD population (MAF >5%); Y-axis: number of haplotypes inferred by PHASE per gene in the ED population (MAF >5%).
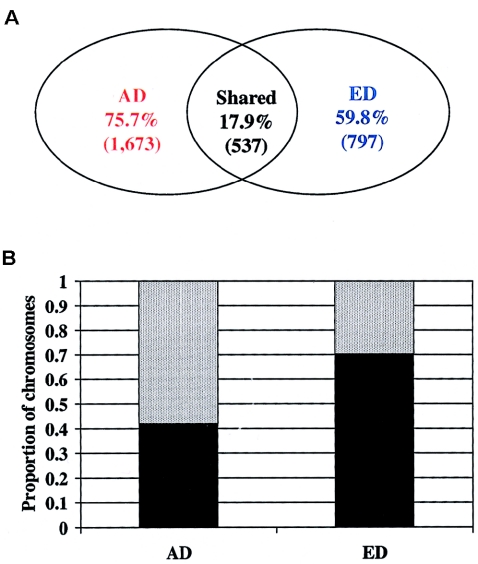
Recent surveys of candidate genes have suggested that even though the AD and ED populations vary with respect to the number and frequency of haplotypes, the populations share haplotypes that account for the majority of chromosomes surveyed (Stephens et al. 2001a; Bonnen et al. 2002; Schneider et al. 2003). To explore this possibility in our survey, we first examined the number of shared and nonshared haplotypes between the AD and ED populations. We chose to base this analysis on haplotypes inferred from the combined population because the set of SNPs with MAF >5% was different in the AD and ED populations, making direct comparisons between populations difficult. Among the 3,007 haplotypes identified using SNPs at MAF >5% in the combined population (table 2), only 537 (17.9%) unique haplotypes were shared between the AD and ED populations (fig. 3a). The majority of haplotypes observed in the AD population (75.7%) or the ED population (59.8%) were specific to a single population (fig. 3a). When the fraction of chromosomes represented by shared haplotypes was considered, 42% and 70% of AD and ED chromosomes, respectively, were represented by shared haplotypes (fig. 3b). Similarly, shared haplotypes represented 61% of the chromosomes in the AD population and 85% of the chromosomes in the ED population when haplotypes were inferred using SNPs with MAF >20% (data not shown). Considering similar as well as identical haplotypes inferred using SNPs with MAF >5% (0.02% divergent; see the “Statistical Methods” section), the fraction of chromosomes representing shared haplotypes increased to 64% in the AD population and 90% in the ED population (data not shown).
Figure 3.
a, Percentage (number) of shared haplotypes and population specific haplotypes inferred from SNPs with MAF >5% (red, AD population–specific haplotypes; blue, ED population–specific haplotypes). b, The frequency of chromosomes represented among the shared (blackened bars) and nonshared (gray bars) haplotypes inferred from SNPs with MAF >5%, by population. X-axis: population; Y-axis: proportion of chromosomes represented.
Haplotypes Inferred from Coding Variation
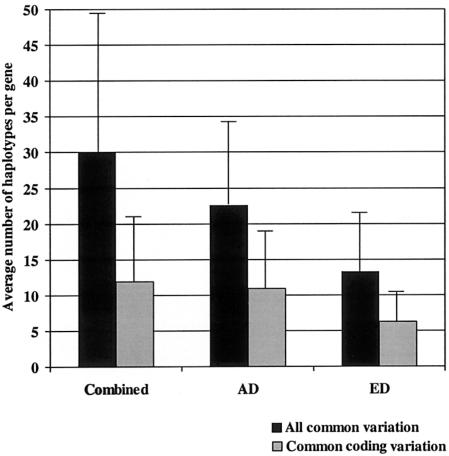
Other large resequencing surveys have targeted coding regions for variation discovery rather than the entire gene (Cambien et al. 1999; Cargill et al. 1999; Stephens et al. 2001a). To compare our results with these previous studies, we selected a subset of SNPs from our resequencing survey of the entire gene to simulate a resequencing strategy that targets coding regions of the gene (see the “Material and Methods” section). In this coding variation subset, we observed an average of 9, 10, and 7 SNPs per gene (MAF >5%), corresponding to a density of 0.55, 0.60, and 0.41 SNPs per kb in the combined sample and the AD and ED populations, respectively. This is roughly one-fourth of the number of SNPs detected when the entire gene sequence is considered. One gene in the AD population (IFNG) and four genes in the ED population (DCN, IL12A, KEL, and TNFAIP1) did not have any sites in the coding subset. Compared with the average number of inferred haplotypes per gene when all available common genetic variation is used, the average number of inferred haplotypes per gene from coding variation at MAF >5% was considerably lower: 12, 11, and 6 versus 30, 23, and 13 in the combined sample and the AD and ED populations, respectively (fig. 4). The observed difference between the average number of haplotypes per gene when all common variation is used, versus common coding variation, is likely an accurate extrapolation, since the observed average number of haplotypes when mostly coding variation is used was similar to the data from 313 other genes presented by Stephens et al. (2001a).
Figure 4.
The average number of haplotypes per gene (MAF >5%) using all common variation (blackened bars) and common coding variation (gray bars), by population. X-axis: population; Y-axis: the average number of haplotypes per gene (MAF >5%). The thin bars represent the standard deviation.
