Abstract
Two disorders, periventricular nodular heterotopia (PVNH) and a group of skeletal dysplasias belonging to the oto-palato-digital (OPD) spectrum, are caused by FLNA mutations. They are considered mutually exclusive because of the different presumed effects of the respective FLNA gene mutations, leading to loss of function (PVNH) and gain of function (OPD), respectively. We describe here the first patient manifesting PVNH in combination with frontometaphyseal dysplasia, a skeletal dysplasia of the OPD-spectrum. A novel de novo mutation, 7315C→A in exon 45 of the FLNA gene, was identified. It leads to two aberrant transcripts, one full-length transcript with the point mutation causing a substitution of a highly conserved leucine residue (L2439M) and a second shortened transcript lacking 21 bp due to the creation of an ectopic splice donor site in exon 45. We propose that the dual phenotype is caused by two functionally different, aberrant filamin A proteins and therefore represents an exceptional model case of allelic gain-of-function and loss-of-function phenotypes due to a single mutational event.
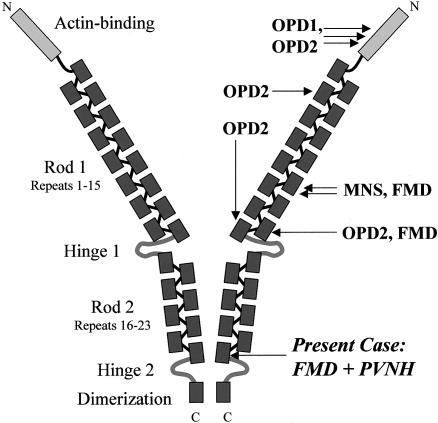
Filamin A is a ubiquitous actin-binding protein involved in cytoskeletal organization. It has three major functional domains: N-terminal actin–binding domain, rod domain consisting of 24 immunoglobulinlike repeat units interrupted by two flexible hinge structures, and a C-terminal dimerization site (fig. 1) (Stossel et al. 2001; van der Flier and Sonnenberg 2001). Mutations in FLNA, the human filamin A gene, were first detected in periventricular nodular heterotopia (PVNH [MIM 300049]), an X-linked neuronal migration disorder that affects predominantly females and leads to early prenatal lethality in males (Fox et al. 1998). The majority of FLNA mutations associated with PVNH are truncating mutations, indicating that loss of filamin A function causes PVNH (Fox et al. 1998; Sheen et al. 2001; Moro et al. 2002; Human Gene Mutation Database). More recently, it was demonstrated that mutations in FLNA are also associated with a group of X-linked skeletal dysplasias subsumed as oto-palato-digital (OPD) syndrome spectrum disorders comprising OPD1 (MIM 311300), OPD2 (MIM 304120), Melnick-Needles syndrome (MNS [MIM 309350]), and frontometaphyseal dysplasia (FMD [MIM 305620]) (Robertson et al. 2003). In contrast to PVNH, all FLNA mutations detected in OPD-spectrum disorders are missense mutations or small deletions/insertions conserving the reading frame and apparently clustering in certain domains of the gene. It was proposed that OPD-spectrum disorders result from gain of function or altered binding affinity of filamin A for unidentified protein partners (Robertson et al. 2003). This concept renders these phenotypes almost incompatible in the same individual. Exceptions to that rule, as described in this report, should therefore be of particular interest and provide further insights into FLNA mutational effects.
Figure 1.
Diagram showing the structure of the filamin A homodimer, functional domains (left), and localization of the previously described and the novel mutations (right). Adapted from Stossel et al. (2001).
