Abstract
The Saami are regarded as extreme genetic outliers among European populations. In this study, a high-resolution phylogenetic analysis of Saami genetic heritage was undertaken in a comprehensive context, through use of maternally inherited mitochondrial DNA (mtDNA) and paternally inherited Y-chromosomal variation. DNA variants present in the Saami were compared with those found in Europe and Siberia, through use of both new and previously published data from 445 Saami and 17,096 western Eurasian and Siberian mtDNA samples, as well as 127 Saami and 2,840 western Eurasian and Siberian Y-chromosome samples. It was shown that the “Saami motif” variant of mtDNA haplogroup U5b is present in a large area outside Scandinavia. A detailed phylogeographic analysis of one of the predominant Saami mtDNA haplogroups, U5b1b, which also includes the lineages of the “Saami motif,” was undertaken in 31 populations. The results indicate that the origin of U5b1b, as for the other predominant Saami haplogroup, V, is most likely in western, rather than eastern, Europe. Furthermore, an additional haplogroup (H1) spread among the Saami was virtually absent in 781 Samoyed and Ob-Ugric Siberians but was present in western and central European populations. The Y-chromosomal variety in the Saami is also consistent with their European ancestry. It suggests that the large genetic separation of the Saami from other Europeans is best explained by assuming that the Saami are descendants of a narrow, distinctive subset of Europeans. In particular, no evidence of a significant directional gene flow from extant aboriginal Siberian populations into the haploid gene pools of the Saami was found.
Introduction
Although not homogeneous, the European genetic landscape has been characterized by relatively short genetic distances between individual populations. Classic genetic markers have revealed a few clearly pronounced outliers, like the Sardinians, the Greeks, the Basques, the Finns and, in particular, the Saami (Cavalli-Sforza and Piazza 1993; Cavalli-Sforza et al. 1994). The Saami (Lapps) are aboriginal inhabitants of Fennoscandia. The Saami language belongs to the Finno-Ugric branch of the Uralic language family and can be divided into 10 dialects (Sammallahti 1998). The closest linguistic neighbors of the Saami are the Finns, the Karelians, and the Estonians. At present, ∼60,000 Saami live in the northern regions of Norway, Sweden, and Finland, as well as in the Russian Kola Peninsula (Haetta 1996). Soon after the beginning of the retreat of the ice sheets covering the area, in the 8th–10th millennia before present (BP), populations of hunters and fishermen, the producers of the Mesolithic Komsa and Fosna-Hensbacka cultures, inhabited the coastal region of Scandinavia, extending well into Finland and to the Kola Peninsula (Kozlowski and Bandi 1984; Nygaard 1989; Sumkin 1990). The linguistic affiliation of these pioneer settlers of the north is largely unknown, but it has been suggested that they are the descendants of the Ahrensburgian population, which migrated toward the north from western Europe, along the Atlantic coast of Norway. It has been proposed that they might have been the ancestors of the present-day Saami (e.g., Sumkin 1990). Another presumably important component in the postglacial recolonization of northern Fennoscandia came from the east, via Karelia and Finland. It has been associated with the movement of Mesolithic populations, carriers of post-Swiderian cultures, to the north. Starting in the Neolithic period, the northern population came into contact with tribes of territories lying to the south (e.g., Sumkin 1990). Thus, according to archeological data, the present-day Saami population might have been shaped in different times both by the eastern and western influences.
Analyses of classic chromosomal marker variation have demonstrated that the genetic distances between the Saami and other European populations are significantly larger than between any other pair of European populations (Cavalli-Sforza et al. 1994). This distinctive pattern has suggested to some that the genetic composition of the Saami population arose from extensive admixture between Caucasoid and Mongoloid populations. Estimates of the relative contribution of each have varied from an equal input to ∼80% Caucasoid and 20% Mongoloid genes (Guglielmino et al. 1990; Cavalli-Sforza et al. 1994, pp 272–273). However, not all classic genetic studies support an idea of an extensive Caucasoid-Mongoloid admixture in the Saami genetic background (e.g., Beckman et al. 1988, 1993).
Similarly, studies of mtDNA have identified large genetic distances between the Saami and other Europeans, including the Finns (Sajantila and Pääbo 1995; Sajantila et al. 1995). Likewise, Lahermo et al. (1996) found no overlap between Saami and the remaining European mtDNA patterns and concluded that the Saami and the Finns must have different genetic histories. One alternative hypothesis to explain the presence of genetic differences and language similarities in the Finns and the Saami involves a language shift by the Finns from Indo-European to Finno-Ugric (Sajantila and Pääbo 1995).
The combined sequence information from the first hypervariable segment (HVS-I) in the D-loop of mtDNA and the variations in the mtDNA coding region of European populations have been exploited to generate a common phylogenetic nomenclature for mtDNA variants in Europe (Torroni et al. 1996; Macaulay et al. 1999). Classification of the Saami mtDNA lineages revealed that the absolute majority of these are clustered in a subset of the European mtDNA pool (Torroni et al. 1998; Villems et al. 1998), where two haplogroups—V and U—cover ∼80% of the variability of this subset (Torroni et al. 1998; Tambets et al. 2000).
In contrast to the predominance of European mtDNA haplogroups observed among the Saami, nearly half of their Y chromosomes share a TatC allele (haplogroup N3, according to the nomenclature of the Y Chromosome Consortium [YCC 2002]) with most Finno-Ugric and Siberian populations. This variant is found at high frequencies among Siberian populations, such as the Yakuts and the Buryats, but is virtually absent in western and Mediterranean Europe; even among the Norwegians and the Swedes, populations that have historically lived in close proximity to the Saami, it is found at frequencies of only 4%–8% (Zerjal et al. 2001; Passarino et al. 2002). High frequencies of the TatC allele have also been observed in Baltic (30%–40%) and Volga-Finnic–speaking populations (20%–50%) (Zerjal et al. 1997; Rootsi et al. 2000; Rosser et al. 2000; Semino et al. 2000; Laitinen et al. 2002). These findings have been interpreted according to the classic view that a substantial element of the Saami (and other European Finno-Ugric–speaking populations) genetic lineages originated in a recent migration from Asia (Zerjal et al. 1997, 2001).
Another half of the Saami paternal lineages have primarily mutations M170, SRY-1532, or M173 (Semino et al. 2000; Raitio et al. 2001; Wells et al. 2001). Accordingly, they are identified as haplogroups I, R1a, and R1b (YCC 2002). Y chromosomes possessing those mutations are widely spread in European populations (Rosser et al. 2000; Semino et al. 2000). Whereas SRY-1532 and M173 are present at moderate frequencies in some Siberian populations as well, M170 Y chromosomes are very rare there (Wells et al. 2001; Karafet et al. 2002).
Here, we first analyze mtDNA variation in the Swedish Saami and improve the resolution level of their Y-chromosomal haplotypes. Second, we examine the hypothetical descent of the Saami gene pool from Siberian ancestors. We exploit extensive data on mtDNA and Y-chromosome variation in Eurasia, in particular among Uralic-, Indo-European–, and Altaic-speaking populations in eastern Europe and western Siberia. Finally, we refine the phylogenetic topology of a frequent Saami mtDNA haplogroup, U5b, to identify the founder haplotypes. We draw conclusions about the possible origin of mtDNA and NRY haplotypes present in the Saami gene pool.
Subjects and Methods
Population Samples
We analyzed mtDNA sequences from 17,541 Eurasian individuals sampled from 65 different Eurasian populations (table 1). These data consist of previously published sequences (n=9,154) and new sequences analyzed in this study (n=8,387). Comparisons of different Saami subpopulations are presented separately in table 2. Y-chromosomal markers were also analyzed from 32 different populations. Data from 1,598 of a total of 2,967 individuals reported in table 3 were taken from published sources, whereas the remaining 1,369 DNA samples were extracted and analyzed in the present study. Further details about the populations other than the Saami will be published elsewhere. Blood samples were collected from healthy unrelated individuals after obtaining informed consent. DNA was extracted using the phenol-chloroform method, as used by Sambrook (1989).
Table 1.
Frequencies (%) of the Predominant mtDNA Haplogroups of the Saami among Other Eurasian Populations
|
Frequency of Haplogroup(%) |
||||||||
| Region andPopulationa | LanguageFamilyb | SampleSize | V | U5b1b1c | H1d | D5e | Z | Reference(s) |
| Scandinavia: | ||||||||
| *Saami | U-FU | 445 | 41.6 | 47.6 | 2.5 | 3.1 | 1.3 | Sajantila et al. 1995, corrected by Bandelt et al. 2001; Dupuy and Olaisen 1996; Delghandi et al. 1998; present study |
| *Swedes | IE-G | 503 | 3.2 | .8 | 3.4 | 0 | .4 | Sajantila et al. 1996; present study |
| *Norwegians | IE-G | 641 | 3.4 | 1.2 | 1.9 | 0 | .6 | Dupuy and Olaisen 1996; Opdal et al. 1998; Helgason et al. 2001; Passarino et al. 2002 |
| Northern Europe: | ||||||||
| *Karelians | U-FU | 83 | 6.0 | 6.0 | 6.0 | 1.2 | 0 | Sajantila et al. 1995 |
| *Finns | U-FU | 581 | 6.4 | 6.7 | 3.6 | .2 | 1.5 | Pult et al. 1994; Sajantila et al. 1995; Lahermo et al. 1996; Kittles et al. 1999; Meinilä et al. 2001 |
| *Estonians | U-FU | 545 | 3.3 | .7 | 4.8 | .2 | 0 | Sajantila et al. 1995, 1996; present study |
| Latvians | IE-B | 299 | 3.0 | 0 | 1.0 | 0 | 0 | Present study |
| Lithuanians | IE-B | 45 | 2.2 | 2.2 | 2.2 | 0 | 0 | Present study |
| North-Russians | IE-S | 134 | 0 | 3.0 | 2.2 | 5.2 | 0 | Present study |
| Eastern Europe: | ||||||||
| *Russians | IE-S | 761 | 3.8 | 1.2 | 1.2 | 0 | .3 | Orekhov et al. 1999; Malyarchuk et al. 2001, 2002; present study |
| Ukrainians | IE-S | 686 | 4.8 | .6 | 1.5 | 0 | .1 | Malyarchuk et al. 2001; present study |
| *Poles | IE-S | 583 | 4.5g | .7 | 1.0 | 0 | 0 | Richards et al. 2000; Malyarchuk et al. 2002; present study |
| *Northwestern Europe | IE-C/G | 1,851 (1,547)f | 2.8g | 0 | 1.6 | 0 | 0 | Piercy et al. 1993; Richards et al. 1996, 2000; Helgason et al. 2000, 2001 |
| Volga-Ural region: | ||||||||
