Abstract
Alcoholism is a complex disease with both genetic and environmental risk factors. To identify genes that affect the risk for alcoholism, we systematically ascertained and carefully assessed individuals in families with multiple alcoholics. Linkage and association analyses suggested that a region of chromosome 4p contained genes affecting a quantitative endophenotype, brain oscillations in the beta frequency range (13–28 Hz), and the risk for alcoholism. To identify the individual genes that affect these phenotypes, we performed linkage disequilibrium analyses of 69 single-nucleotide polymorphism (SNPs) within a cluster of four GABAA receptor genes, GABRG1, GABRA2, GABRA4, and GABRB1, at the center of the linked region. GABAA receptors mediate important effects of alcohol and also modulate beta frequencies. Thirty-one SNPs in GABRA2, but only 1 of the 20 SNPs in the flanking genes, showed significant association with alcoholism. Twenty-five of the GABRA2 SNPs, but only one of the SNPs in the flanking genes, were associated with the brain oscillations in the beta frequency. The region of strongest association with alcohol dependence extended from intron 3 past the 3′ end of GABRA2; all 43 of the consecutive three-SNP haplotypes in this region of GABRA2 were highly significant. A three-SNP haplotype was associated with alcoholism, with P=.000000022. No coding differences were found between the high-risk and low-risk haplotypes, suggesting that the effect is mediated through gene regulation. The very strong association of GABRA2 with both alcohol dependence and the beta frequency of the electroencephalogram, combined with biological evidence for a role of this gene in both phenotypes, suggest that GABRA2 might influence susceptibility to alcohol dependence by modulating the level of neural excitation.
Introduction
Alcohol dependence (alcoholism [MIM 103780]) is a common, complex disease with both genetic and environmental risk factors. Both twin studies and adoption studies demonstrate a substantial heritable component to the risk for alcoholism (Goodwin 1979; Pickens et al. 1991; Kendler et al. 1994; Heath et al. 1997). The increased risk for alcoholism among first-degree relatives of alcohol-dependent individuals is in the range of three- to eightfold (Reich et al. 1998). However, there is no simple pattern of inheritance, suggesting that multiple genes and their interaction with each other and with the environment are involved. Therefore, identification of genes that affect the risk for alcoholism has been a difficult endeavor.
To identify genes that affect the risk for alcoholism, the Collaborative Study on the Genetics of Alcoholism (COGA) systematically ascertained and studied a large collection of families containing at least three alcoholic members (Begleiter et al. 1995; Foroud et al. 2000; Edenberg 2002). Our strategy involved focusing genetic efforts on families containing at least three alcoholic members. These families are likely to have a greater genetic contribution to susceptibility and, thus, may provide increased power to detect genes contributing to this complex phenotype. A whole-genome survey using sibling-pair linkage analysis identified several chromosomal regions for which there was evidence of a gene or genes affecting alcohol dependence (Reich et al. 1998; Foroud et al. 2000; Edenberg 2002). There was evidence for increased allele sharing at a microsatellite marker in the GABRB1 locus on chromosome 4p (Reich et al. 1998). Studies of a different population also provided evidence for linkage of alcohol dependence with a marker very near the GABRB1 gene (Long et al. 1998). Linkage disequilibrium (LD) analyses of the COGA families supported the association of alcoholism with a microsatellite marker in GABRB1, although the evidence was modest (Song et al. 2003).
The definition of alcohol dependence includes serious dysfunction in different domains, including tolerance to the effects of ethanol, a withdrawal syndrome during abstinence, craving for ethanol, and persistent drinking in the face of adverse consequences (American Psychiatric Association 1987, 1994; World Health Organization 1993). Given the heterogeneity inherent in the diagnosis of alcohol dependence, we collected extensive phenotypic data on the available individuals in these families, so that complementary analyses of alcoholism-related endophenotypes could be employed in the search for genes affecting alcoholism (Edenberg 2002).
Biological endophenotypes related to neurological function may help identify predisposing factors that increase the vulnerability to alcohol dependence (Gottesman and Gould 2003). We focused on brain oscillations, as measured by electroencephalography (EEG), as key endophenotypes (Porjesz et al. 2002b). Alcoholics differed from controls by having increased power in the beta frequency band (13–28 Hz) of the EEG (Costa and Bauer 1997; Rangaswamy et al. 2002). The offspring of male alcoholics also showed this difference (Bauer and Hesselbrock 1993; Rangaswamy et al. 2004). These data suggested that the EEG power in the beta frequency band provided a heritable, relevant endophenotype for analysis. Another advantage of this quantitative endophenotype is that we could use data from all individuals in the family, whether or not they met the criteria for alcohol dependence. We found particularly strong linkage for an EEG-β phenotype (see the “Methods” section) in a region of chromosome 4p around a microsatellite marker in GABRB1, with a LOD score of 5.0 (Porjesz et al. 2002a). The evidence for this localization increased to LOD 6.5 when association was also analyzed (Porjesz et al. 2002a). A nonparametric linkage analysis (Ghosh et al. 2003) of EEG-β also showed linkage to this region of chromosome 4 (P<.000001).
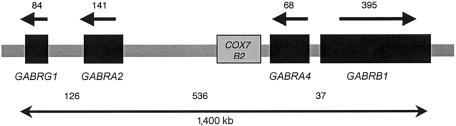
The very strong evidence that this region of chromosome 4p contains genes affecting the beta frequency of the EEG, combined with the evidence for an effect upon the risk for alcoholism, led us to pursue the identification of individual genes that affect these phenotypes. We used public databases to identify genes predicted to lie within the 16-cM region of chromosome 4p that represented a 1-LOD interval around the peak of the EEG linkage (Porjesz et al. 2002a). There is a cluster of genes encoding subunits of the GABAA receptor centered within this region: GABRG1, GABRA2, GABRA4, and GABRB1 (fig. 1). γ-Amino butyric acid (GABA) is the principal inhibitory neurotransmitter in the vertebrate brain (Barnard et al. 1998). GABAA receptors are pentameric assemblies of subunits; 19 mammalian subunits are known, which are classified into α, β, γ, δ, ɛ, π, and ρ types (Barnard et al. 1998). Most GABA receptors contain α, β, and γ subunits (Barnard et al. 1998; Sieghart et al. 1999). Binding of GABA to ionotropic GABAA receptors opens an integral chloride-ion channel that alters the membrane potential of neurons. GABAA receptors mediate fast synaptic inhibition, and beta oscillations involve GABAA-receptor action (Haenschel et al. 2000), making the genes encoding receptor subunits strong candidates for the genes within this region that affect brain oscillations.