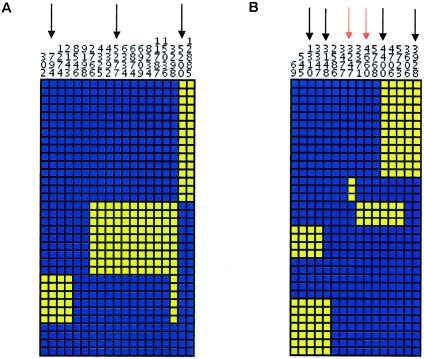
Because SNP density is related to the observed number of inferred haplotypes per gene, it is expected that the average number of haplotypes per gene inferred from the coding variation data set would be smaller than it would if all common sites were used to infer haplotypes. However, it is unknown to what extent haplotype structure is preserved when fewer sites are used to infer haplotypes. For some genes, an increase in the number of SNPs used to infer haplotypes may not necessarily result in an increase in the number of haplotypes because many sites may be correlated (in strong linkage disequilibrium) with one another. To explore the extent of haplotype structure preservation between discovery strategies, we first determined the proportion of genes whose common haplotypes (>5% frequency) were found among the haplotypes inferred from common coding variation (MAF >5%). To do this, sites that uniquely identify the common haplotypes were identified either manually or by SNPtagger (Ke and Cardon 2003) and mapped to the haplotypes inferred from coding variation. To illustrate, in figure 5, four common haplotypes were inferred from all common variation in the gene IL1B in the ED population. In this example, three sites that uniquely identify the common haplotypes from all common variation were also found in the coding variation data set for IL1B and resolve the same common haplotypes (fig. 5a). In contrast, for CSF2 in the AD population, only four of the six sites that distinguish common haplotypes were found among the coding variation data set (fig. 5b). Overall, the profile identified in IL1B was in the minority, since only 19 genes in the AD population and 27 genes in the ED population resolved all common haplotypes when coding variation was used.
Figure 5.
Visual depiction of the common haplotypes inferred from SNPs with MAF >5% in IL1B from the ED population (a) and CSF2 from the AD population (b). Each column represents a SNP, with the minor allele colored yellow and the common allele colored blue. Each row represents a chromosome. Sites within the gene were clustered on the basis of allelic similarity, and haplotypes were clustered using the unweighted pair group method with arithmetic averages. a, Four common haplotypes (>5% frequency) were observed in the ED population for IL1B from all common variation at MAF >5%. The black arrows represent tagSNPs that distinguish the four common haplotypes whose corresponding sites were also found among the coding variation data set. b, Six common haplotypes (>5% frequency) were observed in the AD population for CSF2 from all common variation at MAF >5%. Six tagSNPs were identified that distinguish common haplotypes. The black arrows represent the tagSNPs that distinguish common haplotypes whose corresponding sites were also found among the coding variation data set. The red arrows represent the tagSNPs that distinguish common haplotypes whose corresponding sites were not found among the coding variation data set.
For an alternative look at haplotype structure between the discovery strategies, we examined the preservation of haplotype block structure across all genes in both populations. Blocks were defined using the four-gamete test implemented by HaploBlockFinder (Zhang and Jin 2003). The four-gamete test is a strict definition of a haplotype block, such that there is no evidence of historical recombination between a pair of sites and D′=1 (Wang et al. 2002). By use of this definition for haplotype blocks, a total of 1,470 and 974 blocks—averaging 1.6 kb and 2.9 kb in size, respectively—were identified from haplotypes inferred in the AD and ED populations, respectively, at MAF >5%. When restricted to common coding variation (MAF >5%), there were considerably fewer blocks: 325 and 204 blocks, averaging 3.5 kb and 4.1 kb in size, in the AD and ED populations, respectively. Only 6 genes in the AD population and 21 genes in the ED population had the same number of blocks across the gene when the two discovery strategies were used to infer haplotypes and define blocks. Similar results were observed when block boundaries were defined using less stringent criteria (minimal D′ range set to 0.80): only 12 genes in the AD population and 23 genes in the ED population had the same number of blocks across the gene when the two discovery strategies were compared. Thus, only a small fraction of genes are adequately described using coding variation, regardless of the exact definition used to define the block boundaries.
Discussion
We present here a comprehensive and unbiased catalog of genetic variation and haplotype diversity in 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. In general, the data demonstrated greater genetic variation in the AD population than in the ED population, as has been shown in studies using microsatellites (e.g., Jorde et al. 1997) and SNPs (e.g., Stephens et al. 2001a). This pattern of greater genetic variation extended to the comparison of haplotype similarities and differences between the populations: only a fraction of all haplotypes were shared between the two populations examined here, and these shared haplotypes represented fewer of the chromosomes from the AD population than was expected from previous surveys. More importantly, the number and diversity of haplotypes varied greatly from gene to gene, making generalities about haplotypes across candidate genes in the genome difficult to identify. Finally, the complete resolution of common haplotypes and block structure was dependent on SNP density, with complete SNP discovery leading to different inferences of haplotype structure than incomplete SNP discovery. These results provide a glimpse of true haplotype diversity across candidate genes in the human genome and give insights into prospects for the use of haplotypes in genetic association studies of human diseases.
Haplotype Diversity across the Human Genome
Several preliminary studies have taken steps towards characterizing the genetic variation and haplotype diversity across populations and how this diversity is organized across candidate genes in the human genome. In one such study, Stephens et al. (2001a) undertook a large survey of coding variation discovery in 82 unrelated individuals in four populations for 313 genes hypothesized to be involved in human disease or targets of drug therapies. In contrast to variation-discovery methods described by Stephens et al. (2001a), we studied only genes that were completely resequenced for variation discovery. Thus, our data set included genetic variation discovered in introns as well as repetitive sequences not included in other surveys, which provided a more detailed picture of a gene’s recombinational and mutational history. Similar to our survey, other comprehensive sequence-based variation studies of single genes or regions such as LPL (Clark et al. 1998), β-globin (Harding et al. 1997), and ACE (Rieder et al. 1999) have revealed a level of diversity not appreciated in studies limited to selected markers across the gene (reviewed by Jorde et al. [2001]).