| *Maris | U-FU | 147 | 10.2 | .7 | .7 | 0 | 2.7 | Sajantila et al. 1995; Bermisheva et al. 2002; present study |
| *Mordvin | U-FU | 111 | 3.6 | 2.7 | 0 | 0 | 0 | Sajantila et al. 1995; Bermisheva et al. 2002 |
| Komis | U-FU | 340 | .6 | 1.8 | 2.1 | 1.8 | 1.8 | Bermisheva et al. 2002; present study |
| Udmurts | U-FU | 182 | .5 | 0 | 8.2 | 0 | 7.1 | Bermisheva et al. 2002; present study |
| Chuvashes | A-T | 89 | 5.6 | 1.1 | 2.2 | 0 | 0 | Richards et al. 2000; Bermisheva et al. 2002 |
| Tatars | A-T | 176 | 4.5 | 0 | 0 | 0 | 0 | Bermisheva et al. 2002; present study |
| Bashkirs | A-T | 209 | 3.3 | 2.4 | 0 | 0 | 1.0 | Bermisheva et al. 2002 |
| Southern Europe: | ||||||||
| *Italians | IE-R | 397 (240)f | 1.7g | 0 | 1.0 | 0 | 0 | Francalacci et al. 1996; Richards et al. 2000; Mogentale-Profizi et al. 2001; Tagliabracci et al. 2001; present study |
| Sardinians | IE-R | 115 (133)f | 6.0g | 0 | .9 | 0 | 0 | Richards et al. 2000; Cali et al. 2001 |
| *Sicilians | IE-R | 732 (634)f | 2.2g | 0 | .8 | 0 | 0 | Di Rienzo and Wilson 1991; Richards et al. 2000; Cali et al. 2001; Forster et al. 2002; present study |
| Cypriots | IE | 188 | 0 | 0 | 1.6 | 0 | 0 | Present study |
| Greeks (mainland and Crete) | IE | 399 (414)f | .7g | 0 | 0 | 0 | 0 | Richards et al. 2000; present study |
| Albanians | IE | 199 | .5 | 0 | 0 | 0 | 0 | Present study |
| Croats | IE-S | 440 | 2.7 | .5 | .5 | 0 | 0 | Present study |
| Bosnians | IE-S | 395 | 3.3 | .8 | .8 | 0 | .3 | Malyarchuk et al. 2003; present study |
| Western Europe: | ||||||||
| *French | IE-R | 577 | 3.1 | .3 | 2.3 | 0 | 0 | Rousselet and Mangin 1998; Danan et al. 1999; Richards et al. 2000; Cali et al. 2001; present study |
| *Spaniards | IE-R | 479 (293)f | 2.0g | 0 | .4 | 0 | 0 | Corte-Real et al. 1996; Salas et al. 1998; Crespillo et al. 2000 |
| *Portugese | IE-R | 295 (54)f | 3.7g | 0 | 1.4 | 0 | 0 | Corte-Real et al. 1996; Pereira et al. 2000 |
| *Basques | Isolate | 106 (97)f | 12.4g | 0 | 0 | 0 | 0 | Bertranpetit et al. 1995; Corte-Real et al. 1996 |
| *Central Europe: | ||||||||
| *Germans | IE-G | 582 | 4.3g | 0 | 2.9 | 0 | 0 | Richards et al. 1996; Hofmann et al. 1997; Baasner et al. 1998; Lutz et al. 1998; Pfeiffer et al. 1999 |
| *Swiss | IE-G | 230 | 3.9 | 0 | .9 | 0 | 0 | Pult et al. 1994; Dimo-Simonin et al. 2000 |
| Austrians | IE-G | 101 | 1.0 | 0 | 0 | 0 | 0 | Parson et al. 1998 |
| Hungarians | U-FU | 116 | 2.6 | .9 | 1.7 | 0 | 0 | Present study |
| Czechs | IE-S | 177 | 4.0 | 0 | .6 | 0 | 0 | Richards et al. 2000; present study |
| Slovaks | IE-S | 129 | 2.3 | 0 | 4.7 | 0 | 0 | Present study |
| Slovenes | IE-S | 110 | 5.5 | 1.8 | 3.6 | 0 | 0 | Present study |
| Caucasus | IE, CA-N/S; A-M/T | 1,383 | .5 | .4 | .5 | 0 | .7 | Richards et al. 2000; present study |
| Siberia: | ||||||||
| Khants | U-FU | 255 | 0 | 0 | 0 | 0 | 0 | Present study |
| Mansis | U-FU | 138 | .7 | 0 | 0 | .7 | 0 | Derbeneva et al. 2002b; present study |
| Nganasans | U-SA | 131 | 0 | 0 | .8 | 0 | 2.3 | Derbeneva et al. 2002a; present study |
| Nenets | U-SA | 137 | 0 | 0 | 0 | 0 | 0 | Saillard et al. 2000; present study |
| Selkups | U-SA | 120 | 0 | 0 | 0 | 0 | 0 | Present study |
| Kets | Isolate | 104 | 0 | 0 | 0 | 0 | 2.9 | Derbeneva et al. 2002a; present study |
| Dolgans | A-T | 130 | 0 | 0 | .8 | 0 | 1.5 | Present study |
| *Yakuts | A-T | 395 | 0 | .5 | 0 | 0 | 0 | Fedorova et al. 2003; Pakendorf et al. 2003; Puzyrev et al. 2003 |
| *Buryats | A-M | 126 | 0 | 0 | 0 | 0 | 1.6 | Pakendorf et al. 2003 |
| Evenks | A-TN | 105 | 0 | 0 | 0 | 0 | 1.0 | Present study |
| Altaians | A-T | 339 |
0 | 0 | 0 | .3 | 5.0 | Present study |
| Total | 17,541 | |||||||
Populations for which frequencies were deduced from published HVS-I sequences are indicated by an asterisk (*). Populations are divided into geographic regions; for northwestern Europe (consists of Scots [n=891], English and Welsh [n=426], Irish [n=101], and Icelanders [n=433]) and for the Caucasus region (consists of Nogays [n=183], Adygeis [n=159], Abazins [n=64], Kabardins [n=66], Karachays [n=106], Kumyks [n=109], Kalmyks [n=120], Georgians [n=138], Southern Ossetians [n=198], Armenians [n=192], and Azeris [n=48]), only summary data are shown.
Language-family codes used for each population are as follows: U = Uralic (FU = Finno-Ugric; SA = Samoyedic); IE = Indo-European (B = Baltic; C = Celtic; G = Germanic; R = Romance; S = Slavic); A = Altaic (M = Mongolic; T = Turkic, TN = Tungusic); CA = Caucasian (N = North; S = South).
Only haplotypes with HVS-I motif 16144-16189-16270 and its derivatives have been taken into account.
Only haplotypes with HVS-I motif 16162 and its derivatives have been taken into account.
Only haplotypes with HVS-I motif 16126-16136-16189-16223-16360-16362 and its derivatives have been taken into account.
In the estimation of haplogroup V proportions, data from Torroni et al. (2001) have been used, and the corresponding sample size is given in parentheses after the sample size analyzed for calculating other haplogroup frequencies (in case of German population, the pooled data of different German subpopulations from Torroni et al. [2001] have been used).
Frequencies of haplogroup V are from Torroni et al. (2001).
Table 2.
Frequencies (%) of mtDNA and Y-Chromosomal Haplogroups in Different Saami Subpopulations
|
Frequency (95% CR)a(%) |
||||
| Haplogroup | Swedish Saamib(n=98/35) | Finnish Saamic(n=69/69) | Norwegian Saamid(n=278/⋅⋅⋅) | Kola Saamie(n=⋅⋅⋅/23) |
| mtDNA: | ||||
| H: | 3.1 (1.1–8.6) | 2.9 (.9–9.9) | 4.7 (2.8–7.8) | … |
| H1 | .0 (.0–3.0) | .0 (.0–4.2) | 4.0 (2.2–6.9) | … |
| W | 1.0 (.2–5.5) | .0 (.0–4.2) | 1.4 (.6–3.6) | … |
| T | .0 (.0–3.0) | .0 (.0–4.2) | .4 (.1–2.0) | … |
| U: | 26.5 (18.8–36.1) | 43.5 (32.4–55.3) | 57.6 (51.7–63.2) | … |
| U5a | .0 (.0–3.0) | 2.9 (.9–9.9) | .7 (.2–2.6) | … |
| U5b | 26.5 (18.8–36.1) | 40.6 (29.8–52.4) | 56.8 (51.0–62.5) | … |
| V | 68.4 (58.6–76.7) | 37.7 (27.2–49.5) | 33.1 (27.8–38.8) | … |
| M: | 1.0 (.2–5.5) | 15.9 (9.2–26.4) | 2.9 (1.5–5.6) | … |
| D5 | .0 (.0–3.0) | 8.7 (4.1–17.7) | 2.9 (1.5–5.6) | … |
| Z | 1.0 (.2–5.5) | 7.2 (3.2–15.9) | .0 (.0–1.1) | … |
| NRY: | ||||
| E | .0 (.0–8.0) | .0 (.0–4.2) | … | 8.7 (2.7–27.0) |
| F* | 5.7 (1.8–18.7) | … | .0 (.0–11.7) | |
| I | 31.4 (18.6–48.1) | 40.6 (29.8–52.4)f | … | 17.4 (7.1–37.4) |
| J | .0 (.0–.8) | … | 4.3 (1.0–21.1) | |
| N3 | 37.1 (23.1–53.8) | 55.1 (43.3–66.3) | … | 39.1 (22.1–59.4) |
| R1b | 5.7 (1.8–18.7) | 1.4 (.3–7.7) | … | 8.7 (2.7–27.0) |
| R1a | 20.0 (10.1–36.0) | 2.9 (.9–9.9) | … | 21.7 (9.8–42.2) |
Sample size (n) is given separately for mtDNA/NRY data sets, respectively.
mtDNA data are from this study and inferred from Sajantila et al. (1995); NRY data from Rosser et al. (2000) are further analyzed for markers M89, M52, M130 (RSP4Y), M170, M173, M178, M201, and M269 (YCC 2002) in the present study.
mtDNA data are inferred from Sajantila et al. (1995); NRY data are inferred from Raitio et al. (2001).
mtDNA data are inferred from Sajantila et al. (1995), Dupuy and Olaisen (1996), and Delghandi et al. (1998).
NRY data are from Wells et al. (2001).
Frequency is that of haplogroups F, I, and J combined.
Table 3.
Frequencies (%) of Selected Y-Chromosomal Haplogroups in the Saami and in Other Eurasian Populations
|
Frequency of Haplogroup(%) |
||||||||||
| Region andPopulationa | LanguageFamilyb | SampleSize | Ic | R1bd | R1a | N3 | N2 | Q | C | Reference(s) |
| Scandinavia: | ||||||||||
| *Saamie | U-FU | 127 (58)f | 25.9f | 3.9 | 11.0 | 47.2 | 0 | 0 | 0 | Raitio et al. 2001; Wells et al. 2001; present study |
| Swedes | IE-G | 141 | 48.2 | 22.0 | 18.4 | 2.8 | 0 | 0 | 0 | Present study |
| Norwegians | IE-G | 72 | 40.3 | 27.8 | 23.6 | 6.9 | 0 | 0 | 0 | Passarino et al. 2002 |
| Danes | IE-G | 194 | 38.7 | 36.1 | 16.5 | .5 | 0 | 0 | 0 | Sanchez et al. 2003 |
| Northern Europe: | ||||||||||
| *Finns | U-FU | 38 | 28.9 | 0 | 7.9 | 63.2 | 0 | 0 | 0 | Zerjal et al. 2001 |
| Estonians | U-FU | 209 | 18.2 | 9.1 | 33.5 | 30.6 | 0 | 0 | 0 | Present study |
| Latvians | IE-B | 86 | 7.0 | 9.3 | 38.4 | 41.9 | 0 | 0 | 0 | Present study |
| North Russians | IE-S | 77 | 20.8 | 0 | 40.3 | 28.6 | 0 | 0 | 0 | Wells et al. 2001 |
| Eastern Europe: | ||||||||||
| Russians | IE-S | 61 | 13.1 | 21.3 | 42.6 | 8.2 | 8.2 | 0 | 0 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Ukrainians | IE-S | 50 | 18.0 | 2.0 | 54.0 | 6.0 | 0 | 0 | 0 | Semino et al. 2000 |
| Polese | IE-S | 93 | 16.1 | 13.4 | 55.9 | 3.2 | 0 | 0 | 0 | Present study |
| Volga-Ural region: | ||||||||||
| Maris | U-FU | 111 | 8.1 | 2.7 | 47.7 | 31.5 | 9.9 | 0 | 0 | Semino et al. 2000; present study |
| Mordvin | U-FU | 83 | 19.3 | 13.3 | 26.5 | 16.9 | 2.4 | 0 | 0 | Present study |
| Komis | U-FU | 94 | 5.3 | 16.0 | 33.0 | 22.3 | 12.8 | 0 | 0 | Present study |
| Udmurts | U-FU | 87 | 1.1 | 2.3 | 10.3 | 56.3 | 28.7 | 0 | 0 | Present study |
| Chuvashes | A-T | 79 | 11.4 | 3.8 | 31.6 | 17.7 | 10.1 | 0 | 1.3 | Present study |
| Tatars | A-T | 126 | 4.0 | 8.7 | 34.1 | 18.3 | 4.8 | .8 | 1.6 | Present study |
| Southern Europe: | ||||||||||
| Croats | IE-S | 109 | 37.6 | 15.6 | 33.9 | 0 | 0 | 0 | 0 | Barac et al. 2003 |
| Western Europe: | ||||||||||
| French | IE-R | 61 | 16.4 | 59.0 | 0 | 0 | 0 | 0 | 0 | Semino et al. 2000; present study |
| Central Europe: | ||||||||||
| Germans | IE-G | 16 | 37.5 | 0 | 0 | 0 | 0 | 0 | 0 | Semino et al. 2000 |
| Hungarians | U-FU | 113 | 28.3 | 20.4 | 20.4 | .9 | 0 | 2.6 | .9 | Present study |
| Siberia: | ||||||||||
| Khants | U-FU | 47 | 0 | 19.1 | 4.3 | 38.3 | 38.3 | 0 | 0 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Nganasans | U-SA | 38 | 0 | 0 | 0 | 0 | 92.1 | 0 | 5.3 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Nenets | U-SA | 148 | 0 | 0 | 0 | 40.5 | 56.8 | 1.4 | 0 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Selkups | U-SA | 131 | 0 | 6.1 | 19.1 | 0 | 6.9 | 66.4 | 1.5 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Kets | Isolate | 48 | 0 | 0 | 0 | 0 | 0 | 93.7 | 6.2 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Dolgans | A-T | 67 | 1.5 | 1.5 | 16.4 | 22.4 | 11.9 | 0 | 37.3 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Yakuts | A-T | 155 | 1.3 | 1.9 | 1.9 | 88.4 | 0 | 0 | 3.2 | Karafet et al. 2002 (supplementary data obtained by request from the author); present study |
| Buryats | A-M | 81 | 0 | 1.2 | 1.2 | 28.4 | 2.5 | 0 | 60.5 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Evenks | A-TN | 96 | 5.2 | 0 | 1.0 | 16.7 | 3.1 | 4.2 | 67.7 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Evens | A-TN | 31 | 3.2 | 0 | 6.5 | 12.9 | 0 | 0 | 74.2 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Altaians | A-T | 98 |
0 | 0 | 46.9 | 1.0 | 2.0 | 17.3 | 22.4 | Karafet et al. 2002 (supplementary data obtained by request from the author) |
| Total | 2,967 | |||||||||
Populations for which data are deduced from published sources are indicated with an asterisk.