Figure 1.
GABAA receptor gene cluster on chromosome 4, based on NCBI human genome build 33. Distances are in kilobases.
Important effects of alcohol, including disruption of motor coordination, anxiolysis, sedation, symptoms related to withdrawal, and ethanol preference, are mediated by GABA transmission (Buck 1996; Grobin et al. 1998; Korpi et al. 1998). GABAA agonists increase ethanol intake in rats, whereas GABAA antagonists decrease intake (Boyle et al. 1993; Tomkins and Fletcher 1996; Nowak et al. 1998). The effects vary in different brain regions, perhaps because of the presence of receptors composed of different groups of subunits (Barnard et al. 1998; Grobin et al. 1998). A variation in the Gabrg2 gene in mice correlates with the severity of acute alcohol withdrawal (Buck and Hood 1998) and with ethanol-induced motor incoordination, hypothermia, and ethanol-conditioned taste aversion (Hood and Buck 2000). These lines of biological evidence make GABA receptor genes excellent candidates to contribute to differences in risk for alcoholism.
Therefore, the four genes in this region of chromosome 4p that encode subunits of the GABAA receptor were our highest-priority candidates for study. We report detailed analysis of 69 SNPs across the 1.4-Mb cluster of four GABAA receptor subunit genes. We analyzed LD between the markers and determined that the effects of individual genes could be discriminated. We found very strong evidence that one of these genes, GABRA2, is strongly associated with both alcohol dependence and brain oscillations.
Material and Methods
Subjects
A large collection of families containing multiple alcoholics was systematically ascertained by the COGA through probands in treatment for alcohol dependence (Reich et al. 1998; Foroud et al. 2000). For acceptance into the genetic part of this study, at least two other first-degree relatives of the proband also had to be alcohol dependent, as assessed by direct interview. Informed consent was obtained after the nature and possible consequences of the studies were explained. Diagnostic data were obtained by direct interview using the Semi-Structured Assessment for the Genetics of Alcoholism (Bucholz et al. 1994).
Electrophysiology
EEG recordings were obtained with noninvasive scalp electrodes in awake individuals with their eyes closed. The filtered artifact-free data were transformed into horizontal bipolar derivations (Porjesz et al. 2002a). Absolute power over the range between 3 and 28 Hz was subdivided into theta (3.0–7.0 Hz), alpha 1 (7.5–9.0 Hz), alpha 2 (9.5–12.0 Hz), beta 1 (12.5–16.0 Hz), beta 2 (16.5–20.0 Hz), and beta 3 (20.5–28.0 Hz) frequency bands. A singular value decomposition procedure (Wang et al. 2000) was utilized to obtain phenotypic data for each of the six EEG bands. Since the beta 2 phenotype yielded highly significant linkage and combined linkage/LD (Almasy et al. 1999) with the GABRB1 microsatellite marker on chromosome 4 (Porjesz et al. 2002a), we assessed differences in this EEG phenotype (hereafter referred to as “EEG-β”) by SNP genotype through use of a measured genotype test (Boerwinkle et al. 1986) implemented in SOLAR (Almasy and Blangero 1998). An additive model was assumed, with heterozygotes having a genotypic mean halfway between that of the two homozygotes, and differences in genotype-specific trait means were tested using a likelihood-ratio comparison.
SNP Genotyping and Analysis
SNPs were identified from public databases, including dbSNP, the National Center for Biotechnology Information (NCBI) Reference Sequence project (for human genome contigs), and LocusLink, as well as by DNA sequencing (below). Contigs containing the GABAA receptor genes in the chromosome 4 cluster were downloaded from NCBI into a database we constructed to facilitate choosing SNPs in and near the genes. Positions shown are based on the NCBI human genome build 33 and dbSNP build 116 (table 1; fig. 1). Rather than examine a limited number of coding SNPs, which are usually of low heterozygosity, we genotyped SNPs of high allele frequency across GABRG1, GABRA2, GABRA4, and GABRB1. When our initial data strongly suggested that variations in GABRA2 were associated with both alcohol dependence and EEG-β, we sequenced exons of this gene to find additional variants (below), genotyped several in the full sample, and genotyped additional SNPs in GABRA2.
Table 1.