Given the comprehensive and unbiased nature of this survey, we were able to describe haplotype diversity among 100 genes at a level unexplored up to this point. Not surprisingly, we found a greater number of SNPs per gene compared with previous reports that analyzed mostly coding-region variation (Cambien et al. 1999; Stephens et al. 2001a; Schneider et al. 2003). With regard to haplotypes, we found that the number and frequency varied from gene to gene both within and between the two populations. In this data set, recombinational history rather than nucleotide diversity had a greater influence on haplotype diversity, as the most haplotype diversity was observed among genes with weak linkage disequilibrium (Wall and Pritchard 2003; Carlson et al. 2004). We also found that only a fraction of the observed haplotypes were shared between the two populations, and that these shared haplotypes represented fewer chromosomes surveyed in the AD population than expected based on previous surveys. These results are contrary to those published for haplotypes inferred from coding variation, where the estimates of chromosomes represented by shared haplotypes exceeded 75% (Stephens et al. 2001a; Schneider et al. 2003). However, our results are similar to a recent survey of haplotype diversity across the MHC class II region (Kauppi et al. 2003), which suggests that patterns of haplotype diversity across the genome in global populations may be more complex than previously described.
One specific force that has been proposed to shape haplotype diversity is the presence of hotspots of recombination (Jeffreys et al. 2001; Gabriel et al. 2002). In areas where hotspots of recombination exist, blocks of limited haplotype diversity are flanked by areas of rapid decay in linkage disequilibrium (Jeffreys et al. 2001). It is unclear, however, how frequently these hotspots of recombination occur across the genome, especially given the observation that random drift can generate similar patterns of linkage disequilibrium (Subrahmanyan et al. 2001; Wang et al. 2002; Phillips et al. 2003; Zhang et al. 2003). To date, only a handful of human genes have been identified as containing hotspots either experimentally (MHC class II region and beta-globin: see Jeffreys et al. 2001; Schneider et al. 2002) or through linkage disequilibrium studies (LPL and CD36: see Clark et al. 1998; Templeton et al. 2000; Omi et al. 2003). It is possible that hotspots of recombination exist within a proportion of the genes examined here, generating greater haplotype diversity than would be observed in a data set with genes that do not have hotspots of recombination. As statistical tools are being developed to identify hotspots of recombination (Li and Stephens 2003), additional studies will be necessary to more accurately assess the impact of recombination hotspots on haplotype diversity and linkage disequilibrium across the genome.
One caveat for the findings presented here is the fact that we based our analyses on statistically inferred haplotypes rather than on haplotypes constructed from pedigrees or molecular techniques. However, recent studies that applied PHASE to molecularly determined haplotypes demonstrated that inferred haplotypes are reasonably accurate when singleton SNPs are not included (Lin et al. 2002) and that the overall estimates of haplotype frequencies were reasonably accurate (Xu et al. 2002). We based most of our analyses on haplotypes inferred from biallelic polymorphisms with MAF >5%, which excludes singletons from the data set. Also, we found that the correlation between inferred haplotype counts and true haplotype counts among four genes that we tested was high (see Methods and Materials). Another caveat is that PHASE assumes no recombination and that we treated the “best guess” as correct, both of which will tend to lead to an under estimate of the actual number of haplotypes (M. Stephens, personal communication). A version of PHASE (v2.0) that takes recombination into account in its estimation of haplotypes was recently released (Stephens and Donnelly 2003). The overall results reported here are not expected to differ significantly from one version of PHASE to the next as a preliminary analysis revealed very little difference in the overall haplotype distribution inferred from PHASE v2.0 compared with PHASE v1.01 (data not shown).
Potential Impact of Haplotype Diversity on Genetic Association Studies
The use of haplotypes has been suggested as a tool to capture more information across the region of interest compared with single markers examined one at a time (Daly et al. 2001). Also, some have suggested that haplotypes rather than single SNPs may be exerting a concerted effect on the phenotype of interest (Drysdale et al. 2000). Direct comparison of haplotypes across studies is difficult due to differences in the haplotype inference parameters (such as minor allele frequency thresholds) as well as software used to infer haplotypes. Thus, our comparison of inferred haplotypes from the complete variation data set to the coding variation data set represents a direct comparison not possible with other large surveys. The data presented here indicate that many candidate genes important in the inflammation, lipid metabolism, and blood pressure regulation pathways have a large number of haplotypes in the complete variation data set in these two populations. For many genes of medical interest, such as APOB and LDLR, the efforts to capture these haplotype patterns in a genetic association study can be daunting with respect to the number of sites that require genotyping. The use of tagSNPs, SNPs useful in uniquely identifying or “tagging” common haplotype patterns, has been proposed to alleviate the costs of genotyping redundant sites while retaining sites that contain information relevant to particular haplotypes (Johnson et al. 2001). This type of algorithm may not be appropriate for all genes, since common haplotypes represented only a small fraction of chromosomes in the population surveyed for many of the genes presented here. Another method for choosing SNPs to be genotyped relies on the presumed biological relevance of SNPs found in coding regions (e.g., Stephens et al. 2001a). We demonstrate, however, that limiting a study to coding variation haplotypes incompletely resolves common haplotypes and haplotype block structures that would have been observed if all common variation was used to infer haplotypes. Defining haplotypes solely on the basis of coding variation could therefore reduce the power to detect a causal variant, if the variant is found only on a haplotype that is not uniquely resolved by coding SNPs but rather is grouped with other similar haplotypes that do not carry the causal variant. Our data demonstrate that, for most candidate genes, knowledge of complete variation is required to design an association study in which the sites chosen for genotyping represent all common haplotypes present in the population. Thus, methods of choosing how many and which SNPs to genotype for an association study should be based on complete knowledge of genetic variation of the gene or region, since haplotype diversity is not uniform across candidate genes in the human genome and can be quite complex and challenging to represent in a study designed to identify loci contributing to common, complex human disorders.