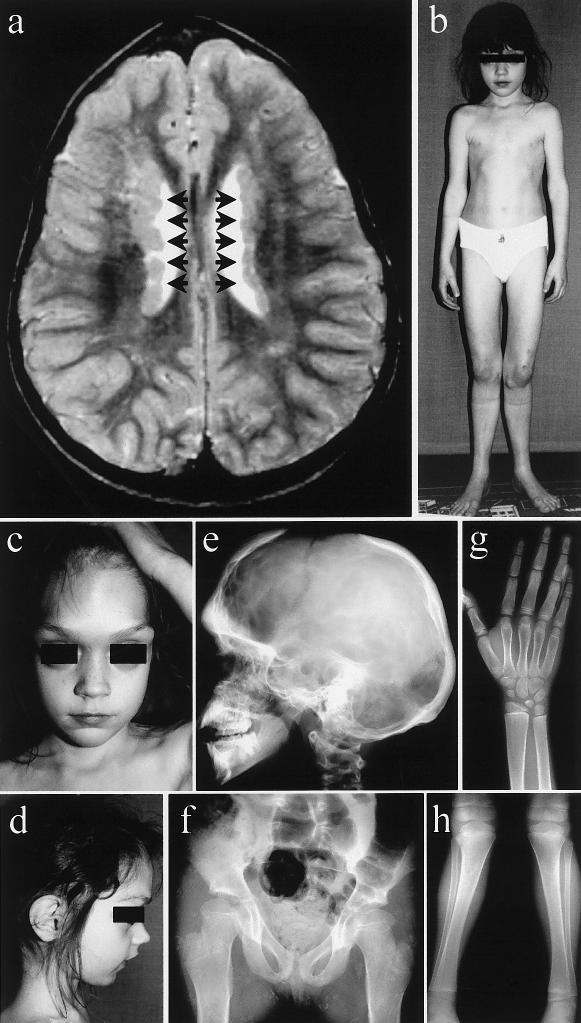
The proposita is the second daughter of healthy, unrelated parents with an unremarkable family history. Skeletal abnormalities became evident during childhood: delayed closure of the anterior fontanel, craniofacial anomalies, and marked genua valga necessitating repeated surgical corrections. Motor development and mental development were normal. Body height developed in the range of the 3rd–10th percentiles. Cranial magnetic resonance imaging (MRI) performed at age 5 years incidentally showed bilateral PVNHs (fig. 2a). The patient never had seizures. When examined first in our department at age 8 years, the girl presented with typical clinical features of FMD (fig. 2b–2d). Radiological anomalies confirmed the diagnosis of FMD (fig. 2e–2h). The patient’s mother had a normal clinical phenotype and cranial MRI.
Figure 2.
a, Cranial MRI of the patient at age 5 years, showing typical PVNHs (arrows). b–d, Clinical photographs of the patient at age 7 years: massively prominent supraorbital ridges; large-appearing eyes with hypertelorism; receding, pointed chin; slender, deformed thorax; genua valga et recurvata; and extension deficit in the elbows. e–h, Radiological findings of the patient at age 7 years. e, Skull radiograph showing increased density of the skull, with deficient pneumatization of mastoid and sinuses and a widely open anterior fontanel. f, Radiograph of the pelvis and hips, showing coxa valga et antetorta, with widened femoral necks. Radiographs of the right distal forearm and hand (g) and of the knees and lower legs (h), showing metaphyseal widening of long bones, short terminal phalanges, genua valga, and slight bending of tibiae and fibulae.
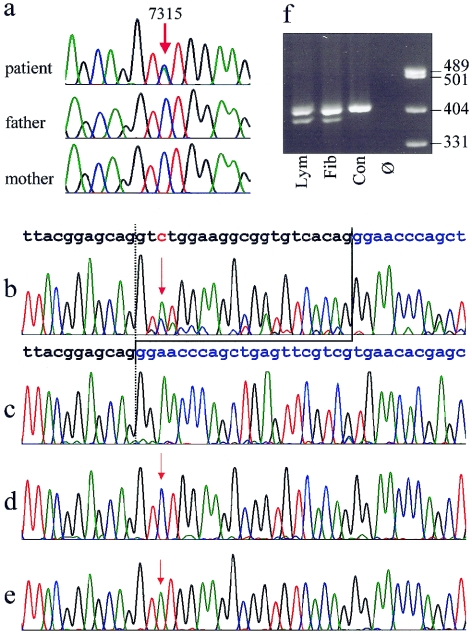
The patient’s karyotype was normal. The whole coding region of FLNA was analyzed by bidirectional direct sequencing by use of an ABI 3730 automated sequencer (Applied Biosystems). Primer sequences and PCR conditions are available on request. We identified a heterozygous mutation, 7315C→A, in exon 45, predicting a substitution of leucine by methionine (L2439M). The mutation was not present in either of the parents (fig. 3a). We confirmed paternity by testing the segregation of 16 microsatellite markers from different chromosomes (PowerPlex 16 [Promega]). PCR with allele-specific forward primers for a polymorphism in intron 45 (IVS45-64T→C [dbSNP accession number rs2070819]), for which the index patient was heterozygous, and an opposite primer designed to include the site of the mutation in exon 45 disclosed that the mutation had emerged on the paternally inherited X-chromosome (details available on request). The mutation 7315C→A was not found in 192 control chromosomes.