Association of SNPs with DSM-IV Alcohol Dependence and EEG[Note]
| Markera | Gene | Function | Numberb | Positionc | AlcoholDependenceP Valued | EEG-βP Valuee |
| rs1497570 | GABRG1 | IVS8 | 45896957 | .65 | .68 | |
| rs1948609 | GABRG1 | IVS5 | 45912695 | .88 | .86 | |
| rs1391175 | GABRG1 | IVS1 | 45951087 | .050* | .41 | |
| rs2221020 | GABRG1 | IVS1 | 45953488 | .068 | .98 | |
| rs1391168 | GABRG1 | IVS1 | 45965082 | .45 | .85 | |
| rs904154 | GABRG1 | 5′ | 1 | 45976294 | .19 | .18 |
| rs490434 | GABRA2 | 3′ | 2 | 46043202 | .0052* | .64 |
| rs576666 | GABRA2 | 3′ | 3 | 46052300 | .095 | .067 |
| rs531460 | GABRA2 | 3′ | 4 | 46060193 | .022* | .024* |
| rs561779 | GABRA2 | 3′ | 5 | 46088701 | .048* | .044* |
| rs495818 | GABRA2 | 3′ | 6 | 46097821 | .022* | .034* |
| rs497068 | GABRA2 | 3′ | 46100600 | .0069* | .26 | |
| rs572227 | GABRA2 | 3′ | 46101316 | .038* | .019* | |
| rs573400 | GABRA2 | 3′UTR | 46101989 | .062 | .27 | |
| rs541418 | GABRA2 | IVS9 | 46103139 | .020* | .10 | |
| rs481311 | GABRA2 | IVS9 | 46104305 | .076 | .17 | |
| rs507788 | GABRA2 | IVS9 | 7 | 46107381 | .031* | .068* |
| rs532780 | GABRA2 | IVS9 | 46111289 | .079 | .016* | |
| rs548583 | GABRA2 | IVS9 | 46113267 | .012* | .028* | |
| ss15649713 | GABRA2 | IVS9 | 46113787 | .103 | .56 | |
| rs496650 | GABRA2 | IVS8 | 8 | 46114308 | .054 | .75 |
| rs540363 | GABRA2 | IVS8 | 9 | 46124169 | .044* | .49 |
| rs526752 | GABRA2 | IVS8 | 46126552 | .12 | .07 | |
| rs530329 | GABRA2 | IVS8 | 46131042 | .034* | .048* | |
| rs483160 | GABRA2 | IVS8 | 10 | 46136998 | .15 | .036* |
| rs279871f | GABRA2 | IVS7 | 11 | 46155656 | .0004* | .049* |
| rs279867 | GABRA2 | IVS6 | 46158226 | .24 | .05* | |
| rs279866 | GABRA2 | IVS6 | 46159687 | .029* | .037* | |
| rs279863 | GABRA2 | IVS5 | 46162945 | .017* | .011* | |
| rs279861 | GABRA2 | IVS5 | 46163248 | .037* | .045* | |
| rs279858 | GABRA2 | Exon 5 | 12 | 46164516 | .0087* | .22 |
| rs175931 | GABRA2 | IVS4 | 46166246 | .10 | .071* | |
| rs279843 | GABRA2 | IVS4 | 13 | 46175127 | .049* | .30 |
| rs279845f | GABRA2 | IVS4 | 14 | 46179646 | .013* | .011* |
| rs279846 | GABRA2 | IVS4 | 46179809 | .017* | .012* | |
| rs183961 | GABRA2 | IVS4 | 46180951 | .038* | .014* | |
| rs1440130 | GABRA2 | IVS4 | 46183176 | .013* | .017* | |
| rs279826 | GABRA2 | IVS4 | 15 | 46184132 | .0008* | .25 |
| ss15649712 | GABRA2 | IVS4 | 46184417 | .014* | .70 | |
| rs279827 | GABRA2 | IVS3 | 46184625 | .0068* | .016* | |
| rs279828 | GABRA2 | IVS3 | 46184733 | .0086* | .02* | |
| rs279834 | GABRA2 | IVS3 | 46188222 | .015* | .027* | |
| rs279836f | GABRA2 | IVS3 | 16 | 46188993 | .0071* | .0066* |
| rs279837 | GABRA2 | IVS3 | 46189246 | .035* | .064 | |
| rs279841 | GABRA2 | IVS3 | 46190686 | .038* | .018* | |
| rs189957 | GABRA2 | IVS3 | 17 | 46196602 | .053* | .27 |
| rs1442059 | GABRA2 | IVS3 | 18 | 46206875 | .034* | .018* |
| rs1442061 | GABRA2 | IVS3 | 46221143 | .37 | .24 | |
| rs1442062 | GABRA2 | IVS3 | 19 | 46226999 | .22 | .13 |
| ss15649711 | GABRA2 | IVS3 | 20 | 46237967 | .76 | .57 |
| ss15649710 | GABRA2 | IVS 1 | 46240788 | .91 | .97 | |
| rs3756007 | GABRA2 | IVS 1 | 46240987 | .99 | .98 | |
| rs894269 | GABRA2 | 5′ | 21 | 46243535 | .097 | .84 |
| rs2165607 | GABRA2 | 5′ | 46250144 | .44 | .67 | |
| rs1545234 | GABRA2 | 5′ | 22 | 46254336 | .41 | .62 |
| rs2036943 | Intergenic | 5′ | 23 | 46726482 | .85 | .030* |
| rs2055943 | GABRA4 | IVS7 | 46817202 | .72 | .70 | |
| rs1512135 | GABRA4 | IVS6 | 46823811 | .63 | .38 | |
| rs1877400 | GABRA4 | IVS5 | 46827224 | .90 | 1.00 | |
| rs2280072 | GABRA4 | IVS1 | 46845093 | .53 | .79 | |
| rs2280071 | GABRA4 | 5′UTR | 46845289 | .98 | .96 | |
| rs2055940 | GABRA4 | 5′UTR | 46847836 | .36 | .29 | |
| rs2119780 | GABRB1 | IVS4 | 47014133 | .33 | 1.00 | |
| rs989808 | GABRB1 | IVS4 | 47016704 | .77 | .50 | |
| rs1372496 | GABRB1 | IVS4 | 47057731 | .99 | .53 | |
| rs1372497 | GABRB1 | IVS4 | 47072261 | .53 | .81 | |
| rs6284 | GABRB1 | IVS5 | 47172142 | .98 | .54 | |
| rs2070922 | GABRB1 | IVS7 | 47255971 | .22 | .61 | |
| rs6289 | GABRB1 | IVS8 | 47258632 | .63 | .66 |
Note.— Significant P values are indicated with an asterisk (*).
Markers are shown as rs numbers (or ss numbers, for those we submitted that are not yet included) from the dbSNP database.
Refers to the numbering in figure 2.
Position is in nucleotides from chromosome 4pter, as estimated in the dbSNP database (build 116) or by blasting against the NCBI Human Genome assembly (build 33).
P value for alcohol dependence, based on the average PDT (Martin et al. 2000).
P value for EEG-β, based on a measured genotype test (Boerwinkle et al. 1986) implemented in SOLAR (Almasy and Blangero 1998).
Marker used for the haplotype analysis in table 2.
