Abstract
We explored the phylogeography of human Y-chromosomal haplogroup E3b by analyzing 3,401 individuals from five continents. Our data refine the phylogeny of the entire haplogroup, which appears as a collection of lineages with very different evolutionary histories, and reveal signatures of several distinct processes of migrations and/or recurrent gene flow that occurred in Africa and western Eurasia over the past 25,000 years. In Europe, the overall frequency pattern of haplogroup E-M78 does not support the hypothesis of a uniform spread of people from a single parental Near Eastern population. The distribution of E-M81 chromosomes in Africa closely matches the present area of distribution of Berber-speaking populations on the continent, suggesting a close haplogroup–ethnic group parallelism. E-M34 chromosomes were more likely introduced in Ethiopia from the Near East. In conclusion, the present study shows that earlier work based on fewer Y-chromosome markers led to rather simple historical interpretations and highlights the fact that many population-genetic analyses are not robust to a poorly resolved phylogeny.
The human Y-chromosome haplogroup E is characterized by the mutations SRY4064, M96, and P29, on a background defined by the insertion of an Alu element (YAP+) (Y Chromosome Consortium 2002; Jobling and Tyler-Smith 2003). Two of the three branches of haplogroup E, the major clades E1 and E2, have been observed almost exclusively on the African continent, where their distribution has been analyzed in detail (Underhill et al. 2000; Cruciani et al. 2002). The third branch, the clade E3, defined by the mutation P2, is the only one that has also been observed in Europe and in western Asia, where it has generally been found at frequencies <25% (Hammer et al. 2000, 2001; Semino et al. 2000; Scozzari et al. 2001; Cinnioğlu et al. 2004).
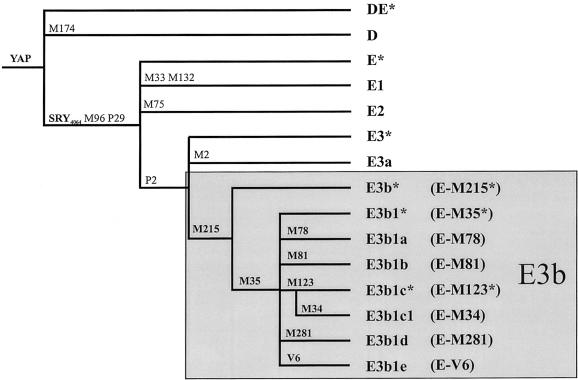
On the basis of the previously published phylogeny (Y Chromosome Consortium 2002; Jobling and Tyler-Smith 2003), the mutations M2/P1/M180, on the one hand, and M35/M215, on the other, further subdivide E3 in two monophyletic haplogroups: E3a and E3b. Both haplogroups are frequent in Africa (Underhill et al. 2000; Cruciani et al. 2002), although, to date, only E3b has also been observed in Europe (Semino et al. 2000) and western Asia (Underhill et al. 2000; Cinnioğlu et al. 2004). Recently, it has been proposed that E3b originated in sub-Saharan Africa and expanded into the Near East and northern Africa at the end of the Pleistocene (Underhill et al. 2001). E3b lineages would have then been introduced from the Near East into southern Europe by immigrant farmers, during the Neolithic expansion (Hammer et al. 1998; Semino et al. 2000; Underhill et al. 2001).
The three main subclades of haplogroup E3b (E-M78, E-M81, and E-M34) and the paragroup E-M35* are not homogeneously distributed on the African continent: E-M78 has been observed in both northern and eastern Africa, E-M81 is restricted to northern Africa, E-M34 is common only in eastern Africa, and E-M35* is shared by eastern and southern Africans (Cruciani et al. 2002). Given the strong geographic structuring observed for the four subsets of E3b within Africa, it is possible that different E3b lineages also have different frequency profiles in western Eurasia and that the evolutionary events underlying the introduction of E3b chromosomes in this area from Africa were not as simple (Rosser et al. 2000; Richards et al. 2002; Jobling and Tyler-Smith 2003) as previously proposed (Hammer et al. 1998; Semino et al. 2000; Underhill et al. 2001).
In the present study, we address the question of the origin and dispersal of haplogroup E3b subclades within and outside of Africa by analyzing 3,401 individuals from five continents. These include 1,510 individuals analyzed here for the first time for Y-chromosome markers (see also footnotes “b,” “c,” and “d” of table 1).