Acknowledgments
We would like to thank members of the SeattleSNPs team (M. Ahearn, T. Armel, E. Calhoun, M. Chung, C. Hastings, P. Keyes, P. Lee, S. Kuldanek, M. Montoya, C. Poel, E. Toth, and N. Rajkumar) for cataloguing the variation data. We would also like to thank M. Stephens, J. Akey, and K. Weiss for critical reading of this manuscript. This work was supported by grants from the National Heart Lung and Blood Institute Program for Genomic Applications (HL66682 to D.A.N. and M.J.R. and HL66642 to L.K.), the National Institute of Mental Health (MH59520 to L.K.), and the National Institutes of Health Pharmacogenetics Research Network (U01 HL69757 to D.A.N.). L.K. is a James S. McDonnell Centennial Fellow.
Appendix A
Table A1.
Supplemental Data
| Gene | GenBankAccessionNumber | ChromosomalLocation | kbResequenced | No. ofAD SNPs(>5% MAF) | No. of ADHaplotypes | AD Ne | No. of ADHaplotypes >5%in Frequency | AD ExpectedHaplotypeHeterozygosity | No. of EDSNPs (>5% MAF) | No. of EDHaplotypes | ED Ne | No. of EDHaplotypes>5% inFrequency | ED ExpectedHaplotypeHeterozygosity |
| ACE | AY436326 | 17q23 | 22.12 | 57 | 26 | 15.36 | 6 | .93 | 30 | 8 | 4.15 | 5 | .76 |
| AGT | AY436323 | 1q42-q43 | 15.59 | 56 | 25 | 15.57 | 5 | .94 | 58 | 12 | 6.19 | 7 | .84 |
| AGTR1 | AY436325 | 3q21-q25 | 45.93 | 154 | 46 | 44.31 | 0 | .98 | 107 | 24 | 14.90 | 4 | .93 |
| APOB | AY324608 | 2p24-p23 | 43.00 | 75 | 35 | 25.04 | 2 | .96 | 39 | 20 | 12.30 | 5 | .92 |
| BDKRB2 | AF378542 | 14q32.1-32.2 | 14.05 | 33 | 17 | 10.87 | 7 | .91 | 31 | 12 | 7.50 | 7 | .87 |
| BF | AF551848 | 6p21.3 | 9.96 | 16 | 13 | 8.47 | 7 | .88 | 20 | 10 | 5.37 | 6 | .81 |
| CCR2 | AF545480 | 3p21 | 10.07 | 21 | 13 | 7.63 | 6 | .87 | 19 | 7 | 6.12 | 6 | .84 |
| CD36 | AY095373 | 7q11.2 | 29.92 | 91 | 35 | 26.18 | 1 | .96 | 53 | 23 | 10.69 | 2 | .91 |
| CEBPB | AY193834 | 20q13.1 | 4.51 | 9 | 8 | 4.48 | 5 | .78 | 4 | 4 | 2.50 | 4 | .60 |
| CRF | AF410771 | 17q21 | 9.29 | 22 | 26 | 19.86 | 7 | .95 | 17 | 8 | 2.84 | 5 | .65 |
| CRP | AF449713 | 1q21-23 | 6.72 | 17 | 10 | 6.30 | 5 | .84 | 9 | 4 | 3.36 | 4 | .70 |
| CSF2 | AF373868 | 5q31.1 | 5.99 | 16 | 15 | 8.29 | 6 | .88 | 9 | 5 | 3.08 | 3 | .68 |
| CSF3 | AF388025 | 17q11.2-q12 | 5.53 | 13 | 11 | 5.73 | 8 | .83 | 10 | 6 | 3.03 | 3 | .67 |
| CSF3R | AY148100 | 1p35-p34.3 | 18.84 | 62 | 31 | 24.51 | 4 | .96 | 23 | 13 | 4.46 | 3 | .78 |
| DCN | AF491944 | 12q23 | 34.95 | 100 | 21 | 12.13 | 5 | .92 | 9 | 8 | 5.51 | 6 | .82 |
| F10 | AF503510 | 13q34 | 28.94 | 49 | 44 | 39.72 | 1 | .97 | 43 | 33 | 24.05 | 1 | .96 |
| F11 | AY191837 | 4q35 | 26.60 | 67 | 25 | 17.45 | 6 | .94 | 48 | 23 | 13.92 | 5 | .93 |
| F12 | AF538691 | 5q33-qter | 10.62 | 20 | 18 | 10.11 | 6 | .90 | 13 | 14 | 7.40 | 6 | .86 |
| F2 | AF478696 | 5q13 | 20.41 | 44 | 18 | 15.57 | 11 | .94 | 45 | 7 | 3.53 | 6 | .72 |
| F2R | AF391809 | 11q11-q12 | 24.23 | 56 | 29 | 19.86 | 6 | .95 | 33 | 18 | 10.58 | 6 | .91 |
| F2RL1 | AF400075 | 5q13 | 17.72 | 26 | 17 | 10.77 | 8 | .91 | 25 | 13 | 7.25 | 6 | .86 |
| F2RL2 | AF374726 | 5q13 | 9.27 | 37 | 26 | 18.29 | 6 | .95 | 24 | 13 | 8.82 | 7 | .89 |
| F2RL3 | AF384819 | 19p12 | 10.21 | 26 | 21 | 13.71 | 8 | .93 | 17 | 11 | 5.01 | 5 | .80 |
| F3 | AF540377 | 1p22-p21 | 16.11 | 32 | 27 | 20.95 | 6 | .95 | 22 | 15 | 6.01 | 3 | .83 |
| FGA | AF361104 | 4q28 | 9.95 | 12 | 11 | 7.07 | 7 | .86 | 9 | 6 | 4.34 | 5 | .77 |
| FGB | AF388026 | 4q28 | 11.60 | 29 | 9 | 5.70 | 7 | .82 | 28 | 6 | 4.77 | 6 | .79 |
| FGG | AF350254 | 4q28 | 10.17 | 8 | 9 | 5.94 | 6 | .83 | 7 | 5 | 3.01 | 4 | .67 |
| FGL2 | AF468959 | 7q | 6.38 | 7 | 9 | 6.00 | 7 | .83 | 6 | 5 | 3.37 | 5 | .70 |
| FSBP | AF487652 | 8 | 9.85 | 15 | 8 | 4.78 | 6 | .79 | 12 | 7 | 2.91 | 5 | .66 |
| GP1BA | AF395009 | 17pter-p12 | 6.24 | 19 | 13 | 6.44 | 6 | .84 | 12 | 8 | 4.28 | 5 | .77 |
| HMGCR | AY321356 | 5q13.3q14 | 27.81 | 29 | 15 | 8.53 | 8 | .88 | 33 | 16 | 4.58 | 4 | .78 |
| ICAM1 | AY225514 | 19p13 | 17.73 | 29 | 30 | 23.51 | 7 | .96 | 20 | 20 | 10.37 | 4 | .90 |
| IFNG | AF375790 | 12q14 | 7.67 | 8 | 9 | 6.44 | 7 | .84 | 8 | 8 | 4.27 | 5 | .77 |
| IL10 | AF418271 | 1q31-q32 | 7.88 | 18 | 10 | 5.07 | 6 | .80 | 20 | 9 | 5.51 | 6 | .82 |