Assays were designed using SpectroDESIGNER software (Sequenom) and performed using a modified single-nucleotide extension reaction with allele discrimination by mass spectrometry (Sequenom MassArray system) (Jurinke et al. 2001). Genotypes were tested for Mendelian inheritance, and inconsistent genotypes (an average of 14 of 3,560; 23 SNPs had ⩽10 inconsistencies) were discarded.
LD between markers was analyzed using the program GOLD (Abecasis and Cookson 2000). To examine association between the SNPs and the phenotype of alcohol dependence, we used the pedigree disequilibrium test (PDT) (Martin et al. 2000), which uses information from the entire pedigree rather than just the affected subject and his/her two parents and thus takes advantage of the extended families that we are studying. Family-based association studies avoid the problems of false positive results arising from population stratification, which can occur in population-based association approaches (Spielman and Ewens 1996). We analyzed two haplotypes constructed from three SNPs each: one based on rs279871, rs279826, and rs279836 (chosen on the basis of their significant association with alcohol dependence; table 1), and one based on rs279871, rs279845, and rs279836 (chosen because of their more significant association with EEG-β; tables 1 and 2). We then systematically examined all consecutive three-SNP haplotypes of GABRA2 (table 3).
Table 2.
Association of Haplotypes with Alcohol Dependence
|
Parental Contribution |
No. of Discordant Sibs |
||||||
| Haplotypea | Frequency | % of AllHaplotypes | No. ofTransmittedAlleles | No. ofNontransmittedAlleles | Affected | Unaffected | P Valueb |
| 1 | 287 | 5.5 | 76 | 69 | 67 | 61 | .66 |
| 2 | 226 | 4.3 | 57 | 61 | 44 | 39 | .94 |
| 3 | 74 | 1.4 | 11 | 19 | 8 | 17 | .25 |
| 4 | 2,531 | 48.5 | 728 | 589 | 549 | 518 | .000000022 |
| 5 | 1,724 | 33.1 | 454 | 437 | 354 | 395 | .67 |
| 6 | 68 | 1.3 | 8 | 15 | 8 | 11 | .70 |
| 7 | 54 | 1.0 | 9 | 17 | 3 | 5 | .96 |
| 8 | 252 | 4.8 | 59 | 77 | 47 | 60 | .17 |
Table 3.
Association between Consecutive Three-SNP Haplotypes in GABRA2 and Alcohol Dependence[Note]
| Marker | OverallSignificancea | Significance ofHigh-RiskHaplotypeb |
| rs490434 | … | … |
| rs576666 | … | … |
| rs531460 | .0066 | .000034 |
| rs561779 | .0030 | .000075 |
| rs495818 | .0064 | .000077 |
| rs497068 | .0075 | .00010 |
| rs572227 | .0037 | .000035 |
| rs573400 | .0113 | .00050 |
| rs541418 | .0088 | .0011 |
| rs481311 | .0164 | .00010 |
| rs507788 | .0127 | .00020 |
| rs532780 | .0428 | .00070 |
| rs548583 | .0028 | .000039 |
| ss15649713 | .0258 | .00050 |
| rs496650 | .0187 | .0010 |
| rs540363 | .0427 | .0010 |
| rs526752 | .0156 | .00020 |
| rs530329 | .0060 | .00020 |
| rs483160 | .0071 | .000084 |
| rs279871 | .00010 | .000003 |
| rs279867 | .000000 | .000001 |
| rs279866 | .000003 | .000001 |
| rs279863 | .00090 | .000004 |
| rs279861 | .0052 | .000050 |
| rs279858 | .0086 | .00030 |
| rs175931 | .0030 | .00010 |
| rs279843 | .0082 | .000096 |
| rs279845 | .00080 | .000013 |
| rs279846 | .0340 | .0014 |
| rs183961 | .0042 | .000051 |
| rs1440130 | .0018 | .000015 |
| rs279826 | .0042 | .000061 |
| ss15649712 | .0318 | .0062 |
| rs279827 | .0240 | .0068 |
| rs279828 | .0114 | .0029 |
| rs279834 | .0141 | .000041 |
| rs279836 | .0011 | .000004 |
| rs279837 | .0025 | .000019 |
| rs279841 | .0019 | .000023 |
| rs189957 | .00080 | .000027 |
| rs1442059 | .031 | .00020 |
| rs1442061 | .022 | .0030 |
| rs1442062 | .013 | .00010 |
| ss15649712 | .026 | .00040 |
| ss15649710 | .059 | .0040 |
| rs3756007 | .35 | .0275 |
| rs894269 | .15 | .0026 |
| rs2165607 | .026 | .0023 |
| rs1545234 | .24 | .0865 |
Note.— Each set of three consecutive SNPs through the GABRA2 gene was analyzed as an eight-allele marker, named by the third SNP in the set.
P value based on the average PDT (Martin et al. 2000), treating each haplotype as a marker in an eight-allele system; the value is shown at the third of the three SNPs used to create each haplotype.
Significance of the high-risk haplotype within each set of three consecutive SNPs.
DNA Sequencing
To examine whether there were coding or splice-site polymorphisms within the GABRA2 gene, the entire coding region of the GABRA2 gene was sequenced in DNAs from 48 individuals, 25 of whom were homozygous for a strongly associated haplotype (P=.000024) consisting of rs279871, rs279826, and rs279836; 21 of whom were homozygous for the most common nonassociated haplotype; and 2 of whom were homozygous for uncommon haplotypes. The coding regions plus at least 60 bp of each flanking intronic sequence were amplified by PCR and sequenced in both directions, using the ABI PRISM 3100 Genetic Analyzer capillary DNA sequencer with Big Dye chemistry (Applied Biosystems). Additional SNPs were found, several of which were genotyped in the full sample; these have been submitted to dbSNP (the ss numbers are given in table 1).