Table 1.
Y-Chromosome Haplogroup E Percent Frequencies in the Populations Studied
|
Frequency of Haplogroup(%) |
|||||||||||||
| Region and Population | N | E(xE3b) | E-M215* | E-M35* | E-M78a | E-M78α | E-M78β | E-M78γ | E-M78δ | E-M81 | E-M123* | E-M34 | E-V6 |
| Europe: | |||||||||||||
| Northern Portuguese | 50 | 4.0 | … | 2.0 | 4.0 | 2.0 | … | … | … | 4.0 | … | … | … |
| Southern Portuguese | 49 | 2.0 | … | … | 4.1 | 4.1 | … | … | … | 12.2 | … | … | … |
| Pasiegos from Cantabriab | 56 | … | … | 1.8 | … | … | … | … | … | 41.1 | … | … | … |
| Asturiansb | 90 | … | … | … | 10.0 | 5.6 | … | … | 4.4 | 2.2 | … | 1.1 | … |
| Southern Spaniardsb | 62 | … | … | 1.6 | 3.2 | … | … | … | 3.2 | 1.6 | … | … | … |
| Spanish Basquesb | 55 | … | … | … | … | … | … | … | … | 3.6 | … | … | … |
| Frenchb,c | 85 | … | … | … | 4.7 | 3.5 | … | … | 1.2 | 3.5 | … | … | … |
| French Basquesc | 16 | … | … | … | 6.3 | … | … | … | 6.3 | … | … | … | … |
| Corsicansb | 140 | … | … | 0.7 | 4.3 | 4.3 | … | … | … | … | … | 1.4 | … |
| Orkney Islandersc | 7 | … | … | … | … | … | … | … | … | … | … | … | … |
| Danishb | 35 | … | … | … | 2.9 | 2.9 | … | … | … | … | … | … | … |
| Northern Italiansb,c | 67 | … | … | … | 9.0 | 7.5 | … | … | 1.5 | 1.5 | … | 1.5 | … |
| Central Italiansb,c | 89 | … | … | … | 11.2 | 6.7 | … | … | 3.4 | 2.2 | … | … | … |
| Southern Italiansb | 87 | … | … | … | 11.5 | 6.9 | … | … | 2.3 | … | … | 2.3 | … |
| Siciliansb | 136 | … | … | … | 14.0 | 7.4 | 0.7 | … | 5.9 | 0.7 | … | 6.6 | … |
| Sardiniansb,c | 367 | 1.6 | … | 1.1 | 3.5 | 1.1 | 1.1 | … | 1.1 | 0.3 | … | 3.5 | … |
| Polishb | 38 | … | … | … | 2.6 | 2.6 | … | … | … | … | … | … | … |
| Estoniansb | 74 | … | … | 1.4 | 4.1 | 4.1 | … | … | … | … | … | … | … |
| Russiansd | 42 | … | … | … | … | … | … | … | … | … | … | … | … |
| Rumanians | 14 | … | … | … | 21.4 | 21.4 | … | … | … | … | … | … | … |
| Bulgarians | 116 | … | … | … | 20.7 | 19.8 | … | … | … | … | 0.9 | … | … |
| Albanians | 19 | … | … | … | 31.6 | 31.6 | … | … | … | … | … | … | … |
| Northern Africa: | |||||||||||||
| Moroccan Arabsb | 54 | 1.9 | … | … | 38.9 | … | 31.5 | 1.9 | 3.7 | 31.5 | … | … | … |
| Moyen Atlas Berbersb | 69 | 5.8 | … | … | 10.1 | … | 10.1 | … | … | 71.0 | … | … | … |
| Marrakesh Berbers | 29 | 3.4 | … | 3.4 | 6.9 | … | … | … | 3.4 | 72.4 | … | 3.4 | … |
| Mozabite Berbersc | 20 | 10.0 | … | … | … | … | … | … | … | 80.0 | … | … | … |
| Northern Egyptians | 21 | … | … | … | 28.6 | 4.8 | … | 4.8 | 19.0 | 4.8 | … | 4.8 | … |
| Southern Egyptians | 34 | … | … | … | 17.6 | … | … | … | 5.9 | … | … | … | … |
| Eastern Africa: | |||||||||||||
| Ethiopian Jewsb | 22 | 18.2 | … | 9.1 | 9.1 | … | … | 9.1 | … | … | … | 13.6 | … |
| Ethiopian Amhara | 34 | 5.9 | 5.9 | 2.9 | 8.8 | … | … | 8.8 | … | … | … | 23.5 | 14.7 |
| Ethiopian Oromo | 25 | 8.0 | … | 12.0 | 32.0 | … | … | 32.0 | … | … | … | 8.0 | 4.0 |
| Ethiopian Wolayta | 12 | 16.7 | … | 16.7 | 16.7 | … | … | 8.3 | 8.3 | … | … | 8.3 | 16.7 |
| Mixed Ethiopians | 12 | 16.7 | … | 8.3 | 33.3 | … | … | 8.3 | 25.0 | … | … | … | 8.3 |
| Somali | 23 | … | … | 17.4 | 52.2 | … | … | 47.8 | 4.3 | … | … | … | 4.3 |