Figure 3.
a, Sequence traces from genomic DNA of the patient and her clinically normal parents, showing the heterozygous C→A exchange position, 7315, in the patient and absence of the mutation in both parents. b–e, Demonstration of the expression of two different aberrant FLNA transcripts in fibroblasts. b, Electropherogram derived from sequencing of the RT-PCR products from fibroblast mRNA, showing the presence of a shortened alternative transcript lacking 21 bases from exon 45. The normal sequence of the exons 45 and 46 are indicated above in black and blue, respectively. The position of the point mutation 7315C→A is depicted in red and marked by red arrows. Note that the T peak at position of the mutation is higher than the C representing the wild-type allele. Predominance of the mutant allele reflects the X-inactvation pattern. c–e, Sequences from different clones of the RT-PCR product cloned in E. coli. All three transcripts predicted from the sequencing results in panel b could be detected: 21-bp deletion (c), wild-type allele (d), and point mutation (e). f, Electrophoresis of a 409-bp RT-PCR fragment on a 2.5% agarose gel, showing presence of a shortened fragment in the patient's lymphocytes (lym) and fibroblasts (fib) but not in a normal male control (con). Ø = no template control.
We excluded larger genomic deletions of the second FLNA allele with Southern blot analysis, using PCR-derived genomic probes from the FLNA gene, as well as metaphase FISH with the cosmid probe Scos10 containing the entire FLNA gene (Patrosso et al. 1994) (details available on request). Both gave normal results. In addition, FLNA sequence analysis showed heterozygosity in the patient for known polymorphisms in the introns 5, 33, 43, and 45 and exon 36, thereby excluding deletions of the respective regions.
FLNA expression was studied on total RNA extracted from blood cells and skin fibroblasts. A 409-bp cDNA fragment containing the entire exon 45 and parts of the flanking exons was amplified by RT-PCR (details available on request). Besides confirmation of the expression of the mutant 7315C→A, the cDNA sequence indicated the presence of a second aberrant transcript lacking 21 nucleotides (fig. 3b). Analysis of sequence motifs revealed that the mutation 7315C→A creates an ectopic-splice donor site in exon 45. The consensus sequence for a 5′ splice donor is A58G78g100t100a/g96a71g84t47, in which exon nucleotides are shown in uppercase and intron nucleotides in lowercase letters, and the numbers indicate the percentage occurrence of the different nucleotides (Ketterling et al. 1999). The C→A exchange corresponds to position +3 of the newly created splicing signal AGgtatgg that matches the consensus sequence almost perfectly. The splice-prediction program Splice-View, which uses the weight matrix in accordance with the method of Shapiro and Senapathy (1987), predicts the ectopic-donor splice site with a high probability (.87). This is only slightly less than the score of .95 calculated for the authentic 5′ splice site of exon 45. Premature 5′ splicing of exon 45 predicts an in-frame deletion of seven amino acid residues in the filamin A protein (del L2439-G2445). For confirmation, RT-PCR products derived from fibroblasts were cloned in Escherichia coli by use of the TOPO TA Cloning kit (Invitrogen) and One Shot (Invitrogen) (fig. 3c–3e). In 25 successfully transfected clones, the wild-type allele and the shortened transcript (FLNAΔ7) were both found nine times, and the transcript with the point mutation (FLNAL2439M) was found seven times. In addition, presence of the shortened transcript in lymphocytes and fibroblasts was demonstrated by agarose gel electrophoresis (fig. 3f).