Results
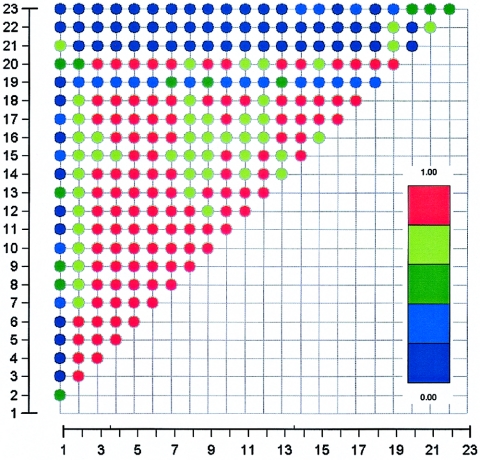
To test whether individual gene(s) within the region of linkage on chromosome 4p affect either the neurophysiological or alcohol-dependence phenotype, we initially genotyped and analyzed 20 SNPs across the 1.4-Mb cluster that contains GABRG1, GABRA2, GABRA4, and GABRB1 (fig. 1). LD between markers within each gene was high (fig. 2). LD between genes was considerably lower than that within a gene; in particular, LD drops at both ends of GABRA2. This is consistent with other data on LD across the human genome (Daly et al. 2001; Goldstein 2001; Reich et al. 2001; Gabriel et al. 2002). Thus, our analysis of the association of alcoholism with the GABAA receptor genes can differentiate among the genes, even though they are physically close together.
Figure 2.
Pattern of LD within GABRA2. The numbers on the X- and Y-axes correspond to 21 markers selected to cover the GABRA2 gene relatively evenly at an average spacing of 10.6 kb, plus two flanking markers; the markers are numbered in table 1. Each colored circle represents the LD between two markers, as measured by D′ (Abecasis and Cookson 2000). Higher values of D′ indicate higher LD.
We used a family-based association method, the PDT (Martin et al. 2000), to examine association between the SNPs and the phenotype of alcohol dependence, as defined by DSM-IV criteria (American Psychiatric Association 1994). Our initial analyses showed that eight of nine SNPs in the GABRA2 gene were significantly associated with alcohol dependence. In contrast, SNPs in the three other GABAA receptor genes in this cluster, including genes on either side of GABRA2, did not show a pattern of association with alcohol dependence. Therefore, additional SNPs in GABRA2 were genotyped and continued to show association with alcohol dependence and with EEG-β (table 1). Thirty-one SNPs within or closely flanking GABRA2 were associated with alcohol dependence, at P values ⩽.05 (table 1). The region associated with alcohol dependence spanned 164 kb, extending from intron 3 to 58 kb past the 3′ end of the gene.
Twenty-five of the SNPs within GABRA2 and one near it were associated with EEG-β (table 1), the endophenotype previously analyzed in the whole-genome survey (Porjesz et al. 2002a). SNPs in the other flanking GABAA receptor genes were not associated with this phenotype.
To further analyze the association of the GABRA2 gene with alcohol dependence, a set of three SNPs that showed association with both alcohol dependence and EEG-β was analyzed as a haplotype (table 2). The global PDT was significant (P=.0001). The higher-risk haplotype, consisting of the alleles that were overtransmitted in the individual SNP analyses, was associated with alcohol dependence at P=.00000002. We then systematically examined, by the global PDT test, all consecutive three-SNP haplotypes within GABRA2 and found that 1 of the 5 haplotypes at the 5′ end of the gene and all 43 of the haplotypes starting within exon 3 and extending to the 3′ end of the gene were significantly associated with alcohol dependence (table 3). The median global significance of the 43 haplotypes from intron 3 to the end of the gene was .007; the most significant were ⩽.000001. The high-risk haplotypes themselves had a median significance of .000098 (table 3).
We examined the prevalence of the higher-risk haplotype by sex. Among alcohol-dependent individuals, the same proportion of males (50%) and females (53%) have the higher-risk haplotype. Looking at the data in a different way, 46% of males who have the high-risk haplotype are affected, whereas only 25% of the females who have the high-risk haplotype are affected; this is reasonable, given the lower prevalence of alcoholism in females.
None of the SNPs tested affect the amino acid sequence of the encoded α2 subunit. To examine whether there were coding polymorphisms in LD with the tested SNPs, we sequenced DNA from 48 individuals, 25 of whom were homozygous for a strongly associated high-risk haplotype (P=.000024) consisting of rs279871, rs279826, and rs279836; 21 of whom were homozygous for the most common nonassociated haplotype; and 2 of whom were homozygous for uncommon haplotypes. No polymorphisms that altered the amino acids encoded by the GABRA2 gene were found.
Discussion
Our results provide extremely strong evidence that variations in the GABRA2 gene, and not in the flanking GABAA receptor genes, affect both brain oscillations in the beta frequency range (EEG-β) and alcohol dependence. We were led to examine genes in this region by strong linkage with EEG-β, demonstrated by two different methods of analysis: a variance-component method with additional evidence for association (Porjesz et al. 2002a) and a nonparametric analysis (Ghosh et al. 2003). The location and function of a set of GABAA receptor genes in the center of this linkage peak made them excellent candidate genes. Rather than examine a single SNP in each gene, we analyzed a total of 69 SNPs in four genes. Only one gene was consistently associated with alcohol dependence and with the EEG endophenotype: GABRA2.
Thirty-one SNPs in GABRA2 were associated with alcohol dependence, and 25 were associated with EEG-β. The P value for association of a three-SNP haplotype with alcohol dependence, .000000022, surpasses the level of genomewide significance. The higher-risk haplotype is common (48.5%). Because so many haplotypes were associated (43 consecutive three-SNP haplotypes out of 47 tested and both selected haplotypes), we do not think there is any reasonable likelihood that the findings are artifacts of multiple testing. The significance of the high-risk haplotype remains after even the most conservative correction for multiple testing (Bonferroni, which assumes independence, an assumption clearly not true in this region of high LD, making the correction overly conservative). A Bonferroni correction would require P<.001, given that we examined 49 three-SNP haplotypes (47 systematically derived from three adjacent SNPs [table 3] and 2 selected from the individual SNPs with the greatest evidence of transmission distortion [table 2 and data not shown]). In fact, 8 of the 49 haplotype analyses surpassed that level of overall significance, and 23 of the high-risk haplotypes surpassed a significance level of .0001 (table 3).