Language-family codes for each population are as follows: U = Uralic (FU = Finno-Ugric; SA = Samoyedic); IE = Indo-European (B = Baltic; G = Germanic; R = Romance; S = Slavic); A = Altaic (M = Mongolic; T = Turkic, TN = Tungusic).
Haplogroup I is, in the case of data deduced from literature, defined by the characteristic STR pattern of haplogroup 2 Y chromosomes (Barac et al. 2003; present study).
Haplogroup R1b is, in the case of data deduced from literature, defined here by M173(xSRY-1532) as studied western Europeans (except three Danes, who belong to R1* [Sanchez et al. 2003]) from this clade have been shown to share the additional mutation M269 (Cruciani et al. 2002; present study).
35 Swedish Saami and 93 Poles from the study by Rosser et al. (2000) were further analyzed for markers M52, M89, M130, M170, M173, M178, M201, and M269.
In the estimation of haplogroup I proportions, only the data from the present study and from Wells et al. (2001) were used. The corresponding sample sizes are given in parentheses.
mtDNA Analysis
HVS-I was sequenced between nucleotide positions (nps) 16024 and 16383 of the revised Cambridge Reference Sequence (Andrews et al. 1999) in 73 Swedish Saami and 8,314 other mtDNA samples. RFLP analysis of diagnostic mtDNA positions was performed, and mtDNA haplogroups were assigned to each sample by use of published criteria (Torroni et al. 1992, 1994, 1996; Richards et al. 1998; Macaulay et al. 1999; Schurr et al. 1999; Finnilä et al. 2000, 2001; Herrnstad et al. 2002; Kivisild et al. 2002). In addition, all U5b genomes (n=160) from 31 populations were typed by sequencing for the np 5656 A→G mutation that differentiates between individuals belonging to haplogroups U5b* and U5b1. All identified U5b1 individuals (5656G) were further sequenced to assess the variation at nps 7385 (A or G) and 10927 (T or C). A summary of these data is given in table 4.
Table 4.
Phylogeographic Distribution of Haplogroup U5b Subclades (Excluding U5b1b1)[Note]
|
No. of Individuals in Populationc |
|||||||||||||||||||||||||||||||||||
| WSE |
SC |
EE |
Other |
Base at npa |
|||||||||||||||||||||||||||||||
| HVS-ISequencea | U5b Sub HGb | al | bo | br | cr | cz | fr | gr | hu | it | si | sk | sl | sw | ba | es | ko | la | mo | po | ru | ta | ud | uk | mc | ng | go | tu | sh | ku | td | nd | 5656 | 7385 | 10927 |
| 270 | U5b* | 1 | 2 | 3 | A | ND | ND | ||||||||||||||||||||||||||||
| 051-168-189-270-362 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 086-192-270-304 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 093-172-189-270-362 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 093-234-270-325 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 093-258-270-292-362 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 145-189-270 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 169-172-189-270 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 169CA-192-235-270-304 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-192-201-270-398 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-192-217-234-270-398 | U5b* | 2 | 2 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-192-234-256-270-311-362-398 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-192-266-270-398 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-192-270 | U5b* | 1 | 1 | 1 | 1 | 1 | 5 | A | ND | ND | |||||||||||||||||||||||||
| 189-192-270-398 | U5b* | 1 | 1 | 1 | 3 | A | ND | ND | |||||||||||||||||||||||||||
| 189-239CA-270 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-261-270 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-270 | U5b* | 1 | 3 | 2 | 6 | A | ND | ND | |||||||||||||||||||||||||||
| 189-270-288-292 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 189-270-311 | U5b* | 1 | 1 | 1 | 3 | A | ND | ND | |||||||||||||||||||||||||||
| 189-292-311 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 192-212-270 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 192-224-261 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 192-235-270-304 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 192-257-270-304 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 192-270 | U5b* | 1 | 1 | 1 | 3 | A | ND | ND | |||||||||||||||||||||||||||
| 192-270-278-304 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 192-270-304 | U5b* | 1 | 1 | 2 | A | ND | ND | ||||||||||||||||||||||||||||
| 192-311 | U5b* | 1 | 1 | 1 | 4 | 2 | 9 | A | ND | ND | |||||||||||||||||||||||||
| 222-224-270-311 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 224-270-311 | U5b* | 1 | 1 | 2 | A | ND | ND | ||||||||||||||||||||||||||||
| 234-270 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 258-270-292-362 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 261-270 | U5b* | 1 | 1 | 1 | 3 | A | ND | ND | |||||||||||||||||||||||||||
| 261-270-390 | U5b* | 1 | 1 | A | ND | ND | |||||||||||||||||||||||||||||
| 051-189-270 | U5b1* | 2 | 2 | G | A | T | |||||||||||||||||||||||||||||
| 086-189-192-270-311-336 | U5b1* | 3 | 3 | G | A | T | |||||||||||||||||||||||||||||
| 092-189-270-294 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 093-182-189-192-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 093-189-270 | U5b1* | 1 | 1 | 2 | G | A | T | ||||||||||||||||||||||||||||
| 093-189-270-362 | U5b1* | 2 | 2 | G | A | T | |||||||||||||||||||||||||||||
| 129-189-223-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 129-189-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 129-192-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 147-189-270 | U5b1* | 1 | 3 | 1 | 5 | G | A | T | |||||||||||||||||||||||||||
| 174-189-223-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 174-189-270-311 | U5b1* | 1 | 1 | 2 | G | A | T | ||||||||||||||||||||||||||||
| 182-189-192-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 186-189-270-293 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 189-192-218-270-320 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 189-192-270 | U5b1* | 6 | 3 | 1 | 10 | G | A | T | |||||||||||||||||||||||||||
| 189-192-270-292 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 189-192-270-311 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 189-221-223-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 189-221-270 | U5b1* | 1 | 1 | 2 | G | A | T | ||||||||||||||||||||||||||||
| 189-223-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 189-270 | U5b1* | 8 | 1 | 1 | 1 | 5 | 2 | 1 | 5 | 2 | 26 | G | A | T | |||||||||||||||||||||
| 189-270-311-336 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 192-224-261-270 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 192-270 | U5b1* | 1 | 1 | 1 | 1 | 1 | 5 | G | A | T | |||||||||||||||||||||||||
| 192-270-296 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 192-270-319 | U5b1* | 1 | 1 | G | A | T | |||||||||||||||||||||||||||||
| 074-172-189-270 | U5b1b | 1 | 1 | G | G | C | |||||||||||||||||||||||||||||
| 074-189-270 | U5b1b | 1 | 1 | G | G | C | |||||||||||||||||||||||||||||
| 093-129-189-270 | U5b1b | 1 | 2 | 1 | 4 | G | G | C | |||||||||||||||||||||||||||
| 093-189-270 | U5b1b | 1 | 1 | 2 | G | G | C | ||||||||||||||||||||||||||||
| 093-189-270-301 | U5b1b | 1 | 1 | G | G | C | |||||||||||||||||||||||||||||
| 093-189-270-301-311 | U5b1b | 1 | 1 | G | G | C | |||||||||||||||||||||||||||||
| 173-189-270-235 | U5b1b | 1 | 1 | G | G | C | |||||||||||||||||||||||||||||
| 189-192-270 | U5b1b | 1 | 1 | 2 | G | G | C | ||||||||||||||||||||||||||||
| 189-270 | U5b1b | 1 | 1 | 1 | 1 | 1 | 5 | G | G | C | |||||||||||||||||||||||||
| 189-270-325 | U5b1b | 1 | 1 | G | G | C | |||||||||||||||||||||||||||||
| Total | 3 | 5 | 1 | 20 | 4 | 13 | 3 | 2 | 1 | 9 | 1 | 1 | 9 | 4 | 14 | 11 | 6 | 5 | 1 | 16 | 5 | 1 | 13 | 3 | 1 | 2 | 2 | 1 | 1 | 2 | 160 | ||||
Note.— mtDNAs belonging to subclade U5b1b1 were screened selectively for the presence of the mutations in nps 5656, 7385, and 10927. All of those (18 Saami, 5 individuals from eastern Europe, and 5 individuals from western and southern Europe) possessed aforementioned transitions.
Mutations in sequences are relative to the revised Cambridge Reference Sequence (Andrews et al. 1999). HVS-I sequences are given minus 16,000; only tranversions are further indicated. ND = not determined.
SubHG = subhaplogroup.
Geographic subdivision codes are as follows: WSE = western and southern Europe; SC = Scandinavian Europe; EE = eastern Europe. Population codes are as follows: al = Albanians; bo = Bosnians; br = Bretons; cr = Croats; cz = Czechs; fr = French; gr = Greeks; hu = Hungarians; it = Italians; si = Sicilians; sk = Slovaks; sl = Slovenians; sw = Swedes; ba = Bashkirs; es = Estonians; ko = Komis; la = Latvians; mo = Mordvin; po = Poles; ru = Russians; ta = Tatarians; ud = Udmurts; uk = Ukrainians; mc = Moroccans; ng = Nogays; go = Georgians; tu = Turks; sh = Shors; ku = Kumyks; td = Tadjik. In addition to population data from table 1, data of Bretons, Moroccans, Shors, Tadjiks, and Turks are shown (authors' unpublished data).
Number of subjects analyzed in haplotype.
Y-Chromosome Analysis
Sixteen Y-chromosomal biallelic markers were assayed in 1,369 DNA samples. Nomenclature of haplogroups is as defined by the YCC (2002). Markers were analyzed as follows. The polymorphic SNPs underlying markers M52, M130 (RSP4Y), M170, M173, M178, M201 (Underhill et al. 2001), M269 (Cruciani et al. 2002), and M242 (Seielstad et al. 2003) were assayed after PCR amplification and sequencing. Markers M9, Tat, SRY-1532, 92R7, M89, and P43 were assayed through restriction digest analysis by use of published protocols (Mathias et al. 1994; Whitfield et al. 1995; Zerjal et al. 1997; Akey et al. 2001; Raitio et al. 2001; Karafet et al. 2002). The YAP and 12f2 polymorphisms were identified following the procedures of Hammer and Horai (1995) and Casanova et al. (1985), respectively. Furthermore, 35 Swedish Saami DNA samples that have previously been analyzed for Y-chromosomal variation by Rosser et al. (2000) were also typed in the present study for the biallelic markers M89, M52, M130 (RSP4Y), M170, M173, M178, M201, and M269 (YCC 2002).
Sequencing
PCR-amplified products were purified using shrimp alkaline phosphatase and exonuclease treatment following Kaessmann (1999). These were sequenced using the DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia Biotech) on an ABI 377 DNA Sequencer. Sequences were aligned and analyzed using the Genetics Computer Group Wisconsin Package.