| IL10RA | AY195619 | 11q23 | 19.94 | 42 | 14 | 6.36 | 6 | .84 | 33 | 17 | 6.87 | 3 | .85 |
| IL11 | AY207429 | 19q13.3-13.4 | 8.96 | 27 | 25 | 15.36 | 4 | .93 | 19 | 12 | 5.09 | 5 | .80 |
| IL12A | AF404773 | 3p12-q13.2 | 11.33 | 30 | 16 | 10.02 | 7 | .90 | 23 | 7 | 3.77 | 4 | .73 |
| IL12B | AF512686 | 5q31.1-33.1 | 14.90 | 31 | 12 | 5.54 | 5 | .82 | 27 | 7 | 4.56 | 5 | .78 |
| IL13 | AF377331 | 5q31 | 6.92 | 18 | 26 | 14.40 | 4 | .93 | 9 | 9 | 2.25 | 4 | .55 |
| IL17B | AF386077 | 5q32-34 | 9.08 | 8 | 6 | 3.95 | 6 | .75 | 17 | 6 | 3.49 | 5 | .71 |
| IL19 | AF390905 | 1q32.2 | 11.00 | 20 | 10 | 4.97 | 6 | .80 | 17 | 7 | 2.76 | 5 | .64 |
| IL1A | AF536338 | 2q14 | 17.85 | 55 | 17 | 9.14 | 7 | .89 | 46 | 18 | 11.14 | 5 | .91 |
| IL1B | AY137079 | 2q14 | 17.45 | 31 | 26 | 16.46 | 6 | .94 | 19 | 14 | 5.66 | 4 | .82 |
| IL1R1 | AF531102 | 2q12 | 27.86 | 68 | 40 | 36.00 | 0 | .97 | 57 | 30 | 25.19 | 4 | .96 |
| IL1R2 | AY124010 | 2q12-q22 | 23.16 | 103 | 33 | 22.15 | 4 | .95 | 86 | 13 | 7.01 | 5 | .86 |
| IL1RN | AY196903 | 2q14.2 | 19.68 | 59 | 28 | 19.53 | 6 | .95 | 82 | 15 | 10.08 | 6 | .90 |
| IL2 | AF359939 | 4p26-27 | 6.75 | 5 | 5 | 3.81 | 5 | .74 | 6 | 7 | 3.56 | 4 | .72 |
| IL20 | AF402002 | 1q32.2 | 6.63 | 12 | 17 | 11.76 | 8 | .91 | 8 | 9 | 4.64 | 6 | .78 |
| IL21R | AY064474 | 16p11 | 25.84 | 57 | 37 | 27.43 | 1 | .96 | 49 | 35 | 30.23 | 2 | .97 |
| IL22 | AF387519 | 12q15 | 8.39 | 23 | 15 | 8.17 | 8 | .88 | 20 | 9 | 4.92 | 5 | .80 |
| IL24 | AY062931 | 1q32.2 | 10.63 | 22 | 13 | 10.11 | 9 | .90 | 13 | 4 | 2.66 | 3 | .62 |
| IL2RB | AF517934 | 22q13.1 | 26.03 | 99 | 45 | 42.67 | 1 | .98 | 69 | 34 | 29.39 | 1 | .97 |
| IL3 | AF365976 | 5q31.1 | 6.39 | 8 | 12 | 4.68 | 6 | .79 | 5 | 6 | 3.41 | 4 | .71 |
| IL4 | AF395008 | 5q31.1 | 22.85 | 61 | 34 | 22.59 | 3 | .96 | 41 | 15 | 4.92 | 4 | .80 |
| IL4R | AF421855 | 16p11.2-12.1 | 25.92 | 101 | 36 | 28.80 | 1 | .97 | 100 | 28 | 21.59 | 4 | .95 |
| IL5 | AF353265 | 5q31.1 | 5.19 | 9 | 10 | 5.82 | 6 | .83 | 1 | 2 | 1.57 | 2 | .36 |
| IL6 | AF372214 | 7p21 | 7.53 | 21 | 18 | 12.94 | 9 | .92 | 17 | 13 | 6.41 | 7 | .84 |
| IL8 | AF385628 | 4q13-q21 | 7.04 | 15 | 10 | 6.16 | 7 | .84 | 7 | 2 | 1.83 | 2 | .45 |
| IL9 | AF361105 | 5q31.1 | 6.68 | 12 | 10 | 4.61 | 7 | .78 | 10 | 5 | 2.70 | 3 | .63 |
| IRAK4 | AY186092 | 12q12 | 33.03 | 81 | 21 | 10.47 | 6 | .90 | 46 | 6 | 3.30 | 4 | .70 |
| JAK3 | AF513860 | 19q13.1 | 19.07 | 72 | 42 | 37.16 | 1 | .97 | 43 | 27 | 17.34 | 3 | .94 |
| KEL | AY228336 | 7q33 | 25.85 | 66 | 35 | 21.74 | 2 | .95 | 10 | 7 | 2.99 | 3 | .67 |
| KLK1 | AY094609 | 19 | 9.92 | 42 | 19 | 9.37 | 6 | .89 | 29 | 8 | 5.32 | 6 | .81 |
| KLKB1 | AY190920 | 4 | 31.67 | 122 | 37 | 29.54 | 2 | .97 | 100 | 17 | 7.40 | 5 | .86 |
| LDLR | AY324609 | 19p13.3 | 42.76 | 103 | 43 | 39.72 | 0 | .97 | 79 | 41 | 37.79 | 0 | .97 |
| LTA | AY070490 | 6p21.3 | 5.03 | 14 | 12 | 5.88 | 5 | .83 | 14 | 12 | 6.41 | 6 | .84 |
| LTB | AY070219 | 6p21.3 | 4.41 | 5 | 4 | 1.55 | 3 | .36 | 4 | 4 | 1.84 | 4 | .46 |
| MC1R | AF514787 | 16q24.3 | 6.55 | 21 | 10 | 8.17 | 9 | .88 | 18 | 10 | 5.04 | 6 | .80 |
| MMP3 | AF405705 | 11 | 11.90 | 30 | 17 | 11.18 | 7 | .91 | 28 | 9 | 2.86 | 5 | .65 |
| NOS3 | AF519768 | 7q36 | 23.31 | 46 | 35 | 27.43 | 2 | .96 | 39 | 27 | 19.24 | 5 | .95 |
| PLAU | AF377330 | 10q24 | 9.27 | 20 | 11 | 6.66 | 6 | .85 | 15 | 9 | 3.40 | 3 | .71 |
| PLAUR | AY194849 | 19q13 | 23.19 | 94 | 43 | 38.40 | 1 | .97 | 70 | 36 | 30.23 | 2 | .97 |
| PON1 | AF539592 | 7q21 | 29.05 | 110 | 41 | 37.16 | 0 | .97 | 69 | 26 | 17.93 | 4 | .94 |
| PON2 | AY210982 | 7q21.3 | 32.49 | 84 | 34 | 23.04 | 3 | .96 | 77 | 28 | 21.16 | 4 | .95 |
| PROC | AF378903 | 2q13-q14 | 12.88 | 34 | 17 | 7.25 | 6 | .86 | 31 | 8 | 3.85 | 4 | .74 |
| PROCR | AF375468 | 20q11.2 | 6.97 | 11 | 7 | 4.22 | 5 | .76 | 8 | 3 | 2.66 | 3 | .62 |
| PROZ | AF440358 | 13q34 | 14.37 | 36 | 23 | 5.65 | 3 | .82 | 28 | 6 | 3.57 | 5 | .72 |
| REN | AY434324 | 1q32 | 13.64 | 47 | 36 | 30.32 | 2 | .97 | 35 | 11 | 5.66 | 7 | .82 |
| SCYA2 | AF519531 | 17q11.2-q21.2 | 9.07 | 24 | 22 | 13.40 | 6 | .93 | 19 | 12 | 7.72 | 8 | .87 |