Our strategy of genotyping multiple SNPs of high heterozygosity rather than focusing on coding SNPs (which are often of low heterozygosity) was driven by our hypothesis that the variations underlying complex genetic diseases predominantly affect gene regulation rather than the structure of the encoded protein. Gene regulation is the result of the combinatorial action of multiple transcription factors binding at multiple sites in and near a gene and therefore can be affected by multiple SNPs. We did not find any coding difference between the higher-risk and lower-risk haplotypes. The lack of coding SNPs in GABRA2 shows that our strategy was advantageous; the alternative approach would not have allowed us to test the association. The association extends across a significant fraction of the GABRA2 gene. It is perhaps surprising that it is strongest from intron 3 through the 3′ end of the gene rather than the 5′ region in which the promoter lies. Although effects of distal regulatory elements on transcriptional initiation are not ruled out, the data suggest that alternative splicing, mRNA stability, or other posttranscriptional mechanisms might be involved.
There are several lines of evidence suggesting biological links among GABRA2, alcoholism, and the EEG power spectral density in the beta range. GABAA receptors have integral chloride channels that are opened by the binding of GABA, the major inhibitory neurotransmitter in the vertebrate brain (Barnard et al. 1998). The resulting chloride flux alters the membrane potential of neurons, inhibiting firing. GABAA receptors are sensitive to ethanol and are believed to mediate many of its effects, including anxiolysis, sedation, disruption of motor coordination, tolerance, and dependence (Grobin et al. 1998; Harris et al. 1998; Korpi et al. 1998; Buck and Finn 2001; Ueno et al. 2001). In rats, ethanol intake is increased by GABAA agonists and decreased by GABAA antagonists (Boyle et al. 1993; Tomkins and Fletcher 1996; Nowak et al. 1998), although the effects vary in different brain regions, perhaps because of differences in the subunit composition of the receptors (Barnard et al. 1998; Grobin et al. 1998). A variation in the Gabrg2 gene in mice correlates with the severity of alcohol withdrawal, motor incoordination, ethanol-conditioned taste aversion, and hypothermia (Buck and Hood 1998; Hood and Buck 2000).
The α2 subunit of the GABAA receptor is a target of benzodiazepines (Harris et al. 1998; Low et al. 2000; Tobler et al. 2001), which are used in the treatment of alcohol withdrawal symptoms. Mice in which the α2 subunit was made insensitive to diazepam by introducing a point mutation into the Gabra2 gene were insensitive to the anxiolytic effects of diazepam but retained sensitivity to its sedative and amnestic properties (Low et al. 2000); similar alteration to the Gabra3 gene did not affect anxiolysis. This demonstrates that the α2 subunit is critical for anxiolysis.
Benzodiazepines strongly increase EEG beta power (Whittington et al. 1996; Traub et al. 1999), particularly in frontal regions (Traub et al. 1999). The α2, α3, or α5 subunits of the GABAA receptor mediate the effects of benzodiazepines on EEG spectral changes (Tobler et al. 2001). There is a significant increase in beta power in alcohol dependent subjects, particularly over fronto-central leads (Costa and Bauer 1997; Rangaswamy et al. 2002). This higher beta power was also found in offspring of male alcoholics (Bauer and Hesselbrock 1993; Bauer 2001; Rangaswamy et al. 2004). Bauer and Hesselbrock (1993) reported that enhanced high-frequency beta activity, originating from deep anterior regions of the frontal brain, was the best predictor of relapse in substance-dependent patients and may also be related to initial risk for dependence. They proposed that relapse, a key feature of alcoholism, was in part due to a deficit in function of the brain region that normally dampens impulsivity.
In addition to the separate evidence making GABA receptor genes good candidates for affecting both neurophysiological and diagnostic phenotypes, there are links between the phenotypes in these two domains. Begleiter and Porjesz (1999) hypothesized that neural disinhibition is involved in predisposition toward alcoholism. Oscillations in the beta (12.5–28 Hz) and gamma (28.5–50 Hz) frequencies are believed to represent an activated state of the neuronal network that generates them (Haenschel et al. 2000), so differences in these oscillations could affect the overall level of neural excitation and thereby the risk for alcoholism.
Our finding of very strong LD with alcohol dependence and with brain oscillations across the GABRA2 gene, combined with the evidence for an effect of this gene on brain oscillations (Porjesz et al. 2002a) and anxiolysis (Low et al. 2000), draws these lines of evidence together. These data suggest that differences in the expression or function of GABAA receptors can modulate both the beta activity and neural inhibition. Our results also illustrate the power of studying endophenotypes to aid in the identification of genes affecting complex diseases: we were led to this region, in large part, by the very strong linkage to an electrophysiological endophenotype. The GABRA2 gene proved to be associated both with the endophenotype and with alcoholism. The convergence of evidence from different analyses and phenotypes, along with the biological data on its function, provides strong evidence that GABRA2 is a key gene affecting the risk for alcoholism.
Acknowledgments
The COGA (H. Begleiter, State University of New York Health Sciences Center, Brooklyn, principal investigator; T. Reich, Washington University, co–principal investigator; and H. Edenberg, Indiana University, co–principal investigator) includes nine different centers where data collection, analysis, and storage take place. The nine sites and principal investigators and coinvestigators are: Howard University (R. Taylor), Indiana University (H. Edenberg, J. Nurnberger, Jr., P. M. Conneally, and T. Foroud), Rutgers University (J. Tischfield), Southwest Foundation (L. Almasy), State University of New York Health Sciences Center at Brooklyn (B. Porjesz and H. Begleiter), University of California at San Diego (M. Schuckit), University of Connecticut (V. Hesselbrock), University of Iowa (R. Crowe and S. Kuperman), and Washington University in St. Louis (T. Reich, C. R. Cloninger, J. Rice, and A. Goate). Lisa Neuhold serves as the National Institute on Alcohol Abuse and Alcoholism (NIAAA) staff collaborator. This national collaborative study is supported by the National Institutes of Health grant U10AA08403 from the NIAAA. Genotyping facilities were provided by the Center for Medical Genomics at Indiana University School of Medicine, supported, in part, by the Indiana 21st Century Research and Technology Fund and the Indiana Genomics Initiative (INGEN, supported in part by the Lilly Endowment).