Data Analysis
Principal-component (PC) analysis was undertaken using the POPSTR program, kindly provided by H. Harpending (see details described by Richards et al. [2002]). Only those haplogroups that had a noticeable impact on the scatterplot were used for the analysis (the sum of the absolute values for both coordinates of each allele was >0.2 [M, V, U5, H, A, J, and U4 for mtDNA and Q, N2, I, R1b, R1a, and N3 for the Y chromosome]). Bayesian 95% credible regions (CRs) for haplogroup frequencies were calculated with the computer program SAMPLING, kindly provided by V. Macaulay. The diversity of mtDNA haplotypes was estimated as by Nei (1987).
Phylogenetic networks of mtDNA HVS-I haplotypes were constructed using the program Network 3.1.1.1 (Fluxus Engineering Web site). Different weights were assigned to substitutions, as in the study by Richards et al. (1998). Coalescence-age calculations and SDs were estimated following the formulae of Forster et al. (1996) and Saillard et al. (2000).
Results
Saami mtDNA Haplogroup Frequencies
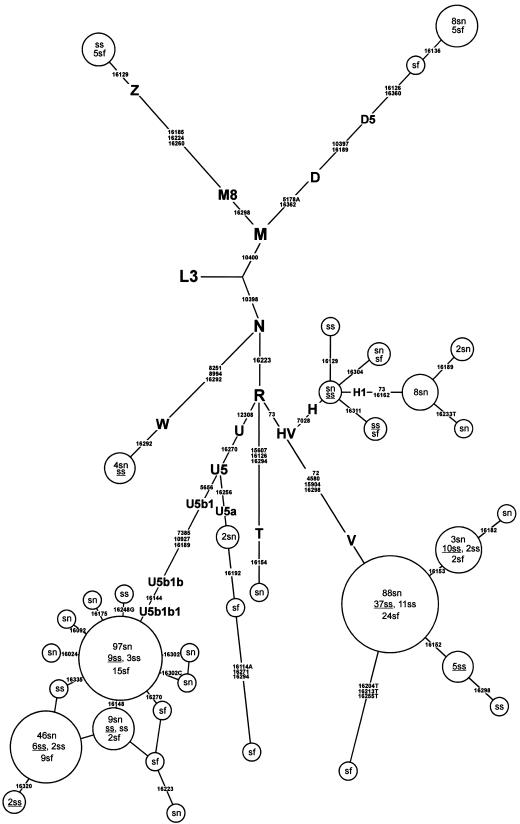
The mtDNA haplogroup frequencies were estimated in Swedish Saami (n=73) and in 8,314 samples from different European and Siberian populations, through use of HVS-I sequencing and RFLP analysis. The combined D-loop and coding region information also enabled us to infer the haplogroup affiliations from previously published data for the Finnish, Swedish, and Norwegian Saami (Sajantila et al. 1995; Dupuy and Olaisen 1996; Delghandi et al. 1998) that has been scored for the presence of the HVS-I polymorphisms (fig. 1; tables 1 and 2).
Figure 1.
Phylogenetic network of 445 Saami mtDNA HVS-I sequences. Only those coding-region markers that have been analyzed in this study are shown (for finer resolution of haplogroup U subbranches, see fig. 3). Observed mutations are numbered according to the revised Cambridge Reference Sequence (Anderson et al. 1981; Andrews et al. 1999). The tree is rooted in haplogroup L3. Variable nps are shown on the links; nucleotide change is specified by suffixes only for transversions. Nodes indicate different haplotypes and have sizes that are proportional to their frequencies. HVS-I sequences are from Sajantila et al. (1995), using the corrections given by Bandelt et al. (2001) (n=114); from Dupuy and Olaisen (1996) (n=197); from Delghandi et al. (1998) (n=61); and from the present study (n=73, underlined samples). The analysis of coding region variation has been performed for only the 73 samples analyzed in the present study; haplogroup affiliations for data published elsewhere are inferred from HVS-I sequences. sf = Saami from Finland, sn = Saami from Norway, ss = Saami from Sweden. Note that the presence of three transversions reported in a single haplotype (sf) from haplogroup V (Sajantila et al. 1995) has not been reconfirmed by an independent study.
The phylogenetic network in figure 1 relates all the HVS-I sequence haplotypes in the three geographic subpopulations of the Saami. It also indicates a limited number of highly frequent haplotypes, which are, in most cases, shared by all analyzed geographic subpopulations (see table 2). Thus, 98% of the Saami mtDNA pool is encompassed within haplogroups V, U5b, H, Z, and D5. The two largest subsets, V and U5b, account for a lion’s share (89%) of the Saami mtDNA pool. The representative proportion of individual haplogroups varies substantially among different Saami subpopulations (table 2). Haplogroup V is by far the most frequent haplogroup in the Swedish Saami and is present at significantly lower frequencies (P<.0001) in Norwegian and Finnish subpopulations. We note that, in all three Saami subpopulations, U5b was virtually the only subclade of the otherwise frequent and divergent western Eurasian haplogroup U5 (Tambets et al. 2003). It is the most frequent haplogroup among Norwegian and Finnish subpopulations (table 2).
The frequencies of the most widely spread haplogroups among the Saami in other analyzed populations are shown in table 1. Both haplogroups V and U5b are spread at moderate frequencies across Europe, from Iberia to the Ural Mountains. In contrast, among 393 Ob-Ugric speakers (including 98 Mansi, published elsewhere by Derbeneva et al. [2002b]) and 388 Samoyeds (including 58 Nenets, published elsewhere by Saillard et al. [2000]), only one haplogroup V–carrying individual was found in the Mansi sample, whereas not a single Saami variant of U5b (U5b1b1) was identified there (table 1). These two variants of maternal lineages are virtually absent in the other 1,199 native Siberians analyzed in this study.
The third-most-frequent haplogroup among the Saami is H, which is present at a frequency of 4%, ∼10-fold lower than that of other North European populations (Richards et al. 2000). The majority of the Saami haplogroup H lineages (61%) contain transitions at nps 73 and 16162. According to information obtained from complete sequences, these lineages belong to subhaplogroup H1, defined by the coding region transition at np 3010 (Finnilä et al. 2001). We notice here that HVS-I haplotypes with a transition at np 16162 in the Volga-Ural area people (Bermisheva et al. 2002) and other European populations studied by us belong exclusively to this branch of subhaplogroup H1 (authors' unpublished data). This variant of mtDNA can be found both in western and eastern Europe, being as frequent in Germanic-speaking Scandinavians and the Germans as among the Norwegian Saami but absent in the Finnish and Swedish Saami (table 2). Though present in several Volga-Finnic populations, H1 haplotypes with mutation 16162 are extremely rare or absent in almost 2,000 Siberian Ugric-, Samoyedic-, and Altaic-speaking people (table 1) as well as in central Asians (Comas et al. 1998; Metspalu et al. 1999)—that is to say that this clade has an overwhelmingly European phylogeographic pattern. Its presence only in the Norwegian Saami sample may have been generated by admixture with the Norwegian population. This inference is supported by the presence of one H1 haplotype, with HVS-I motif 16162–16189, found among the Saami, in the Norwegian sample (Helgason et al. 2001). This haplotype has not been found in any other population analyzed in the present study.
Only a minor portion of the Saami maternal lineages (average ∼5%) that exhibit restricted diversity belong to haplogroups that are characteristic of Asian populations—that is, D5 and Z (table 1). These eastern Eurasian haplogroups are significantly more frequent (P<.05) among the Finnish Saami compared with Norwegian and Swedish Saami samples (table 2). Such fluctuations in haplogroup frequencies could be due to genetic drift or just due to stochastic variation in relatively small samples. At the same time, the similar pattern of mtDNA haplogroup distribution found in different subpopulations of the Saami provides evidence of their common genetic background.
Saami Y-Chromosomal Haplogroup Pattern and Frequencies
The Y-chromosomal variation of 35 Swedish Saami, published by Rosser et al. (2000), was further analyzed to improve the resolution of paternal lineage clusters according to the YCC (2002). For comparison, data from different European and Siberian populations were used (table 3). To include the data published before the high-resolution nomenclature of biallelic markers (YCC 2002) was available, a combination of the results of the analysis of biallelic markers and polymorphisms of STRs was used, when possible (Zerjal et al. 2001), to infer the haplogroup frequencies presented in table 3. In some cases (haplogroups N3 and R1a), the updating of the data was simple and unambiguous. In the case of haplogroups R1b and I, the classification of the samples was more problematic. However, the STR pattern of those haplogroups in combination with the typed biallelic markers is, in most cases, haplogroup-specific enough to make the misclassification of the samples highly unlikely. The probability of misclassification is higher in the data set from Raitio et al. (2001), in which only some informative biallelic markers and no STRs were scored. Therefore, these samples are left at a lower resolution level in table 2. Detailed criteria used in the process of deduction are given in the footnotes of table 3.
Three major haplogroups—N3, I, and R1a—comprise >80% of the Saami Y-chromosomal gene pool (table 2) and appear, likewise, as the major haplogroups in northeastern Europe (table 3). The nearly equal distribution of the Y-chromosomal variants among different Saami populations strongly supports their common paternal history. The comparison of the Y-chromosomal haplogroup frequencies in different subpopulations shows that, similar to mitochondrial haplogroup frequencies, some Y-chromosomal haplogroups (J and R1a) exhibit notable variation between geographic subpopulations of the Saami (table 2).
Haplogroup N3, the most frequent haplogroup in the Saami population, is distributed in eastern European and northern Asian populations but it is rare or absent in western Europe (table 3). All analyzed Swedish Saami N3 lineages fall into subcluster N3a, defined by M178 (YCC 2002). Although N3a is widespread in Siberia, other haplogroups, characteristic of Samoyedic-speaking and other Siberian populations (such as C and Q), are either almost absent in Baltic-Finnic populations, including the Saami, or are only sporadic, as for haplogroup N2, which is found only among Volga region Finnic speakers (table 3).
The second largest haplogroup, I, corresponds to roughly one-third of the Saami Y-chromosomal lineages. It is widespread in Europe but virtually absent in Asian populations (table 3). The defining mutation of haplogroup I, M170, most likely arose in Europe (Semino et al. 2000). It is well dispersed over the continent, as well as among Volga River basin Finnic and Turkic speakers (table 3). At the current level of resolution, the phylogenetic reconstructions do not identify the geographic origin of haplogroup I in Europe. Nevertheless, its virtual absence among Samoyeds as well as among Ugric-speaking Mansis and Khants suggests that these populations have not shared a recent ancestry with the Saami. Indeed, the high frequencies of haplogroup I in the Norwegians, Swedes, Finns, and the Saami (table 3) suggest that haplogroup I represents the heritage from the very first settlers of Fennoscandia. Analyses of microsatellite variation of the Saami haplogroup I Y chromosomes reveals that 9 of 10 Saami M170 chromosomes share the same modal microsatellite haplotype (6/9) or its derivatives with other Nordic haplogroup I chromosomes (authors' unpublished data). The latter corresponds to haplogroup 2 chromosomes with alleles 14-23-10-11-13, defined by DYS19-390-391-392-393, respectively, in the studies by Helgason et al. (2000), Zerjal et al. (2001), Weale et al. (2002), and Capelli et al. (2003). These haplotypes form a subclade of haplogroup I that is different from the variety of equally abundant M170 chromosomes found in the Adriatic coastal region, where the Dinaric Modal Haplotype (16-24-11-11-13) is common (Barac et al. 2003).
Haplogroup R1a, encompassing 11% of the Saami Y-chromosomal gene pool, is a frequent Y-chromosomal variety in eastern Europe, but it is also found in Siberia, the Altai Mountains (table 3), and India (Kivisild et al. 2003). Like haplogroup I, it is absent among Nenets, the largest Samoyed population, living partly in northeast Europe. Haplogroup R1a is present in the Swedish and Kola Saami at frequencies that are comparable to those observed among other Scandinavian and northeastern European populations, but it is relatively rare in Finland, both in the Saami and non-Saami populations (table 3).
Haplogroup R1b is present in the Saami population at a level of 4%. It is the most frequent haplogroup in western Europe, especially among the French, and it is relatively frequent in Scandinavian Germanic-speaking populations, but it has not been found among the Finns. In the Volga-Ural region populations, R1b is present at frequencies similar to those of the Saami (table 3).