Since the androgen receptor gene polymorphism usually used for studying the X-inactivation pattern (Allen et al. 1992) was uninformative, we tested differential methylation of the FMR1 locus with a technique similar to one described elsewhere (Carrel and Willard 1996), as well as expression of coding SNPs within FLNA (dbSNP accession number rs2070825) and TIMP1 (dbSNP accession number rs4898) by RT-PCR. Consistently, in blood cells almost equal inactivation of both alleles was found; in fibroblasts, a distribution of roughly 70:30 in favor of the paternally inherited X-chromosome carrying the mutation was found (data not shown). This is in accordance with the RT-PCR results demonstrated in figure 3b.
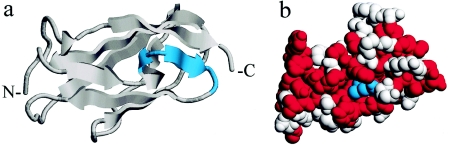
Clinical and molecular findings in this unique patient demonstrate, for the first time (to our knowledge), an overlap of PVNH and OPD-spectrum disorders, and it is tempting to assume that the combination of both phenotypes is caused by the presence of two different aberrant transcripts. In the shortened protein (FLNAΔ7), a central strand in the β-sheet complex of the immunoglobulinlike rod domain unit 23 is deleted (fig. 4a), predicting a significant disruption of its conserved structure. Moreover, owing to sterical interrelationship, the deletion might also influence structure or stability of the following hinge region or repeat 24, which is essential for dimerization (Stossel et al. 2001; van der Flier and Sonnenberg 2001). These considerations justify the assumption that the mutant FLNAΔ7 causes loss of function, which is the presumed mechanism for PVNH. Mutations, published elsewhere, in PVNH leading to loss of the most-carboxyterminal portion of the filamin A protein (Sheen et al. 2001; Moro et al. 2002) indicate that integrity of the C-terminal repeats is essential for filamin A function in neuronal migration. In contrast, the amino acid exchange in the full-length mutant (mutant FLNAL2439M) is not predicted to have significant impact on the filamin A repeat structure, but it leads to substitution of a highly conserved amino acid (fig. 5). In a structural model, the amino acid residue 2439 is imbedded in a large nonpolar surface area, representing a possible candidate for protein-protein interactions (fig. 4b). Moreover, since methionine residues are known to play important roles in recognition and binding of nonpolar surfaces in protein-protein interactions (Gellman 1991), it might be speculated that the mutant FLNAL2439M has the potential to alter binding of unidentified protein partners, in accordance with the proposed effects of mutations in the filamin A rod domain associated with OPD-spectrum disorders (Robertson et al. 2003). Functional experiments, which were out of the scope of the present study, are still needed to clarify the pathogenic mechanisms of mutations associated with OPD-spectrum disorders.
Figure 4.
Three-dimensional models of the filamin A rod domain repeat unit 23, generated on the basis of the homologous structures of the 4th (Fucini et al. 1997) and 5th (McCoy et al. 1999) module of the F-Actin Cross-Linking Gelation Factor (Abp-120) by use of Swiss-Model (Guex and Peitsch 1997; Protein Data Bank). a, Ribbon model of a repeat unit showing, in cyan, the regular position of the strand deleted in the shortened protein. Amino- and carboxyterminal ends of the repeat are marked by N- and -C, respectively. b, Model showing the respective position of the leucine residue corresponding to position 2439 depicted in cyan. It resides in a hydrophobic surface cluster. Nonpolar residues are shown in red.
Figure 5.
Alignment of the beginning of filamin A repeat 23 and filamin A homologues from various species, as well as the human paralogues filamin B and C (GenBank), showing conservation of the leucine residue at position 2439 (white frames on black background). The conserved amino acids are shown with black background and similar amino acids with a gray background. Numbers above indicate amino acid position in the filamin A protein (GenBank accession number NP_001447).
The findings that the mutation described here is not localized in the proximity of the clusters reported elsewhere (fig. 1) and that there is no skewing of X-inactivation against the mutant allele might raise concerns against its pathogenic significance with respect to FMD. The current knowledge on molecular mechanisms in FMD, however, is based on only three patients with mutations in different repeats (10 and 14) of the filamin A rod domain and on two females with documented skewing of X-inactivation (Robertson et al. 2003). On the other hand, the published data shows that negative selection against cells in which the mutant is active is not absolute in female carriers for OPD-spectrum disorders (Robertson et al. 2003) and that a less-skewed X-inactivation pattern may be related to clinical affection status (Robertson et al. 2001). In a further patient with FMD who also lacks significant skewing of X-inactivation, we recently identified a novel de novo missense mutation in repeat 15 (unpublished data). In conclusion, there is obviously no single mutation hotspot in FMD that would bring the findings presented here in substantial conflict with current pathogenic concepts.