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for markers listed in tables 1 and 3, including new SNPs submitted: ss15649710, ss15649711, ss15649712, and ss15649713)
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink/
- NCBI Reference Sequence, http://www.ncbi.nlm.nih.gov/RefSeq/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for alcoholism)
References
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62:1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Williams JT, Dyer TD, Blangero J (1999) Quantitative trait locus detection using combined linkage/disequilibrium analysis. Genet Epidemiol 17 Suppl 1:S31–S36 [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (1987) Diagnostic and statistical manual of mental disorders, 3rd ed, revised. American Psychiatric Association Press, Washington, DC [Google Scholar]
- ——— (1994) Diagnostic and statistical manual of mental disorders, 4th ed. American Psychiatric Association Press, Washington, DC [Google Scholar]
- Barnard EA, Skolnick P, Olsen RW, Mohler H, Sieghart W, Biggio G, Braestrup C, Bateson AN, Langer SZ (1998) International Union of Pharmacology. XV. Subtypes of γ-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol Rev 50:291–313 [PubMed] [Google Scholar]
- Bauer LO (2001) Predicting relapse to alcohol and drug abuse via quantitative electroencephalography. Neuropsychopharmacology 25:332–340 10.1016/S0893-133X(01)00236-6 [DOI] [PubMed] [Google Scholar]
- Bauer LO, Hesselbrock V (1993) EEG, autonomic, and subjective correlates of the risk for alcoholism. J Stud Alcohol 54:577–589 [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B (1999) What is inherited in the predisposition toward alcoholism? A proposed model. Alcohol Clin Exp Res 23:1125–1135 [DOI] [PubMed] [Google Scholar]
- Begleiter H, Reich T, Hesselbrock V, Porjesz B, Li T-K, Schuckit MA, Edenberg HJ, Rice JP (1995) The Collaborative Study on the Genetics of Alcoholism. Alcohol Health Res World 19:228–236 [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF (1986) The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet 50:181–194 [DOI] [PubMed] [Google Scholar]
- Boyle AE, Segal R, Smith BR, Amit Z (1993) Bidirectional effects of GABAergic agonists and antagonists on maintenance of voluntary ethanol intake in rats. Pharmacol Biochem Behav 46:179–182 10.1016/0091-3057(93)90338-T [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr, Reich T, Schmidt I, Schuckit MA (1994) A new semi-structured psychiatric interview for use in genetic linkage studies: a report of the reliability of the SSAGA. J Stud Alcohol 55:149–158 [DOI] [PubMed] [Google Scholar]
- Buck KJ (1996) New insight into the mechanisms of ethanol effects on GABAA receptor function and expression, and their relevance to behavior. Alcohol Clin Exp Res 20:198A–202A [DOI] [PubMed] [Google Scholar]
- Buck KJ, Finn DA (2001) Genetic factors in addiction: QTL mapping and candidate gene studies implicate GABAergic genes in alcohol and barbiturate withdrawal in mice. Addiction 96:139–149 10.1046/j.1360-0443.2001.96113910.x [DOI] [PubMed] [Google Scholar]
- Buck KJ, Hood HM (1998) Genetic association of a GABA(A) receptor γ2 subunit variant with severity of acute physiological dependence on alcohol. Mamm Genome 9:975–978 10.1007/s003359900909 [DOI] [PubMed] [Google Scholar]
- Costa L, Bauer L (1997) Quantitative electroencephalographic differences associated with alcohol, cocaine, heroin and dual-substance dependence. Drug Alcohol Depend 46:87–93 10.1016/S0376-8716(97)00058-6 [DOI] [PubMed] [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES (2001) High-resolution haplotype structure in the human genome. Nat Genet 29:229–232 10.1038/ng1001-229 [DOI] [PubMed] [Google Scholar]
- Edenberg HJ (2002) The collaborative study on the genetics of alcoholism: an update. Alcohol Res Health 26:214–218 [PMC free article] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T (2000) Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res 24:933–945 10.1097/00000374-200007000-00001 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Begleiter H, Porjesz B, Chorlian DB, Edenberg HJ, Foroud T, Goate A, Reich T (2003) Linkage mapping of beta 2 EEG waves via non-parametric regression. Am J Med Genet 118B:66–71 [DOI] [PubMed] [Google Scholar]
- Goldstein DB (2001) Islands of linkage disequilibrium. Nat Genet 29:109–111 10.1038/ng1001-109 [DOI] [PubMed] [Google Scholar]
- Goodwin DW (1979) The cause of alcoholism and why it runs in families. Br J Addict Alcohol Other Drugs 74:161–164 [DOI] [PubMed] [Google Scholar]
- Gottesman, II, Gould TD (2003) The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry 160:636–645 10.1176/appi.ajp.160.4.636 [DOI] [PubMed] [Google Scholar]
- Grobin AC, Matthews DB, Devaud LL, Morrow AL (1998) The role of GABA(A) receptors in the acute and chronic effects of ethanol. Psychopharmacology (Berl) 139:2–19 [DOI] [PubMed] [Google Scholar]
- Haenschel C, Baldeweg T, Croft RJ, Whittington MA, Gruzelier J (2000) Gamma and beta frequency oscillations in response to novel auditory stimuli: a comparison of human electroencephalogram (EEG) data with in vitro models. Proc Natl Acad Sci USA 97:7645–7650 10.1073/pnas.120162397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Mihic SJ, Valenzuela CF (1998) Alcohol and benzodiazepines: recent mechanistic studies. Drug Alcohol Depend 51:155–164 10.1016/S0376-8716(98)00073-8 [DOI] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG (1997) Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med 27:1381–1396 10.1017/S0033291797005643 [DOI] [PubMed] [Google Scholar]