Haplogroups J and E, found solely among Kola Saami (table 2), may have arisen as a recent contribution from the neighboring northern Russian population, since these Y-chromosomal variants are present there (Wells et al. 2001).
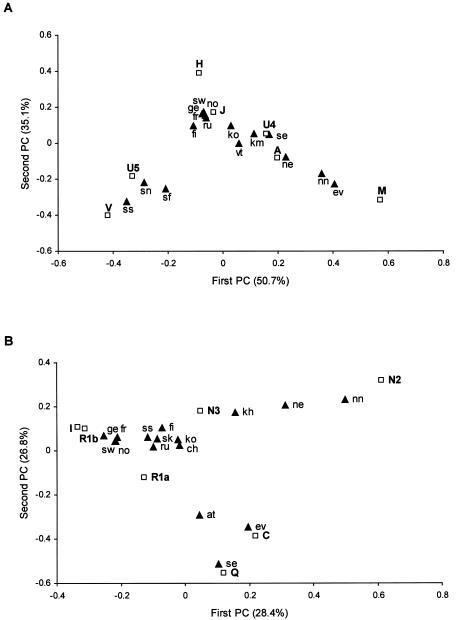
PC Analysis
The results of the PC analysis based on mtDNA haplogroup frequencies (fig. 2A) illustrate why the Saami are identified as mitochondrial genetic “outliers” in Europe. Here, it is the relative haplogroup proportions and not the phylogeographic affiliations that are distinctive. The first PC (fig. 2A) reflects predominantly the differences between frequencies of haplogroups characteristic of the eastern and western parts of Eurasia (H, J, V, and U5 vs. A and M). All Saami subpopulations cluster together as a distinct group, but the distance between the Saami and native Siberians is much greater than between the Saami and other Europeans. Different distributions of the populations along the axis of the second PC arise primarily from the different proportions of haplogroups H, U5, and V in the populations. In addition, the lack of haplogroups J, U4, and A among the Saami plays a relatively important role here.
Figure 2.
PC analysis based on mtDNA (A) and Y-chromosomal (B) haplogroup frequencies in some European and Siberian populations. Only the seven haplogroups (squares) having the main impact on the scatter plot have been used (see the “Subjects and Methods” section). Numbers in parenthesis indicate the proportion of the total genetic information retained by a given PC. The Saami population is subdivided according to the present location of the subpopulations. Population data (triangles) are the same as in tables 1 and 3 and are listed here in alphabetical order as follows: at = Altaians, ch = Chuvashes, ev = Evenks, fi = Finns, fr = French, ge = Germans, kh = Khants, km = Khants and Mansis, ko = Komis, ne = Nenets, nn = Nganasans, no = Norwegians, sf = Saami from Finland, sk = Saami from Kola Peninsula, Russia, sn = Saami from Norway, ss = Saami from Sweden, se = Selkups, sw = Swedes, vt = Volga-Uralic Turkic-speakers (Bashkirs and Chuvashes). Inclusion of all populations listed in tables 1 and 3 did not change the overall outcome (data not shown).
Figure 2B shows a similar analysis undertaken on the Y-chromosomal haplogroup frequencies. Here, the first PC is determined, on the one hand, by the western Eurasian–specific haplogroups I, R1a, and R1b and, on the other hand, by Asian-specific haplogroup C and by the largely Siberian-specific N2, which both separate all Samoyeds and Ob-Ugric-speaking populations (except Selkups) from the Saami. In contrast to that of other Samoyedic-speaking populations, the Selkup Y-chromosomal pool is dominated by eastern Eurasian haplogroup Q, which explains their solitary location on the second dimension of the PC graph. Two Saami subpopulations map close together and are placed in the immediate vicinity of the Finns, Volga-Finnic, and Volga-Turkic populations, as well as the Russians. The latter two are characterized by haplogroup N3 at moderate frequencies (table 3). Furthermore, at this resolution, Germanic-speaking populations (including Scandinavian Norwegians and Swedes) and the French form an additional cluster that is close to other European populations included in the analysis.
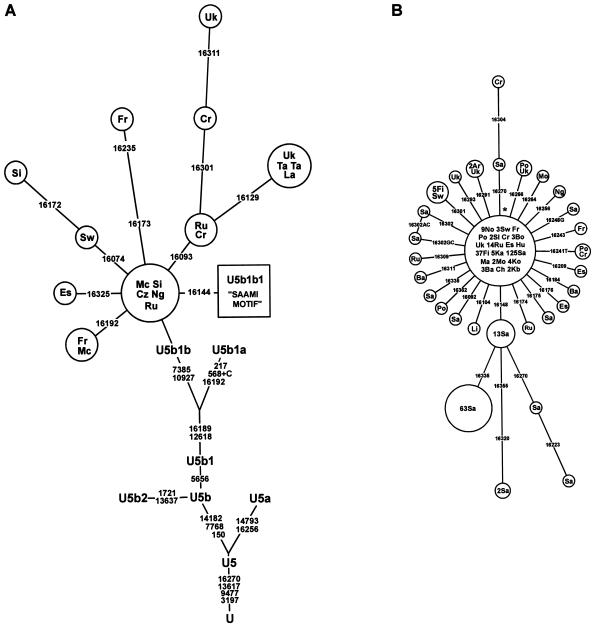
Phylogeography of Haplogroup U5b1b and the “Saami-Specific” Motif (U5b1b1)
Haplogroup U5b is found at low frequencies all over Europe (Richards et al. 1998). A distinct phylogenetic subbranch of U5b carries the “Saami-specific” HVS-I motif defined by 16144-16189-16270 in the study by Sajantila et al. (1995) and has been named “U5b1” by Richards et al. (1998). All U5b sequences share two diagnostic synonymous substitutions at nps 7768 and 14182 (see fig. 3A). The major U5b subset found among the Finns contains a derived G allele at noncoding np 5656 and a synonymous substitution at np 12618 (Finnilä et al. 2001). Through use of complete mitochondrial sequence data, it has previously been demonstrated that the Finnish sequences containing the “Saami-specific” HVS-I motif share an additional synonymous mutation at np 10927, which is often associated with another transition at np 7385 (Finnilä et al. 2001).
Figure 3.
A, Phylogenetic network of U5b1b lineages based on HVS-I sequences and its position in the phylogeny of haplogroup U. Sequence information from Herrnstadt et al. (2002) and Finnilä et al. (2001) has been used for the coding region and HVS-II (see also table 4). The nucleotide positions relative to the revised Cambridge Reference Sequence (Anderson et al. 1981; Andrews et al. 1999), at which two nodes differ, are listed along links. Nucleotide changes are specified by suffixes only for transversions; “+” indicates an insertion. Note that we have redefined subclade U5a of Finnilä et al. (2001) and Herrnstadt et al. (2002) as “U5b2,” on the basis of its position in the phylogenetic tree. U5b1b1 haplotypes are shown as the square labeled as “Saami motif” and are further refined in panel B. B, Phylogenetic network of 330 U5b1b1 lineages based on HVS-I sequences. The star indicates the basal node (transitions in nps 16144, 16189, and 16270). Population sizes and U5b1b1 frequencies are shown in table 1. Ar = Armenians, Ba = Bashkirs, Bo = Bosnians, Ch = Chuvashes, Cr = Croats, Cz = Czechs, Es = Estonians, Fi = Finns, Fr = French, Hu = Hungarians, Ka = Karelians, Kb = Kabardians, Ko = Komis, La = Latvians, Li = Lithuanians, Ma = Maris, Mc = Moroccans, Mo = Mordvin, Ng = Nogays, No = Norwegians, Po = Poles, Sa = Saami, Sl = Slovaks, Ru = Russians, Si = Sicilians, Sw = Swedes, Ta = Tatars, Uk = Ukrainians. For further information, see the legend to figure 1.
To study the phylogeography of the “Saami-specific” branch of U5b, an analysis of np 5656 variation among the relevant genomes was performed. The results reveal (see table 4) that 95 of 160 samples carried the derived 5656G allele and belong, thus, to a subhaplogroup referred to as “U5b1.” We note that this subhaplogroup is different from the U5b1 branch defined by Richards et al. (1998) using HVS-I data alone. Here, “U5b1” refers to a deeper phylogenetic node of U5b (see fig. 3A). The transition at np 7385 was always found to be associated with a transition at np 10927 and is referred to as “U5b1b.” This cluster also includes the “Saami-specific” subclade, which we name here as “U5b1b1” (fig. 3). Notice that the sequence motif 16189-16192-16270 in U5b1 appears to be associated with at least three independent subhaplogroups of U5b: U5b* (5656A), U5b1*/U5b1a (5656G, 7385A, and 10927T), and U5b1b (5656G, 7385G, and 10927C) (fig. 3; table 4). Therefore, reliance on published HVS-I sequences alone, without regard to the relevant coding region information, makes the exploration of the phylogeography of haplogroup U5b1b ambiguous, and these sequences were not included in the analysis.
The 5656G allele, including all major subsets of U5b1, is broadly distributed both in western and eastern Europe (fig. 3A; table 4). The U5b1b subclade is found all over Europe, but it occurs in western and central Europe with notable sequence variation. The haplotype diversity (excluding subclade U5b1b1) is 0.96. We note that U5b1b was also identified in a northwestern African population, among the Moroccans (fig. 3A). In contrast, its diversity in eastern Europe is much lower. There, the haplotype diversity (excluding subclade U5b1b1) is 0.79, whereas most of the U5b1b sequences in eastern Europe belong to the U5b1b1 branch (fig. 3A and 3B). In the Eurasian cohort, U5b1b (other than U5b1b1) is absent from native Siberians and is notably absent from the Ob-Ugric populations and the Samoyeds.
Most importantly, the data indicate that U5b1b1, the only subcluster of U5b in the Saami population, is present outside the Scandinavian-Baltic and the Volga-Uralic regions—namely, in the French, Croatian, Bosnian, Slovenian, Czech, Russian, Ukrainian, Polish, and Hungarian mtDNA pools—and, further, is present even in the Caucasus, among the Nogay, Kabardinian, and Armenian mtDNAs (fig. 3B). Thus, this subhaplogroup is much more widely distributed than was believed previously (Sajantila et al. 1995). Two principal haplotypes, differing by one mutational step, define the Saami U5b1b1 (fig. 3B). One, with transitions at nps 16144, 16189, and 16270, can be identified as the founder haplotype because of its presence in many other populations. The second, containing an additional transition at np 16148, is so far exclusive to the Saami population. This subfounder comprises 38% of their U5b1b1 mtDNAs and is present in all studied subpopulations of the Saami. Furthermore, no other derivatives of the U5b1b1 founder node present in the Saami sample was identified in other Scandinavians, including the Finns, irrespective of the previous observation that the northern-central Finnish population is relatively rich in U5b1b1 (Meinilä et al. 2001). Thus, the “leakage” of the Saami U5b1b1 to neighboring populations seems to be rather limited. The coalescence time of all non-Saami U5b1b1 lineages in Europe (see fig. 3B) is 4,300±1,400 years BP (table 5). This is approximately the same as that for the eastern or western-southern European subsets but is older than that observed for Scandinavians.
Table 5.
Coalescence Ages for Different Subsets of Subhaplogroup U5b1b1
| Set of Sequences Considereda | n | ρb | Tc(years) | ΔTd(years) |
| U5b1b1 (without Sa) | 118 | 25/118 = .212 | 4,300 | 1,400 |
| U5b1b1 in Scandinavia (without Sa) | 55 | 6/55 = .109 | 2,200 | 2,200 |
| U5b1b1 in Scandinavia (with Sa, without subfounder 16148) | 187 | 11/187 = .059 | 1,200 | 700 |
| U5b1b1 in eastern Europe (Es/Li/Ma/Mo/Ko/Ru/Uk/Po/Ba/Ch) | 47 | 13/47 = .277 | 5,600 | 1,700 |
| U5b1b1 in southern and western Europe (Bo/Cr/Sl/Hu/Fr) | 11 | 3/11 = .273 | 5,500 | 3,200 |
Population codes are the same as those used in figure 3.
Average mutational distance to the founder haplotype of the cluster.
Coalescence time, calculated by means of ρ with a mutation rate of 1 transition per 20,180 years in the HVS-I segment between nps 16090 and 16365 (Forster et al. 1996).
SD for ρ, calculated as in the study by Saillard et al. (2000).