| Borana (Oromo) from Kenya | 7 | 14.3 | … | 14.3 | 71.4 | … | … | 71.4 | … | … | … | … | … |
| Bantu from Kenyac | 28 | 78.6 | … | 10.7 | 3.6 | … | … | 3.6 | … | … | … | … | … |
| Nilo-Saharan from Kenya | 18 | 33.3 | … | 11.1 | 11.1 | … | … | … | 11.1 | … | … | … | 5.6 |
| Sub-Saharan Africa: | |||||||||||||
| Mandenka Senegalesec | 16 | 93.8 | … | … | 6.3 | … | … | … | … | … | … | … | … |
| Songhai from Niger | 10 | 80.0 | … | … | … | … | … | … | … | … | … | … | … |
| Tuareg from Niger | 22 | 63.6 | … | … | 4.5 | … | … | 4.5 | … | 9.1 | … | … | … |
| Fulbe from Niger | 7 | 71.4 | … | … | … | … | … | … | … | … | … | … | … |
| Fulbe from Nigeria | 32 | 100.0 | … | … | … | … | … | … | … | … | … | … | … |
| Hausa from Nigeria | 10 | 40.0 | … | … | … | … | … | … | … | … | … | … | … |
| Yoruba from Nigeriac | 21 | 90.5 | … | … | … | … | … | … | … | … | … | … | … |
| Biaka Pygmiesc | 33 | 66.7 | … | … | … | … | … | … | … | … | … | … | … |
| Mbuti Pygmiesc | 13 | 53.8 | … | … | … | … | … | … | … | … | … | … | … |
| San from Namibiac | 7 | … | … | … | … | … | … | … | … | … | … | … | … |
| Southern African !Kungb | 64 | 45.3 | … | 10.9 | … | … | … | … | … | … | … | … | … |
| Southern African Khweb | 26 | 57.7 | … | 30.8 | … | … | … | … | … | … | … | … | … |
| Southern Africa Bantuc | 8 | 75.0 | … | 12.5 | … | … | … | … | … | … | … | … | … |
| Near East: | |||||||||||||
| Sephardi Turkish | 19 | … | … | … | … | … | … | … | … | 5.3 | … | 5.3 | … |
| Istanbul Turkish | 35 | 2.9 | … | … | 8.6 | 2.9 | … | … | 5.7 | 5.7 | … | 2.9 | … |
| Southwestern Turkishb | 40 | … | … | … | 2.5 | 2.5 | … | … | … | 2.5 | … | 2.5 | … |
| Northeastern Turkishb | 41 | … | … | … | … | … | … | … | … | 2.4 | … | … | … |
| Central Anatolianb | 61 | … | … | … | 6.6 | 4.9 | … | … | 1.6 | … | … | 3.3 | … |
| Southeastern Turkishb | 24 | … | … | … | 4.2 | 4.2 | … | … | … | … | … | 4.2 | … |
| Erzurum Turkishb | 25 | … | … | … | 4.0 | … | … | … | 4.0 | … | … | 8.0 | … |
| Turkish Cypriotsb | 46 | 4.3 | … | … | 13.0 | 10.9 | … | … | 2.2 | 8.7 | … | 2.2 | … |
| Bedouinsb | 28 | 3.6 | … | … | 3.6 | … | … | … | 3.6 | 3.6 | … | 7.1 | … |
| Druze Arabsb | 28 | … | … | … | 10.7 | … | … | … | 10.7 | … | … | 3.6 | … |
| Palestiniansb | 29 | 10.3 | … | … | 10.3 | 3.4 | … | … | 6.9 | … | … | 3.4 | … |
| United Arab Emirateb | 41 | 7.3 | … | … | 2.4 | … | … | … | 2.4 | … | … | 4.9 | … |
| Omaniteb | 13 | … | … | … | 7.7 | … | … | … | 7.7 | … | … | 7.7 | … |
| Caucasus: | |||||||||||||
| Azerib | 97 | … | … | … | 2.1 | … | … | … | … | … | … | 2.1 | … |
| Adygeic | 18 | … | … | … | … | … | … | … | … | … | … | … | … |
| Pakistanic | 176 | 0.6 | … | … | 1.1 | … | … | … | 1.1 | … | … | … | … |
| Eastern Asiansb,c | 245 | … | … | … | … | … | … | … | … | … | … | … | … |
| Oceaniansc | 21 | … | … | … | … | … | … | … | … | … | … | … | … |
| Native Americansc | 43 | … | … | … | … | … | … | … | … | … | … | … | … |
E-M78 frequency includes chromosomes belonging to clusters α–δ and 14 additional chromosomes (12 chromosomes excluded from the four clusters in fig. 2B and two Azeri chromosomes for which complete microsatellite data were not available).