- Hood HM, Buck KJ (2000) Allelic variation in the GABA A receptor gamma2 subunit is associated with genetic susceptibility to ethanol-induced motor incoordination and hypothermia, conditioned taste aversion, and withdrawal in BXD/Ty recombinant inbred mice. Alcohol Clin Exp Res 24:1327–1334 10.1097/00000374-200009000-00002 [DOI] [PubMed] [Google Scholar]
- Jurinke C, van den Boom D, Cantor CR, Koster H (2001) Automated genotyping using the DNA MassArray technology. Methods Mol Biol 170:103–116 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Heath AC, Kessler RC, Eaves LJ (1994) A twin-family study of alcoholism in women. Am J Psychiatry 151:707–715 [DOI] [PubMed] [Google Scholar]
- Korpi ER, Makela R, Uusi-Oukari M (1998) Ethanol: novel actions on nerve cell physiology explain impaired functions. News Physiol Sci 13:164–170 [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D (1998) Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from and autosome-wide scan in an American Indian population. Am J Med Genet 81:216–221 [DOI] [PubMed] [Google Scholar]
- Low K, Crestani F, Keist R, Benke D, Brunig I, Benson JA, Fritschy JM, Rulicke T, Bluethmann H, Mohler H, Rudolph U (2000) Molecular and neuronal substrate for the selective attenuation of anxiety. Science 290:131–134 10.1126/science.290.5489.131 [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak KL, McBride WJ, Lumeng L, Li TK, Murphy JM (1998) Blocking GABA(A) receptors in the anterior ventral tegmental area attenuates ethanol intake of the alcohol-preferring P rat. Psychopharmacology (Berl) 139:108–116 [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Lykken DT, Heston LL, Clayton PJ (1991) Heterogeneity in the inheritance of alcoholism: a study of male and female twins. Arch Gen Psychiatry 48:19–28 [DOI] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, Goate A, Rice JP, O’Connor SJ, Rohrbaugh J, Kuperman S, Bauer LO, Crowe RR, Schuckit MA, Hesselbrock V, Conneally PM, Tischfield JA, Li T-K, Reich T, Begleiter H (2002a) Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci USA 99:3729–3733 10.1073/pnas.052716399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Wang K, Almasy L, Chorlian DB, Stimus AT, Kuperman S, O’Connor SJ, Rohrbaugh J, Bauer LO, Edenberg HJ, Goate A, Rice JP, Reich T (2002b) Linkage and linkage disequilibrium mapping of ERP and EEG phenotypes. Biol Psychol 61:229–248 10.1016/S0301-0511(02)00060-1 [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Bauer LO, Kuperman S, O’Connor S, Rohrbaugh J, Reich T, Begleiter H (2002) Beta power in the EEG of alcoholics. Biol Psychiatry 52:831–842 10.1016/S0006-3223(02)01362-8 [DOI] [PubMed] [Google Scholar]
- Rangaswamy M, Porjesz B, Chorlian DB, Wang K, Jones KA, Kuperman S, Rohrbaugh J, O’Connor S, Bauer LO, Reich T, Begleiter H (2004) Resting EEG in offspring of male alcoholics: beta frequencies. Int J Psychophysiol 51:239–251 10.1016/j.ijpsycho.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES (2001) Linkage disequilibrium in the human genome. Nature 411:199–204 10.1038/35075590 [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li T-K, Conneally PM, Nurnberger JI Jr, Tischfield JA, Crowe R, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H (1998) A genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet 81:207–215 [DOI] [PubMed] [Google Scholar]
- Sieghart W, Fuchs K, Tretter V, Ebert V, Jechlinger M, Hoger H, Adamiker D (1999) Structure and subunit composition of GABAA receptors. Neurochem Int 34:379–385 10.1016/S0197-0186(99)00045-5 [DOI] [PubMed] [Google Scholar]
- Song J, Koller DL, Foroud T, Carr K, Zhao J, Rice J, Nurnberger JI Jr, Begleiter H, Porjesz B, Smith TL, Schuckit MA, Edenberg HJ (2003) Association of GABAA receptors and alcohol dependence and the effects of genetic imprinting. Am J Med Genet 117B:39–45 [DOI] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ (1996) The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet 59:983–989 [PMC free article] [PubMed] [Google Scholar]
- Tobler I, Kopp C, Deboer T, Rudolph U (2001) Diazepam-induced changes in sleep: role of the α1 GABAA receptor subtype. Proc Natl Acad Sci USA 98:6464–6469 10.1073/pnas.111055398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins DM, Fletcher PJ (1996) Evidence that GABA(A) but not GABA(B) receptor activation in the dorsal raphe nucleus modulates ethanol intake in Wistar rats. Behav Pharmacol 7:85–93 [PubMed] [Google Scholar]
- Traub RD, Jefferys JGR, Traub RD (1999) Fast oscillations in cortical circuits. MIT Press, Cambridge, MA [Google Scholar]
- Ueno S, Harris RA, Messing RO, Sanchez-Perez AM, Hodge CW, McMahon T, Wang D, Mehmert KK, Kelley SP, Haywood A, Olive MF, Buck KJ, Hood HM, Blednov Y, Findlay G, Mascia MP (2001) Alcohol actions on GABA(A) receptors: from protein structure to mouse behavior. Alcohol Clin Exp Res 25:76S–81S 10.1097/00000374-200105051-00014 [DOI] [PubMed] [Google Scholar]
- Wang K, Begleiter H, Porjesz B (2000) Trilinear modeling of event-related potentials. Brain Topogr 12:263–271 10.1023/A:1023455404934 [DOI] [PubMed] [Google Scholar]
- Whittington MA, Jefferys JG, Traub RD (1996) Effects of intravenous anaesthetic agents on fast inhibitory oscillations in the rat hippocampus in vitro. Br J Pharmacol 118:1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (1993) International classification of disease. World Health Organization, Geneva [Google Scholar]