Discussion
Samoyed/Siberian Heritage of the Saami Population?
Uralic-speaking Samoyeds, Khants, Mansis, and Altaic-speaking Siberians virtually lack the European mtDNA haplogroups V and U5b1b1 that predominate in the Saami mtDNA pool (fig. 1; table 1). Eastern Eurasian mtDNA variants in the Saami are represented by a restricted set of lineages that belong to superhaplogroup M. In this respect, the Saami do not differ markedly from Finnic-speaking Karelians, Maris, Komis, Udmurts, or northern Russians, all of whom possess haplogroups of eastern Eurasian origin at similar frequencies (table 1). This minor part of the Saami mtDNA pool consists of two branches of the eastern Eurasian mtDNA tree—D5 and Z1. According to published data, the frequency of haplogroup D5 is relatively high in China (Yao et al. 2002). D5 is also present among Mongols and Siberians (Kolman et al. 1996; Derbeneva et al. 2002b). However, the Saami haplogroup D5 lineages, with the HVS-I motif 16126-16136-16360 and its derivatives (defined as “D5b” by Derenko et al. 2003), have been identified only in some northern and eastern European populations (among Karelians, Finns, Estonians, North-Russians, and Komis) and in some Siberian populations but not in Samoyeds (table 1). This suggests, again, the lack of gene flow from Samoyeds to eastern Europe.
Haplogroup Z, a subcluster of the M8 clade within the haplogroup M family of mtDNA (Kivisild et al. 2002), is found at highest frequencies in the northeastern Asians: the Itelmens and Koryaks (Schurr et al. 1999). It is also present in several Siberian populations, including the Altaic people (table 1). Though not identified in a large data set of the Yakuts (Fedorova et al. 2003; Pakendorf et al. 2003), haplogroup Z has been observed among several Finnic- and Turkic-speaking populations of the Volga-Ural region (Bermisheva et al. 2002). It is curious that it is more frequent there in Finnic- than in Turkic-speaking populations. The absence of haplogroup Z from most of the Siberian Uralic-speaking populations (Samoyedic-speaking Nenets and Selkups, as well as Siberian Ob-Ugric-speaking Khants and Mansis) (table 1) is therefore striking. We note that all haplogroup Z lineages that are found in eastern Europe belong to a subhaplogroup Z1, characterized by transitions at nps 151, 10325, and 16129 within the Z phylogeny (Kong et al. 2003a, 2003b). A matching HVS-I founder haplotype has been observed in the Koryak and the Itelmen populations (Schurr et al. 1999). The limited diversity of haplogroup Z in Europe suggests its relatively recent spread west of the Urals.
The sister clade of Z—haplogroup C—is far more diverse and frequent than haplogroup Z in eastern Eurasians (Derenko et al. 2003), as well as in the populations of the Volga-Ural region (Bermisheva et al. 2002), but it is absent among the Saami. Here, again, the lack of haplogroup C in the Saami is specifically indicative of the absence of possible genetic links between the Saami and the Samoyeds. Indeed, the only Samoyedic-speaking population that also inhabits northeastern Europe and is thus geographically close to the Saami, the Nenets, has haplogroup C as the most frequent (∼30% frequency) variant of mtDNA.
The predominant Saami Y-chromosomal haplogroup N3 has a nearly uniform circumarctic distribution in Eurasia (table 3). The closely related N2 lineages are frequent in Siberian and Volga-Uralic populations. Thus, it is likely that haplogroup N variation represents a prehistoric link between the Siberian and eastern European/proto-Finnic populations via their paternal heritage. The improved resolution of the Y-chromosomal phylogenetic tree (Jobling and Tyler-Smith 2003) reveals an ancestral node shared by haplogroups N and O, with the latter restricted largely to eastern Asia. This connection is intriguing, but it is still unclear when and where this common ancestor first appeared. Nevertheless, one does not need to postulate a recent Siberian flow of Y chromosomes into the Saami gene pool to explain their high N3 frequency. First, such a flow from Samoyedic-speaking aboriginal Siberians to the Saami Y-chromosomal pool would predict the presence there of haplogroup N2 and/or haplogroup Q, widely spread in Samoyeds (Karafet et al. 2002). Second, the much higher diversity of N3 in eastern Europe than in Siberia (Villems et al. 1998; Rootsi et al. 2000) suggests that eastern Europe, rather than Siberia, is a possible origin of the earliest expansion of this haplogroup in northern Eurasia. Third, the lack of Y-chromosomal haplogroup C in Saami contrasts with its high frequency among Tungusic-speaking native Siberians (such as the Evenks and the Evens) as well as among Mongolic-speaking Mongols, the Buryats, the Kalmyks, and the Turkic-speaking Kazakhs and Uzbeks (Wells et al. 2001; Karafet et al. 2002; authors' unpublished data). Therefore, without introducing specific additional ad hoc scenarios, these observations make it unlikely that there was recent Y-chromosomal flow from these Siberian populations into the gene pool of the Saami.
Origins of the Saami mtDNA and Y Chromosomes
Scenarios involving extremes of genetic drift, such as that due to repeated bottlenecks, could explain how the Saami mtDNA pool evolved as a narrow subset of that found in other European populations. Indeed, there are good reasons to believe that even a much larger Finnish population went through several “bottlenecks” in its demographic history (Nevanlinna 1972; de la Chapelle and Wright 1998; Kittles et al. 1999; Peltonen et al. 2000). Likewise, it is possible that, when the proto-Saami gene pool was in statu nascendi, it was restricted to only a few basic mtDNA haplotypes that were carried by the founding settlers—a plausible scenario during the “Paleolithic isolation.”
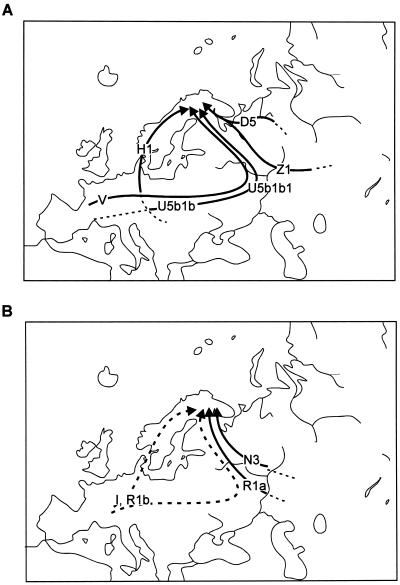
Torroni et al. (1998, 2001) have suggested that the spread of haplogroup V in Scandinavia and in eastern Europe is due to its late Pleistocene/early Holocene expansion from a Franco-Cantabrian glacial refugium. Haplogroup V shows high diversity and the presence of pre-V lineages in Iberia and the islands of Croatia (Tolk et al. 2000; Torroni et al. 2001). However, the new data on Volga River basin and Slavic-speaking populations (Bermisheva et al. 2002; Malyarchuk et al. 2002; present study) show that haplogroup V is also well present in eastern Europeans. Furthermore, haplogroup V lineages with HVS-I transitions 16153 and 16298 (fig. 1) that are frequent in the Saami population are much more widespread in eastern than in western Europe (Torroni et al. 2001; Bermisheva et al. 2002; authors' unpublished data). This indicates that haplogroup V might have reached Fennoscandia via central/eastern Europe (see fig. 4A). Such a scenario is indirectly supported by the absence, among the Saami, of the pre-V mtDNAs that are characteristic of southwestern Europeans and northwestern Africans and that are also present in Germans (Torroni et al. 2001).
Figure 4.
Schematic reconstruction of possible entry routes of the predominant Saami maternal (A) and paternal (B) lineages to Fennoscandia. Broken lines indicate that the exact place of origin/route of spread of the haplogroup is unsolved/not indicated.
The phylogeography and the ancestry of the other components of the Saami mitochondrial profile have so far not been well understood. Here, we showed that mtDNA haplogroup U5b1b1—the set of lineages with the so-called “Saami-specific” motif—is spread, besides among the Saami, mostly in eastern Europe (fig. 3B). This might suggest that haplogroup U5b1b1 may have spread/arisen from eastern Europe. On the other hand, the considerable diversity of the U5b1b cluster in western and southern Europe suggests that these regions, rather than eastern Europe, were the likely place of origin of U5b1b. Thus, the distribution of U5b1b is similar to that suggested above for haplogroup V. Notice that this cluster, like haplogroup V, is also found in northwestern Africa (table 4). Hence, we envision an initial diversification of U5b1b in western Europe, followed by the spread of a particular subhaplogroup in eastern Europe, finally reaching Fennoscandia (but not the Samoyeds or any other aboriginal Siberians) via an eastern route (fig. 4A). Indeed, U5b1b1 is absent from a large sample set of Germans from Lower Saxony (Pfeiffer et al. 2001), and it is detected only in trace frequencies in other western European populations. That makes it less likely that U5b1b1 entered the Saami mtDNA pool (or that the proto-Saami tribes carried it) directly from the west. The wide geographic distribution of both U5b1b1 and U5b1b in western Eurasian populations and the apparent absence of U5b1b “twigs” (except of U5b1b1) in Finno-Ugric speakers suggests that the latter may have originated before the differentiation of the European Finnic-speaking people.
Haplogroup H1 lineages most probably spread to Fennoscandia via a western route (fig. 4A). The diversity of H1 is relatively higher in Norwegian (0.76), Swedish (0.78), German (0.78), and Polish (0.80) populations than in Finnish (0.09), Estonian (0.35), Latvian (0.50), Karelian (0.0), and Volga-Uralic populations (highest among Komis: 0.40). Furthermore, as was mentioned above, the Saami may have obtained their H1 lineages from recent admixture with the Norwegians.
The two eastern Eurasian mtDNA variants, haplogroup Z1 and the particular subbranch of D5, have probably reached northeastern Europe not via the Arctic but via a more southern route across the southern Urals and, passing the Volga River basin (fig. 4A), left their “traces” among the gene pools of the Volga-Ural peoples, although not, as we have already stressed above, among Ob-Ugric and Nenets populations (table 1). Here, history and archeology provide several possible scenarios: not only events of historic times, like migrations of Huns, Avars, and Mongols, but also a likely influx of Asian tribes to eastern Europe during the early Holocene and contributing to the Kama culture of the upper Volga and Petchora basins (Kozlowski and Bandi 1984), could have been behind movements that brought a few selected and specific eastern Asian mtDNA variants to Fennoscandia. Note that northern Fennoscandia became accessible to humans at the very end of the Pleistocene/early Holocene, both from the west and the east (Donner 1995).
The Y-chromosomal haplogroups N3 and R1a, which make up ∼60% of the Saami Y-chromosomal variants, have likely reached Fennoscandia from eastern Europe (fig. 4B), where these haplogroups can be found in high frequencies, as among the Saami. Haplogroups I and R1b, which together make up a third of Saami Y chromosomes, seem to have arisen in western Europe (fig. 4B). For R1b, this scenario is most plausible because it is a characteristically frequent Y-chromosomal variant in western Europe (table 3). In the case of haplogroup I, a specific pattern of STR variation in the Saami is close to that observed among other Scandinavians and western Europeans and is dissimilar to that observed in southern Europe.
We conclude that the phylogeography of mtDNA and Y-chromosome variants that correspond to the maternal and paternal gene pools of the Saami does not provide any evidence for the Saami population arising among the northernmost Uralic-speaking populations—Siberian Ugric and Samoyedic speakers—or among any other aboriginal Siberians. The Samoyeds are the least genetically close to the Saami among the people of the Uralic language family, whereas nearly all of the mtDNA and Y-chromosomal heritage of the Saami can be adequately explained within the European pools of the two haploid genetic systems. This genetics-based reconstruction (fig. 4) is in agreement with the reconstruction of the spread of Ahrensburgian and Swiderian Mesolithic technologies in northern Europe, linking it with population expansion that can be likely traced back to the post–Last Glacial Maximum recolonization of the European north (Torroni et al. 2001; Tambets et al. 2003). The results also stress that the grouping of populations according to language families should be used exclusively only in a linguistic context.