Even if there is convincing evidence that the two aberrant transcripts have divergent functional consequences, it deserves further consideration how their coexpression in those cells, in which the mutant X-chromosome is active, can lead to the manifestation of both phenotypes. It is obvious that different tissues have very different sensitivity to specific filamin A mutants: the virtual lack of extracerebral manifestations in patients with PVNH in the presence of a random X inactivation in blood cells (Fox et al. 1998) clearly indicates that a simple loss of function of filamin A does not result in a selective disadvantage or phenotypic consequences in other tissues. In contrast, skeletal tissue is particularly sensitive to a special kind of functionally abnormal filamin A, whereas the same mutant functions normally in neuronal migration, as evidenced by the absence of cerebral features compatible with a neuronal migration defect even in male patients with OPD-spectrum disorders (Verloes et al. 2000; Robertson et al. 2003). Accordingly, we postulate that in the state of coexpression of the two mutants in skeletal tissue, the functionally defective filamin A mutant (presumably FLNAΔ7) is phenotypically silent, whereas the presence of a protein with abnormal (potentially increased) function (presumably FLNAL2439M) gives rise to the features of FMD. On the other hand, if it is true that filamin A mutants responsible for the manifestations of FMD have the potential to function normally in terms of neuronal migration, shouldn’t the mutant FLNAL2439M be able to compensate for the coexpressed functionally defective mutant in neurons and thereby rescue the neuronal phenotype? The clue may be that the initiation of neuronal migration is a process highly sensitive to filamin A dosage or its dynamic recruitment. It has been shown that a rise in intracellular filamin A availability following the decrease in activity of the filamin A-interacting protein FILIP is the critical physiological impulse for neurons to leave the ventricular zone (Nagano et al. 2002). Cells constitutionally producing a significant proportion of nonfunctional filamin A may not be able to cope with these dynamic requirements.
The present observation highlights the importance of determining the effect of point mutations on mRNA expression. Splicing can be affected by different mechanisms even if mutations are not located within the authentic splice-recognition sites (Cartegni et al. 2002). The presence of multiple aberrant transcripts due to mutations affecting 5′ splicing is not rare (Roca et al. 2003). However, the production of two aberrant transcripts, presumably leading to loss and gain of function due to a single mutational event, renders this mutation an exceptional model for mechanisms that can underlie the overlap of otherwise mutually exclusive allelic phenotypes.
Acknowledgments
We thank the family, for their participation in this study, and Angelika Diem and Andrea Eberwein, for expert technical assistance. We are grateful to Lucia Susani, Istituto di Tecnologie Biomediche Avanzate, Segrate, Italy, for providing the cosmid clone Scos10.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nih.gov/SNP/ (for IVS45-64T→C [accession number rs2070819] and coding SNPs within FLNA [accession number rs2070825] and TIMP1 [accession number rs4898])
- GenBank, http://www.ncbi.nih.gov/Genbank/ (for FLNA [accession numbers: genomic: NT_025965; mRNA: NM_001456; protein: NP_001447; GenBank: X53416], FLNA protein [Mus musculus: gi|13278531|gb|AAH04061.1], filamin A [Bos taurus: gi|18377580|gb|AAL66773.1], for filamin A [Gallus gallus: gi|15341202|dbj|BAB63943.1], filamin C [Homo sapiens: gi|4218955|gb|AAD12245.1], filamin B [H. sapiens: gi|3298597|gb|AAC39842.1], filamin1 [Drosophila melanogaster: gi|6707288|gb|AAF25614.1|AF174492_1], Filamin/ABP280 repeat family member (4B112) [Caenorhabditis elegans: gi|17543918|ref|NP_499911.1])
- Human Gene Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html (for known FLNA mutations in PVNH)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PVNH, OPD1, OPD2, MNS, and FMD)