This sample (or a subset of it) was previously typed for a subset of the markers here analyzed (Scozzari et al. 1997, 2001; Malaspina et al. 2000, 2001; Cruciani et al. 2002).
Sample (or a subset of it) from the Human Genome Diversity Project/CEPH DNA panel (Cann et al. 2002).
This sample includes 16 DNA samples from the Human Genome Diversity Project/CEPH DNA panel and 26 previously reported samples (Scozzari et al. 2001).
All of the subjects were typed for the YAP polymorphism (Hammer and Horai 1995), and those who were YAP+ (haplogroup DE) were analyzed for the SRY4064 (Whitfield et al. 1995), M35, and M215 mutations (Underhill et al. 2000, 2001). Two subjects were found to carry the derived state at M215 and the ancestral state at M35. This modifies the topology of the E3 branch of the tree and the nomenclature of the corresponding haplogroups, as shown in figure 1 (note that “E3b” now refers to all haplogroups with the M215 derived state). Five hundred fifteen haplogroup E3b subjects were identified and further analyzed for the biallelic markers M34, M78, M81, M123, M281 (Underhill et al. 2000; Semino et al. 2002), and V6. The new V6 biallelic marker was discovered in the present survey in the TBL1Y gene by denaturing high-performance liquid chromatography analysis (primer sequences available on request). This marker identifies a subset of chromosomes previously assigned to E-M35* and now classified as “E3b1e” (fig. 1). No individual was found to carry the M281 mutation. We further typed 509 of the 515 E3b subjects for seven GATA STR (A7.1, A7.2, and A10 [White et al. 1999]; DYS19, DYS391, and DYS393 [Roewer et al. 1992, 1996]; and DYS439 [Ayub et al. 2000]) and four CA dinucleotide repeat (YCAIIa, YCAIIb, DYS413a, and DYS413b [Mathias et al. 1994; Malaspina et al. 1997]) polymorphisms. Both tetra- and dinucleotide microsatellites were used to reconstruct haplogroup-specific networks, through use of reduced-median and median-joining procedures (Bandelt et al. 1995, 1999). The seven tetranucleotide repeat polymorphisms were also used for the estimation of the time to the most recent common ancestor (TMRCA) (Goldstein et al. 1995; Slatkin 1995; Thomas et al. 1998) and the time since two populations split from a common ancestor (TD estimator [Zhivotovsky et al. 2004]). For four of the tetranucleotide loci here used, locus-specific mutation rates based on father-son transmissions (μi) are not available (Kayser et al. 2000). Since both TMRCA and TD estimations critically depend on the unknown parameter μi, we used the averaged effective mutation rate described by Zhivotovsky et al. (2004), which is based on a list of markers close to the one used here. CIs for the TMRCA were obtained as described by Scozzari et al. (2001). It should be noted that uncertainties in the mutation rate, in the shape of the genealogy, and in the mutation process would increase the CIs. Since any two chromosomes sampled from two populations have a TMRCA older than the split between populations, and since we considered as null the variance of the ancestral population at the time of its splitting, the figures reported here for the TD estimator represent upper bounds. In all of the analyses, except the networks, the YCAIIa, YCAIIb, DYS413a, and DYS413b dinucleotide repeats were not considered, since univocal assignment of phenotypic patterns to allelic series could not be obtained.