Acknowledgments
We thank Tatyana Karafet and Boris Malyarchuk, for useful information; Henry Harpending, for the program POPSTR; Vincent Macaulay, for the program SAMPLING; Ille Hilpus and Jaan Lind, for technical assistance; and Charles Kurland and Thomas Gilbert, for helpful discussion and comments. We are grateful to two anonymous reviewers for their suggestions and advice. The research of R.V. was supported by Estonian basic research grant 514 and European Commission Directorate General Research grant ICA1CT20070006. The research of T.K. was supported by Estonian basic research grant 5574. The work of E.K. was supported by the Russian Foundation for Basic Research (project number 01-04-48487a) and the Ministry of Sciences and Technology of Russia. M.G., S.Z., and L.O. received support from expedition grants from the Siberian Branch of the Russian Academy of Sciences (1992–1997) and the Russian Foundation of Basic Research (project number 02-06-80524-a), and the research of P.R. received support from project number 0196005 of the Ministry of Science and Technology of the Republic of Croatia.
Electronic-Database Information
The URL for data presented herein is as follows:
- Fluxus Engineering, http://www.fluxus-engineering.com/ (for Network 3.1.1.1)
References
- Akey JM, Sosnoski D, Parra E, Dios S, Hiester K, Su B, Bonilla C, Jin L, Shriver MD (2001) Melting curve analysis of SNPs (McSNP): a gel-free and inexpensive approach for SNP genotyping. Biotechniques 30:358–362, 364, 366–367 [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Baasner A, Schafer C, Junge A, Madea B (1998) Polymorphic sites in human mitochondrial DNA control region sequences: population data and maternal inheritance. Forensic Sci Int 98:169–178 10.1016/S0379-0738(98)00163-7 [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Lahermo P, Richards M, Macaulay V (2001) Detecting errors in mtDNA data by phylogenetic analysis. Int J Legal Med 115:64–69 10.1007/s004140100228 [DOI] [PubMed] [Google Scholar]
- Barac L, Pericic M, Klaric IM, Rootsi S, Janicijevic B, Kivisild T, Parik J, Rudan I, Villems R, Rudan P (2003) Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet 11:535–542 10.1038/sj.ejhg.5200992 [DOI] [PubMed] [Google Scholar]
- Beckman G, Beckman L, Sikstrom C (1993) Serum complement (C3, BF, C4) types in Swedish Saamis. Hum Hered 43:362–365 [DOI] [PubMed] [Google Scholar]
- Beckman L, Beckman G, Nylander PO (1988) Gc subtypes in Finns, Swedes and Swedish Lapps. Hum Hered 38:18–21 [DOI] [PubMed] [Google Scholar]
- Bermisheva M, Tambets K, Villems R, Khusnutdinova E (2002) [Diversity of mitochondrial DNA haplotypes in ethnic populations of the Volga-Ural region of Russia.] Mol Biol (Mosk) 36:990–1001 [PubMed] [Google Scholar]
- Bertranpetit J, Sala J, Calafell F, Underhill PA, Moral P, Comas D (1995) Human mitochondrial DNA variation and the origin of Basques. Ann Hum Genet 59:63–81 [DOI] [PubMed] [Google Scholar]
- Cali F, Le Roux MG, D’Anna R, Flugy A, De Leo G, Chiavetta V, Ayala GF, Romano V (2001) mtDNA control region and RFLP data for Sicily and France. Int J Legal Med 114:229–231 10.1007/s004140000169 [DOI] [PubMed] [Google Scholar]
- Capelli C, Redhead N, Abernethy JK, Gratrix F, Wilson JF, Moen T, Hervig T, Richards M, Stumpf MP, Underhill PA, Bradshaw P, Shaha A, Thomas MG, Bradman N, Goldstein DB (2003) A Y chromosome census of the British isles. Curr Biol 13:979–984 10.1016/S0960-9822(03)00373-7 [DOI] [PubMed] [Google Scholar]
- Casanova M, Leroy P, Boucekkine C, Weissenbach J, Bishop C, Fellous M, Purrello M, Fiori G, Siniscalco M (1985) A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 230:1403–1406 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Cavalli-Sforza LL, Piazza A (1993) Human genomic diversity in Europe: a summary of recent research and prospects for the future. Eur J Hum Genet 1:3–18 [DOI] [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bosch E, Martinez-Arias R, Clarimon J, Facchini F, Fiori G, Luiselli D, Pettener D, Bertranpetit J (1998) Trading genes along the silk road: mtDNA sequences and the origin of Central Asian populations. Am J Hum Genet 63:1824–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corte-Real HB, Macaulay VA, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit J, Sykes BC (1996) Genetic diversity in the Iberian Peninsula determined from mitochondrial sequence analysis. Ann Hum Genet 60:331–350 [DOI] [PubMed] [Google Scholar]
- Crespillo M, Luque JA, Paredes M, Fernandez R, Ramirez E, Valverde JL (2000) Mitochondrial DNA sequences for 118 individuals from northeastern Spain. Int J Legal Med 114:130–132 10.1007/s004140000158 [DOI] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danan C, Sternberg D, Van Steirteghem A, Cazeneuve C, Duquesnoy P, Besmond C, Goossens M, Lissens W, Amselem S (1999) Evaluation of parental mitochondrial inheritance in neonates born after intracytoplasmic sperm injection. Am J Hum Genet 65:463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Chapelle A, Wright FA (1998) Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc Natl Acad Sci USA 95:12416–12423. 10.1073/pnas.95.21.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delghandi M, Utsi E, Krauss S (1998) Saami mitochondrial DNA reveals deep maternal lineage clusters. Hum Hered 48:108–114 10.1159/000022789 [DOI] [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaya EB, Volod’ko NV, Wallace DC, Sukernik RI (2002a) [Mitochondrial DNA variation in Kets and Nganasans and the early peoples of Northern Eurasia]. Genetika 38:1554–1560 [PubMed] [Google Scholar]
- Derbeneva OA, Starikovskaya EB, Wallace DC, Sukernik RI (2002b) Traces of early Eurasians in the Mansi of northwest Siberia revealed by mitochondrial DNA analysis. Am J Hum Genet 70:1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenko MV, Grzybowski T, Malyarchuk BA, Dambueva IK, Denisova GA, Czarny J, Dorzhu CM, Kakpakov VT, Miscicka-Sliwka D, Wozniak M, Zakharov IA (2003) Diversity of mitochondrial DNA lineages in south Siberia. Ann Hum Genet 67:391–411 10.1046/j.1469-1809.2003.00035.x [DOI] [PubMed] [Google Scholar]
- Dimo-Simonin N, Grange F, Taroni F, Brandt-Casadevall C, Mangin P (2000) Forensic evaluation of mtDNA in a population from south west Switzerland. Int J Legal Med 113:89–97 [DOI] [PubMed] [Google Scholar]
- Di Rienzo A, Wilson AC (1991) Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci USA 88:1597–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner J (1995) The quaternary history of Scandinavia. Cambridge University Press, Cambridge [Google Scholar]
- Dupuy BM, Olaisen B (1996) mtDNA sequences in the Norwegian Saami and main population. In: Carracedo A, Brinkmann B, Bär W (eds) Advances in forensic haemogenetics, vol 6. Springer-Verlag, Berlin, Heidelberg, New York, pp 23–25 [Google Scholar]
- Fedorova SA, Bermisheva MA, Villems R, Maksimova NR, Khusnutdinova EK (2003) [Analysis of mitochondrial DNA lineages in Yakuts.] Mol Biol (Mosk) 37:643–653 [PubMed] [Google Scholar]
- Finnilä S, Hassinen IE, Ala-Kokko L, Majamaa K (2000) Phylogenetic network of the mtDNA haplogroup U in northern Finland based on sequence analysis of the complete coding region by conformation-sensitive gel electrophoresis. Am J Hum Genet 66:1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Cali F, Rohl A, Metspalu E, D’Anna R, Mirisola M, De Leo G, Flugy A, Salerno A, Ayala G, Kouvatsi A, Villems R, Romano V (2002) Continental and subcontinental distributions of mtDNA control region types. Int J Legal Med 116:99–108 10.1007/s00414-001-0261-z [DOI] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J (1996) Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet 59:935–945 [PMC free article] [PubMed] [Google Scholar]
- Francalacci P, Bertranpetit J, Calafell F, Underhill PA (1996) Sequence diversity of the control region of mitochondrial DNA in Tuscany and its implications for the peopling of Europe. Am J Phys Anthropol 100:443–460 [DOI] [PubMed] [Google Scholar]
- Guglielmino CR, Piazza A, Menozzi P, Cavalli-Sforza LL (1990) Uralic genes in Europe. Am J Phys Anthropol 83:57–68 [DOI] [PubMed] [Google Scholar]
- Haetta OM (1996) The Sami: an indigenous people of the Arctic. Davvi Girji, Kárásjohka/Karasjoki, Vaasa [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Hickey E, Goodacre S, Bosnes V, Stefansson K, Ward R, Sykes B (2001) mtDNA and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry. Am J Hum Genet 68:723–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigurethardottir S, Nicholson J, Sykes B, Hill EW, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefansson K (2000) Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet 67:697–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JL, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N (2002) Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet 70:1152–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Jaksch M, Bezold R, Mertens S, Aholt S, Paprotta A, Gerbitz KD (1997) Population genetics and disease susceptibility: characterization of central European haplogroups by mtDNA gene mutations, correlation with D loop variants and association with disease. Hum Mol Genet 6:1835–1846 10.1093/hmg/6.11.1835 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Heissig F, von Haeseler A, Pääbo S (1999) DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nat Genet 22:78–81 10.1038/8785 [DOI] [PubMed] [Google Scholar]
- Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF (2002) High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol 74:761–789 [DOI] [PubMed] [Google Scholar]
- Kittles RA, Bergen AW, Urbanek M, Virkkunen M, Linnoila M, Goldman D, Long JC (1999) Autosomal, mitochondrial, and Y chromosome DNA variation in Finland: evidence for a male-specific bottleneck. Am J Phys Anthropol 108:381–399 [DOI] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Golge M, Usanga E, Papiha SS, Cinnioglu C, King R, Cavalli-Sforza L, Underhill PA, Villems R (2003) The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet 72:313–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Tolk H-V, Parik J, Wang Y, Papiha SS, Bandelt H-J, Villems R (2002) The emerging limbs and twigs of the East Asian mtDNA tree. Mol Biol Evol 19:1737–1751 [DOI] [PubMed] [Google Scholar]
- Kolman C, Sambuughin N, Bermingham E (1996) Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics 142:1321–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong QP, Yao YG, Liu M, Shen SP, Chen C, Zhu CL, Palanichamy MG, Zhang YP (2003a) Mitochondrial DNA sequence polymorphisms of five ethnic populations from northern China. Hum Genet 113:391–405 [DOI] [PubMed] [Google Scholar]
- Kong QP, Yao YG, Sun C, Bandelt HJ, Zhu CL, Zhang YP (2003b) Phylogeny of East Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet 73:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski J, Bandi HG (1984) The paleohistory of circumpolar arctic colonization. Arctic 37:359–372 [Google Scholar]
- Lahermo P, Sajantila A, Sistonen P, Lukka M, Aula P, Peltonen L, Savontaus ML (1996) The genetic relationship between the Finns and the Finnish Saami (Lapps): analysis of nuclear DNA and mtDNA. Am J Hum Genet 58:1309–1322 [PMC free article] [PubMed] [Google Scholar]
- Laitinen V, Lahermo P, Sistonen P, Savontaus ML (2002) Y-chromosomal diversity suggests that Baltic males share common Finno-Ugric-speaking forefathers. Hum Hered 53:68–78 10.1159/000057985 [DOI] [PubMed] [Google Scholar]
- Lutz S, Weisser HJ, Heizmann J, Pollak S (1998) Location and frequency of polymorphic positions in the mtDNA control region of individuals from Germany. Int J Legal Med 111:67–77 10.1007/s004140050117 [DOI] [PubMed] [Google Scholar]