| SELE | AF540378 | 1q22 | 13.89 | 41 | 22 | 16.23 | 8 | .94 | 50 | 17 | 8.40 | 6 | .88 |
| SELP | AF542391 | 1q22 | 43.45 | 120 | 44 | 41.14 | 0 | .98 | 76 | 37 | 32.06 | 2 | .97 |
| SERPINA5 | AF361796 | 14 | 7.81 | 43 | 29 | 25.04 | 7 | .96 | 37 | 18 | 11.14 | 6 | .91 |
| SERPINC1 | AF386078 | 1q23-25.1 | 15.21 | 29 | 21 | 12.39 | 6 | .92 | 16 | 7 | 2.21 | 4 | .55 |
| SERPINE1 | AF386492 | 7q21 | 13.21 | 42 | 24 | 12.66 | 7 | .92 | 32 | 16 | 8.89 | 6 | .89 |
| SFTPA1 | AY198391 | 10q22 | 23.13 | 98 | 33 | 24.51 | 3 | .96 | 90 | 17 | 11.14 | 7 | .91 |
| SFTPA2 | AY206682 | 10q22 | 18.04 | 94 | 36 | 26.79 | 2 | .96 | 63 | 18 | 7.78 | 4 | .87 |
| SFTPB | AF400074 | 2p12 | 11.09 | 26 | 28 | 20.21 | 4 | .95 | 14 | 15 | 10.17 | 8 | .90 |
| SFTPD | AY216721 | 10q22 | 23.54 | 98 | 30 | 17.19 | 3 | .94 | 86 | 21 | 9.28 | 5 | .89 |
| SMP1 | AF458851 | 1p | 25.27 | 47 | 25 | 14.58 | 5 | .93 | 22 | 22 | 12.45 | 5 | .92 |
| STAT6 | AF417842 | 12q13 | 18.77 | 28 | 23 | 15.36 | 7 | .93 | 14 | 11 | 5.06 | 5 | .80 |
| TGFB3 | AY208161 | 14q24 | 23.24 | 42 | 39 | 33.88 | 1 | .97 | 34 | 9 | 3.18 | 5 | .69 |
| THBD | AF495471 | 20 | 7.25 | 9 | 11 | 6.86 | 7 | .85 | 5 | 6 | 3.14 | 4 | .68 |
| TNF | AY066019 | 6p21.3 | 4.83 | 7 | 7 | 3.02 | 6 | .67 | 10 | 6 | 2.19 | 4 | .54 |
| TNFAIP1 | AY065346 | 17q22 | 14.33 | 15 | 11 | 5.54 | 6 | .82 | 7 | 6 | 3.20 | 4 | .69 |
| TNFRSF1A | AY131997 | 12p13.2 | 16.21 | 38 | 31 | 23.04 | 4 | .96 | 18 | 10 | 3.65 | 5 | .73 |
| TRPV5 | AY206695 | 7q35 | 29.56 | 106 | 32 | 9.76 | 1 | .90 | 9 | 4 | 1.81 | 2 | .45 |
| TRPV6 | AY225461 | 7 | 27.63 | 105 | 26 | 10.02 | 4 | .90 | 2 | 3 | 1.70 | 3 | .41 |
| VCAM1 | AF536818 | 1p32 | 22.87 | 47 | 41 | 36.00 | 1 | .97 | 23 | 23 | 17.63 | 7 | .94 |
| VEGF | AF437895 | 6p12 | 15.44 | 37 | 35 | 25.60 | 4 | .96 | 35 | 22 | 13.39 | 5 | .93 |
| VTN | AF382388 | 17q11 | 5.56 | 16 | 11 | 5.67 | 7 | .82 | 4 | 3 | 2.35 | 3 | .57 |
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (accession numbers for all genes are listed in [online only])
- HaploBlockFinder V0.6b, http://cgi.uc.edu/cgi-bin/kzhang/haploBlockFinder.cgi/
- Nickerson Group, http://droog.gs.washington.edu
- Pharmacogenetics Research Network, http://www.nigms.nih.gov/pharmacogenetics/
- PHASE, http://www.stat.washington.edu/stephens/software.html
- Phred/Phrap/Consed System Home Page, http://www.phrap.org
- Polyphred 4.0, http://droog.mbt.washington.edu./PolyPhred.html
- SNPtagger, http://www.well.ox.ac.uk/~xiayi/haplotype/index.html
- UW-FHCRC Variation Discovery Resource, http://pga.gs.washington.edu/
References
- Bonnen PE, Wang PJ, Kimmel M, Chakraborty R, Nelson DL (2002) Haplotype and linkage disequilibrium architecture for human cancer-associated genes. Genome Res 12:1846–1853 10.1101/gr.483802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet Suppl 33:228–237 10.1038/ng1090 [DOI] [PubMed] [Google Scholar]
- Cambien F, Poirier O, Nicaud V, Herrmann SM, Mallet C, Ricard S, Behague I, Hallet V, Blanc H, Loukaci V, Thillet J, Evans A, Ruidavets JB, Arveiler D, Luc G, Tiret L (1999) Sequence diversity in 36 candidate genes for cardiovascular disorders. Am J Hum Genet 65:183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–99 10.1038/35052543 [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–238 10.1038/10290 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L, Nickerson DA (2003) Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet 33:518–521 10.1038/ng1128 [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengard J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Green ED, Guttmacher AE, Guyer MS (2003) A vision for the future of genomics research. Nature 422:835–847 10.1038/nature01626 [DOI] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Charkravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278:1580–1581 10.1126/science.278.5343.1580 [DOI] [PubMed] [Google Scholar]
- Collins FS, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L (1998) New goals for the U.S. Human Genome Project: 1998–2003. Science 282:682–689 10.1126/science.282.5389.682 [DOI] [PubMed] [Google Scholar]