- Protein Data Bank, http://www.rcsb.org/pdb/ (for the structures of the 4th [1KSR] and 5th [1QFH] module of the F-actin cross-linking gelation factor [Abp-120])
- Splice-View, http://l25.itba.mi.cnr.it/~webgene/wwwspliceview.html) (for calculating the probability of alternative splicing)
- Swiss-Model, http://www.expasy.org/swissmod/SWISS-MODEL.html (for generating the three-dimensional model of the 23rd domain of human filamin)
References
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW (1992) Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 51:1229–1239 [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF (1996) An assay for X inactivation based on differential methylation at the fragile X locus, FMR1. Am J Med Genet 64:27–30 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, Berkovic SF, Huttenlocher PR, Walsh CA (1998) Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21:1315–1325 [DOI] [PubMed] [Google Scholar]
- Fucini P, Renner C, Herberhold C, Noegel AA, Holak TA (1997) The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat Struct Biol 4:223–230 [DOI] [PubMed] [Google Scholar]
- Gellman SH (1991) On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry 30:6633–6666 [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PDBViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- Ketterling RP, Drost JB, Scaringe WA, Liao DZ, Liu JZ, Kasper CK, Sommer SS (1999) Reported in vivo splice-site mutations in the factor IX gene: severity of splicing defects and a hypothesis for predicting deleterious splice donor mutations. Hum Mutat 13:221–231 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Fucini P, Noegel AA, Stewart M (1999) Structural basis for dimerization of the Dictyostelium gelation factor (ABP120) rod. Nat Struct Biol 6:836–841 10.1038/12296 [DOI] [PubMed] [Google Scholar]
- Moro F, Carrozzo R, Veggiotti P, Tortorella G, Toniolo D, Volzone A, Guerrini R (2002) Familial periventricular heterotopia: missense and distal truncating mutations of the FLN1 gene. Neurology 58:916–921 [DOI] [PubMed] [Google Scholar]
- Nagano T, Yoneda T, Hatanaka Y, Kubota C, Murakami F, Sato M (2002) Filamin A-interacting protein (FILIP) regulates cortical cell migration out of the ventricular zone. Nat Cell Biol 4:495–501 [DOI] [PubMed] [Google Scholar]
- Patrosso MC, Repetto M, Villa A, Milanesi L, Frattini A, Faranda S, Mancini M, Maestrini E, Toniolo D, Vezzoni P (1994) The exon-intron organization of the human X-linked gene (FLN1) encoding actin-binding protein 280. Genomics 21:71–76 10.1006/geno.1994.1226 [DOI] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO, OPD-Spectrum Disorders Clinical Collaborative Group (2003) Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet 33:487–491 10.1038/ng1119 [DOI] [PubMed] [Google Scholar]
- Robertson SP, Walsh S, Oldridge M, Gunn T, Becroft D, Wilkie AOM (2001) Linkage of otopalatodigital syndrome type 2 (OPD2) to distal Xq28: evidence for allelism with OPD1. Am J Hum Genet 69:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Sachidanandam R, Krainer AR (2003) Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res 31:6321–6333 10.1093/nar/gkg830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen VL, Dixon PH, Fox JW, Hong SE, Kinton L, Sisodiya SM, Duncan JS, et al (2001) Mutations in the X-linked filamin 1 gene cause periventricular nodular heterotopia in males as well as in females. Hum Mol Genet 10:1775–1783 10.1093/hmg/10.17.1775 [DOI] [PubMed] [Google Scholar]
- Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS (2001) Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2:138–145 10.1038/35052082 [DOI] [PubMed] [Google Scholar]
- van der Flier A, Sonnenberg A (2001) Structural and functional aspects of filamins. Biochim Biophys Acta 1538:99–117 10.1016/S0167-4889(01)00072-6 [DOI] [PubMed] [Google Scholar]
- Verloes A, Lesenfants S, Barr M, Grange DK, Journel H, Lombet J, Mortier G, Roeder E (2000) Fronto-otopalatodigital osteodysplasia: clinical evidence for a single entity encompassing Melnick-Needles syndrome, otopalatodigital syndrome types 1 and 2, and frontometaphyseal dysplasia. Am J Med Genet 90:407–422 [DOI] [PubMed] [Google Scholar]