Figure 1.
Phylogenetic tree of haplogroup E3b. Markers typed in this study are in boldface letters. Haplogroups are designated according to the Y Chromosome Consortium (2002) and Jobling and Tyler-Smith (2003), by subclade and also by mutation (within parentheses). Note that the discovery of a new mutation (V6) and new samples with an intermediate haplogroup (derived state at M215 and ancestral state at M35) determine a revision in topology and nomenclature of the E3b haplogroup. Subjects belonging to paragroup DE* have recently been reported (Weale et al. 2003).
We obtained an estimate of 25.6 thousand years (ky) (95% CI 24.3–27.4 ky) for the TMRCA of the 509 haplogroup E3b chromosomes, which is close to the 30±6 ky estimate for the age of the M35 mutation reported by Bosch et al. (2001) using a different method. Several observations point to eastern Africa as the homeland for haplogroup E3b—that is, it had (1) the highest number of different E3b clades (table 1), (2) a high frequency of this haplogroup and a high microsatellite diversity, and, finally, (3) the exclusive presence of the undifferentiated E3b* paragroup.
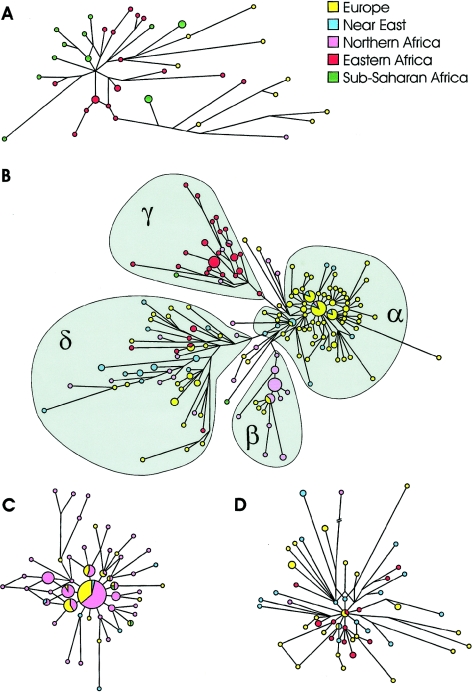
Our data show that haplogroup E3b appears as a collection of subclades with very different evolutionary histories. Haplogroup E-M78 was observed over a wide area, including eastern (21.5%) and northern (18.5%) Africa, the Near East (5.8%), and Europe (7.2%), where it represents by far the most common E3b subhaplogroup. The high frequency of this clade (table 1) and its high microsatellite diversity suggest that it originated in eastern Africa, 23.2 ky ago (95% CI 21.1–25.4 ky). The network of the E-M78 chromosomes reveals a strong geographic structuring, since each of the clusters α, β, and γ (fig. 2B) reaches high frequencies in only one of the regions analyzed. Cluster α is largely characterized by the otherwise rare nine-repeat allele at A7.1 (we found only 3 such alleles out of 800 E[xE3b1] chromosomes analyzed [present study; R.S., unpublished data]), often associated with the uncommon DYS413 24/23 pattern and its one-step neighbors. When compared with the other clusters in the network, it displays marked starlike features, with three central haplotypes accounting for 26% of the entire cluster. This cluster is very common in the Balkans (with frequencies of 20%–32%), and its frequencies decline toward western (7.0% in continental Italy, 7.4% in Sicily, 1.1% in Sardinia, 4.3% in Corsica, 3.0% in France, and 2.2% in Iberia) and northeastern (2.6%) Europe. In the Near East, this cluster is essentially limited to Turkey (3.4%). The relatively high frequency of DYS413 24/23 haplogroup E chromosomes in Greece (A.N., unpublished data) suggests that cluster α of the E-M78 haplogroup is common in the Aegean area, too.
Figure 2.
Microsatellite networks of E3b haplogroups. A, E-M35*. B, E-M78. C, E-M81. D, E-M34. Reduced-median and median-joining procedures (Bandelt et al. 1995, 1999) were applied sequentially. A haplogroup-specific weight proportional to the reciprocal of microsatellite variance was used in the construction of the networks. The E-M78 unweighted network (not shown) gave the same quadripartite structure. Unassigned chromosomes (B) showed an intermediate position between clusters α and δ in the unweighted network. Microsatellite haplotypes are represented by circles, with areas proportional to the number of individuals harboring the haplotype. Branch lengths are proportional to the number of one-step mutations separating two haplotypes.