- Coronini R, de Looze MA, Puget P, Bley G, Ramani SV (2003) Decoding the literature on genetic variation. Nat Biotechnol 21:21–29 10.1038/nbt0103-21 [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 10.1038/ng1001-229 [DOI] [PubMed] [Google Scholar]
- Dawson E, Abecasis GR, Bumpstead S, Chen Y, Hunt S, Beare DM, Pabial J, et al (2002) A first-generation linkage disequilibrium map of human chromosome 22. Nature 418:544–548 [DOI] [PubMed] [Google Scholar]
- Drysdale CM, McGraw DW, Stack CB, Stephens JC, Judson RS, Nandabalan K, Arnold K, Ruano G, Liggett SB (2000) Complex promoter and coding region beta 2-adrenergic receptor haplotypes alter receptor expression and predict in vivo responsiveness. Proc Natl Acad Sci USA 97:10483–10488 10.1073/pnas.97.19.10483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ (1964) The maintenance of alleles by mutation. Genetics 50:891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Haga H, Yamada R, Ohnishi Y, Nakamura Y, Tanaka T (2002) Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190,562 genetic variations in the human genome. Single-nucleotide polymorphism. J Hum Genet 47:605–610 10.1007/s100380200092 [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22:239–247 10.1038/10297 [DOI] [PubMed] [Google Scholar]
- Harding RM, Fullerton SM, Griffiths RC, Bond J, Cox MJ, Schneider JA, Moulin DS, Clegg JB (1997) Archaic African and Asian lineages in the genetic ancestry of modern humans. Am J Hum Genet 60:772–789 [PMC free article] [PubMed] [Google Scholar]
- Hartl DL, Clark AG (1997) Principles of population genetics. 3rd ed. Sinauer Associates, Sunderland, MA [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG (2001) Replication validity of genetic associations studies. Nat Genet 29:306–309 10.1038/ng749 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R (2001) Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet 29:217–222 10.1038/ng1001-217 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 10.1038/ng1001-233 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Rogers AR, Bamshad M, Watkins WS, Krakowiak P, Sung S, Kere J, Harpending HC (1997) Microsatellite diversity and the demographic history of modern humans. Proc Natl Acad Sci USA 94:3100–3103 10.1073/pnas.94.7.3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ (2001) Population genomics: a bridge from evolutionary history to genetic medicine. Hum Mol Genet 10:2199–2207 10.1093/hmg/10.20.2199 [DOI] [PubMed] [Google Scholar]
- Jorde LB, Watkins WS, Bamshad MJ, Dixon ME, Ricker CE, Seielstad MT, Batzer MA (2000) The distribution of human genetic diversity: a comparison of mitochondrial, autosomal, and Y chromosome data. Am J Hum Genet 66:979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Sajantila A, Jeffreys AJ (2003) Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum Mol Genet 12:33–40 10.1093/hmg/ddg008 [DOI] [PubMed] [Google Scholar]
- Ke X, Cardon LR (2003) Efficient selective screening of haplotype tag SNPs. Bioinformatics 19:287–288 10.1093/bioinformatics/19.2.287 [DOI] [PubMed] [Google Scholar]
- Kruglyak L (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet 22:139–144 10.1038/9642 [DOI] [PubMed] [Google Scholar]
- Kruglyak L, Nickerson DA (2001) Variation is the spice of life. Nat Genet 27:234–236 10.1038/85776 [DOI] [PubMed] [Google Scholar]
- Li N, Stephens M (2003) A new multilocus model for linkage disequilibrium, with application to exploring variations in recombination rate. Genetics 165:2213–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Cutler DJ, Zwick ME, Chakravarti A (2002) Haplotype inference in random population samples. Am J Hum Genet 71:1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33:177–182 10.1038/ng1071 [DOI] [PubMed] [Google Scholar]
- Nakajima T, Jorde LB, Ishigami T, Umemura S, Emi M, Lalouel JM, Inoue I (2002) Nucleotide diversity and haplotype structure of the human angiotensinogen gene in two populations. Am J Hum Genet 70:108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omi K, Ohashi J, Patarapotikul J, Hananantachai H, Naka I, Looareesuwan S, Tokunaga K (2003) CD36 polymorphism is associated with protection from cerebral malaria. Am J Hum Genet 72:364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BT, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SP, Cox DR (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719–1723 10.1126/science.1065573 [DOI] [PubMed] [Google Scholar]
- Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, Donaldson MA, Studebaker JF, et al (2003) Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet 33:382–387 10.1038/ng1100 [DOI] [PubMed] [Google Scholar]
- Reich DE, Gabriel SB, Altshuler D (2003) Quality and completeness of SNP databases. Nat Genet 33:457–458 10.1038/ng1133 [DOI] [PubMed] [Google Scholar]
- Rieder MJ, Taylor SL, Clark AG, Nickerson DA (1999) Sequence variation in the human angiotensin converting enzyme. Nat Genet 22:59–62 10.1038/8760 [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K (1996) The future of genetic studies of complex human diseases. Science 273:1516–1517 [DOI] [PubMed] [Google Scholar]
- Risch NJ (2000) Searching for genetic determinants in the new millennium. Nature 405:847–856 10.1038/35015718 [DOI] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, et al (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 10.1038/35057149 [DOI] [PubMed] [Google Scholar]
- Schneider JA, Peto TE, Boone RA, Boyce AJ, Clegg JB (2002) Direct measurement of the male recombination fraction in the human beta-globin hot spot. Hum Mol Genet 11:207–215 10.1093/hmg/11.3.207 [DOI] [PubMed] [Google Scholar]
- Schneider JA, Pungliya MS, Choi JY, Jiang R, Sun XJ, Salisbury BA, Stephens JC (2003) DNA variability of human genes. Mech Ageing Dev 124:17–25 10.1016/S0047-6374(02)00165-3 [DOI] [PubMed] [Google Scholar]
- Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, et al (2001a) Haplotype variation and linkage disequilibrium in 313 human genes. Science 293:489–493 10.1126/science.1059431 [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001b) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyan L, Eberle MA, Clark AG, Kruglyak L, Nickerson DA (2001) Sequence variation and linkage disequilibrium in the human T-cell receptor beta (TCRB) locus. Am J Hum Genet 69:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JG, Tang DC, Savage SA, Leitman SF, Heller SI, Serjeant GR, Rodgers GP, Chanock SJ (2002) Variants in the VCAM1 gene and risk for symptomatic stroke in sickle cell disease. Blood 100:4303–4309 10.1182/blood-2001-12-0306 [DOI] [PubMed] [Google Scholar]
- Templeton AR, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF (2000) Cladistic structure within the human Lipoprotein lipase gene and its implications for phenotypic association studies. Genetics 156:1259–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiret L, Poirier O, Nicaud V, Barbaux S, Herrmann SM, Perret C, Raoux S, Francomme C, Lebard G, Tregouet D, Cambien F (2002) Heterogeneity of linkage disequilibrium in human genes has implications for association studies of common diseases. Hum Mol Genet 11:419–429 10.1093/hmg/11.4.419 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, et al (2001) The sequence of the human genome. Science 291:1304–1351 10.1126/science.1058040 [DOI] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK (2003) Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet 4:587–597 10.1038/nrg1123 [DOI] [PubMed] [Google Scholar]
- Wang N, Akey JM, Zhang K, Chakraborty R, Jin L (2002) Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet 71:1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CF, Lewis K, Cantone KL, Khan P, Donnelly C, White N, Crocker N, Boyd PR, Zaykin DV, Purvis IJ (2002) Effectiveness of computational methods in haplotype prediction. Hum Genet 110:148–156 10.1007/s00439-001-0656-4 [DOI] [PubMed] [Google Scholar]
- Zhang K, Akey JM, Wang N, Xiong M, Chakraborty R, Jin L (2003) Randomly distributed crossovers may generate block-like patterns of linkage disequilibrium: an act of genetic drift. Hum Genet 113:51–59 [DOI] [PubMed] [Google Scholar]
- Zhang K, Jin L (2003) HaploBlockFinder: haplotype block analyses. Bioinformatics 19:1300–1301 10.1093/bioinformatics/btg142 [DOI] [PubMed] [Google Scholar]