- Macaulay VA, Richards MB, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A (1999) The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk BA, Derenko MV (2001) Mitochondrial DNA variability in Russians and Ukrainians: implications to the origin of the Eastern Slavs. Ann Hum Genet 65:63–78 10.1046/j.1469-1809.2001.6510063.x [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Grzybowski T, Derenko MV, Czarny J, Drobnic K, Miscicka-Sliwka D (2003) Mitochondrial DNA variability in Bosnians and Slovenians. Ann Hum Genet 67:412–425 10.1046/j.1469-1809.2003.00042.x [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Grzybowski T, Derenko MV, Czarny J, Wozniak M, Miscicka-Sliwka D (2002) Mitochondrial DNA variability in Poles and Russians. Ann Hum Genet 66:261–283 10.1046/j.1469-1809.2002.00116.x [DOI] [PubMed] [Google Scholar]
- Mathias N, Bayes M, Tyler-Smith C (1994) Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet 3:115–123 [DOI] [PubMed] [Google Scholar]
- Meinilä M, Finnilä S, Majamaa K (2001) Evidence for mtDNA admixture between the Finns and the Saami. Hum Hered 52:160–170 10.1159/000053372 [DOI] [PubMed] [Google Scholar]
- Metspalu E, Kivisild T, Kaldma K, Parik J, Reidla M, Tambets K, Villems R (1999) The trans-Caucasus and the expansion of the Caucasoid-specific human mitochondrial DNA. In: Papiha S, Deka R, Chakraborty R (eds) Genomic diversity: application in human population genetics. Kluwer Academic/Plenum Publishers, New York, pp 121–134 [Google Scholar]
- Mogentale-Profizi N, Chollet L, Stevanovitch A, Dubut V, Poggi C, Pradie MP, Spadoni JL, Gilles A, Beraud-Colomb E (2001) Mitochondrial DNA sequence diversity in two groups of Italian Veneto speakers from Veneto. Ann Hum Genet 65:153–166 10.1046/j.1469-1809.2001.6520153.x [DOI] [PubMed] [Google Scholar]
- Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York, pp 145–163 [Google Scholar]
- Nevanlinna HR (1972) The Finnish population structure. A genetic and genealogical study. Hereditas 71:195–236 [DOI] [PubMed] [Google Scholar]
- Nygaard S (1989) The stone age of Northern Scandinavia: a review. J World Prehistory 3:71–116 [Google Scholar]
- Opdal SH, Rognum TO, Vege A, Stave AK, Dupuy BM, Egeland T (1998) Increased number of substitutions in the D-loop of mitochondrial DNA in the sudden infant death syndrome. Acta Paediatr 87:1039–1044 10.1080/080352598750031347 [DOI] [PubMed] [Google Scholar]
- Orekhov V, Poltoraus A, Zhivotovsky LA, Spitsyn V, Ivanov P, Yankovsky N (1999) Mitochondrial DNA sequence diversity in Russians. FEBS Letters 445:197–201 10.1016/S0014-5793(99)00115-5 [DOI] [PubMed] [Google Scholar]
- Pakendorf B, Wiebe V, Tarskaia LA, Spitsyn VA, Soodyall H, Rodewald A, Stoneking M (2003) Mitochondrial DNA evidence for admixed origins of central Siberian populations. Am J Phys Anthropol 120:211–224 10.1002/ajpa.10145 [DOI] [PubMed] [Google Scholar]
- Parson W, Parsons TJ, Scheithauer R, Holland MM (1998) Population data for 101 Austrian Caucasian mitochondrial DNA d-loop sequences: application of mtDNA sequence analysis to a forensic case. Int J Legal Med 111:124–132 10.1007/s004140050132 [DOI] [PubMed] [Google Scholar]
- Passarino G, Cavalleri GL, Lin AA, Cavalli-Sforza LL, Borresen-Dale AL, Underhill PA (2002) Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms. Eur J Hum Genet 10:521–529 10.1038/sj.ejhg.5200834 [DOI] [PubMed] [Google Scholar]
- Peltonen L, Palotie A, Lange K (2000) Use of population isolates for mapping complex traits. Nat Rev Genet 1:182–190 10.1038/35042049 [DOI] [PubMed] [Google Scholar]
- Pereira L, Prata MJ, Amorim A (2000) Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation. Ann Hum Genet 64:491–506 10.1046/j.1469-1809.2000.6460491.x [DOI] [PubMed] [Google Scholar]
- Pfeiffer H, Brinkmann B, Huhne J, Rolf B, Morris AA, Steighner R, Holland MM, Forster P (1999) Expanding the forensic German mitochondrial DNA control region database: genetic diversity as a function of sample size and microgeography. Int J Legal Med 112:291–298 10.1007/s004140050252 [DOI] [PubMed] [Google Scholar]
- Pfeiffer H, Forster P, Ortmann C, Brinkmann B (2001) The results of an mtDNA study of 1,200 inhabitants of a German village in comparison to other Caucasian databases and its relevance for forensic casework. Int J Legal Med 114:169–172 10.1007/s004140000165 [DOI] [PubMed] [Google Scholar]
- Piercy R, Sullivan KM, Benson N, Gill P (1993) The application of mitochondrial DNA typing to the study of white Caucasian genetic identification. Int J Legal Med 106:85–90 [DOI] [PubMed] [Google Scholar]
- Pult I, Sajantila A, Simanainen J, Georgiev O, Schaffner W, Pääbo S (1994) Mitochondrial DNA sequences from Switzerland reveal striking homogeneity of European populations. Biol Chem Hoppe Seyler 375:837–840 [PubMed] [Google Scholar]
- Puzyrev VP, Stepanov VA, Golubenko MV, Puzyrev KV, Maximova NR, Kharkov VN, Spiridonova MG, Nogovitsyna AN (2003) mtDNA and Y-chromosome lineages in the Yakut population. Genetika 39:975–981 [PubMed] [Google Scholar]
- Raitio M, Lindroos K, Laukkanen M, Pastinen T, Sistonen P, Sajantila A, Syvanen A (2001) Y-chromosomal SNPs in Finno-Ugric-speaking populations analyzed by minisequencing on microarrays. Genome Res 11:471–482 10.1101/gr.156301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Corte-Real H, Forster P, Macaulay V, Wilkinson-Herbots H, Demaine A, Papiha S, Hedges R, Bandelt HJ, Sykes B (1996) Paleolithic and Neolithic lineages in the European mitochondrial gene pool. Am J Hum Genet 59:185–203 [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Torroni A, Bandelt HJ (2002) In search of geographical patterns in European mitochondrial DNA. Am J Hum Genet 71:1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards MB, Macaulay VA, Bandelt H-J, Sykes BC (1998) Phylogeography of mitochondrial DNA in western Europe. Ann Hum Genet 62:241–260 10.1046/j.1469-1809.1998.6230241.x [DOI] [PubMed] [Google Scholar]
- Rootsi S, Kivisild T, Tambets K, Adojaan M, Parik J, Reidla M, Metspalu E, Laos S, Tolk HV, Villems R (2000) On the phylogeographic context of sex-specific genetic markers of Finno-Ugric populations. In: Künnap A (ed) The roots of peoples and languages of Northern Eurasia II and III. University of Tartu, Division of Uralic Languages/Societas Historiae Fenno-Ugricae, Tartu, pp 148–164 [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousselet F, Mangin P (1998) Mitochondrial DNA polymorphisms: a study of 50 French Caucasian individuals and application to forensic casework. Int J Legal Med 111:292–298 10.1007/s004140050174 [DOI] [PubMed] [Google Scholar]
- Saillard J, Evseva I, Tranebjaerg L, Norby S (2000) Mitochondrial DNA diversity among Nenets. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research Monograph Series, Cambridge University Press, Cambridge, pp 255–258 [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S (2000) mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet 67:718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Gedde-Dahl T, Issel-Tarver L, DiRienzo A, Pääbo S (1995) Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res 5:42–52 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Pääbo S (1995) Language replacement in Scandinavia. Nat Genet 11:359–360 [DOI] [PubMed] [Google Scholar]
- Sajantila A, Salem AH, Savolainen P, Bauer K, Gierig C, Pääbo S (1996) Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci USA 93:12035–12039 10.1073/pnas.93.21.12035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A (1998) mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet 6:365–375 10.1038/sj.ejhg.5200202 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sammallahti P (1998) The Saami languages: an introduction. Davvi Girji, Kárásjohka/Karasjoki, Vaasa [Google Scholar]
- Sanchez JJ, Børsting C, Hallenberg C, Buchard A, Hernandez A, Morling N (2003) Multiplex PCR and minisequencing of SNPs—a model with 35 Y chromosome SNPs. Forensic Sci Int 137:74–84 10.1016/S0379-0738(03)00299-8 [DOI] [PubMed] [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC (1999) Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea-Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–39 [DOI] [PubMed] [Google Scholar]
- Seielstad M, Yuldasheva N, Singh N, Underhill P, Oefner P, Shen P, Wells RS (2003) A novel Y-chromosome variant puts an upper limit on the timing of first entry into the Americas. Am J Hum Genet 73:700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Sumkin VJ (1990) On the ethnogenesis of the Saami: an archaeological view. Acta Borealia 7:3–20 [Google Scholar]
- Tagliabracci A, Turchi C, Buscemi L, Sassaroli C (2001) Polymorphism of the mitochondrial DNA control region in Italians. Int J Legal Med 114:224–228 10.1007/s004140000168 [DOI] [PubMed] [Google Scholar]
- Tambets K, Kivisild T, Metspalu E, Parik J, Kaldma K, Laos S, Tolk HV, Gölge M, Demirtas H, Geberhiwot T, Papiha SS, De Stefano GF, Villems R (2000) The topology of the maternal lineages of the Anatolian and Trans-Caucasus populations and the peopling of the Europe: some preliminary considerations. In: Renfrew C, Boyle K (eds) Archaeogenetics: DNA and the population prehistory of Europe. McDonald Institute for Archaeological Research Monograph Series, Cambridge University Press, Cambridge, pp 219–235 [Google Scholar]
- Tambets K, Tolk HV, Kivisild T, Metspalu E, Parik J, Reidla M, Voevoda M, Damba L, Bermisheva M, Khusnutdinova E, Golubenko M, Stepanov V, Puzyrev V, Usanga E, Rudan P, Beckman L, Villems R (2003) Complex signals for population expansions in Europe and beyond. In: Bellwood P, Renfrew C (eds) Examining the farming/language dispersal hypothesis. McDonald Institute for Archaeological Research Monograph Series, Cambridge University Press, Cambridge, pp 449–458 [Google Scholar]
- Tolk HV, Pericic M, Barac L, Klaric IM, Janicijevic B, Rudan I, Parik J, Villems R, Rudan P (2000) mtDNA haplogroups in the populations of Croatian Adriatic Islands. Coll Antropol 24:267–280 [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, et al (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, et al (2001) A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC (1996) Classification of European mtDNAs from an analysis of three European populations. Genetics 144:1835–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Miller JA, Moore LG, Zamudio S, Zhuang J, Droma T, Wallace DC (1994) Mitochondrial DNA analysis in Tibet: implications for the origin of the Tibetan population and its adaptation to high altitude. Am J Phys Anthropol 93:189–199 [DOI] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang CC, Szathmary EJ, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM, Wallace DC (1992) Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130:153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley R, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 10.1046/j.1469-1809.2001.6510043.x [DOI] [PubMed] [Google Scholar]
- Villems R, Adojaan M, Kivisild T, Metspalu E, Parik J, Pielberg G, Rootsi S, Tambets K, Tolk HV (1998) Reconstruction of maternal lineages of Finno-Ugric speaking people and some remarks on their paternal inheritance. In: Wiik K, Julku K (eds) The roots of peoples and languages of Northern Eurasia I. Societas Historiae Fenno-Ugricae, Turku, pp 180–200 [Google Scholar]
- Weale ME, Weiss DA, Jager RF, Bradman N, Thomas MG (2002) Y chromosome evidence for Anglo-Saxon mass migration. Mol Biol Evol 19:1008–1021 [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 10.1073/pnas.171305098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield LS, Sulston JE, Goodfellow PN (1995) Sequence variation of the human Y chromosome. Nature 378:379–380 10.1038/378379a0 [DOI] [PubMed] [Google Scholar]
- Yao YG, Kong QP, Bandelt HJ, Kivisild T, Zhang YP (2002) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, Beckman L, Beckman G, Mikelsaar AV, Krumina A, Kucinskas V, Hurles ME, Tyler-Smith C (2001) Geographical, linguistic, and cultural influences on genetic diversity: Y-chromosomal distribution in Northern European populations. Mol Biol Evol 18:1077–1087 [DOI] [PubMed] [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, Kayser M, Roewer L, Santos FR, Schiefenhovel W, Fretwell N, Jobling MA, Harihara S, Shimizu K, Semjidmaa D, Sajantila A, Salo P, Crawford MH, Ginter EK, Evgrafov OV, Tyler-Smith C (1997) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet 60:1174–1183 [PMC free article] [PubMed] [Google Scholar]