Cluster β, characterized by the DYS413 23/21 pattern and the rare 10-repeat allele at DYS439, is common in northwestern Africa (14.0%), representing 80% of E-M78 chromosomes in that area. Outside this region, E-M78β was observed only in five European subjects.
All of the chromosomes in cluster γ (fig. 2B) are identified by the rare short 11-repeat allele at the DYS19 STR locus. We did not find this allele in >2,000 Y(xE-M78) chromosomes analyzed (present study; R.S., unpublished data), and it is reported in only 9 of 13,447 subjects analyzed for this marker in the European Y-STR reference database (Y-STR Haplotype Reference Database Web site). The cluster E-M78γ was found in eastern Africa at an average frequency of 17.7%, with the highest frequencies in the three Cushitic-speaking groups: the Borana from Kenya (71.4%), the Oromo from Ethiopia (32.0%), and the Somali (52.2%). Outside of eastern Africa, it was found only in two subjects from Egypt (3.6%) and in one Arab from Morocco.
The fourth cluster (cluster δ in fig. 2B) is present, albeit at low frequencies, in all of the regions analyzed (4.0% in eastern and northern Africa, 3.3% in the Near East, and 1.5% in Europe) and shows a notable microsatellite differentiation (fig. 2B). The two E-M78 chromosomes found in Pakistan, at the eastern borders of the area of dispersal of haplogroup E3b, also belong to cluster δ. On the basis of these data, we suggest that cluster δ was involved in a first dispersal or dispersals of E-M78 chromosomes from eastern Africa into northern Africa and the Near East. Time-of-divergence estimates for E-M78δ chromosomes suggest a relatively great antiquity (14.7±2.7 ky) for the separation of eastern Africans from the other populations. A later range expansion from the Near East or, possibly, from northern Africa would have introduced E-M78 cluster δ into Europe. However, given the low frequencies of E-M78δ, it seems to have contributed only marginally to the shaping of the present E-M78 frequency distribution in Africa and western Eurasia. Indeed, later (and previously undetected) demographic population expansions involving clusters α in Europe (TMRCA 7.8 ky; 95% CI 6.3–9.2 ky), β in northwestern Africa (5.2 ky; 95% CI 3.2–7.5 ky), and γ in eastern Africa (9.6 ky; 95% CI 7.2–12.9 ky) should be considered the main contributors to the relatively high frequency of haplogroup E-M78 in the surveyed area.
The present distributions of these clusters also suggest episodes of range expansions. Although E-M78β and E-M78γ show only modest levels of gene flow (from northern Africa to Europe and from eastern to northern Africa, respectively), the clinal frequency distribution of E-M78α within Europe testifies to important dispersal(s), most likely Neolithic or post-Neolithic. These took place from the Balkans, where the highest frequencies are observed, in all directions, as far as Iberia to the west and, most likely, also to Turkey to the southeast. Thus, it appears that, in Europe, the overall frequency pattern of the haplogroup E-M78, the most frequent E3b haplogroup in this region, is mostly contributed by a new molecular type that distinguishes it from the aboriginal E3b chromosomes from the Near East. These data are hard to reconcile with the hypothesis of a uniform spread of a single Near Eastern gene pool into southeastern Europe. On the other hand, they might be consistent with either a small-scale leapfrog migration from Anatolia into southeastern Europe at the beginning of the Neolithic or with an expansion of indigenous people in southeastern Europe in response to the arrival of the Neolithic cultural package. At the present level of phylogenetic resolution, it is difficult to distinguish between these possibilities.
E-M81 is very common in northwestern Africa, with frequencies as high as 80% (Bosch et al. 2001; Cruciani et al. 2002; present study), but its frequency sharply declines on the continent toward the east, and the haplogroup is not found in sub-Saharan Africa. The distribution of E-M81 chromosomes in Africa closely matches the present area of distribution of Berber-speaking populations on the continent, suggesting a close haplogroup–ethnic group parallelism: in northwestern Africa, the lowest frequencies for this haplogroup have been reported in two Arab-speaking Moroccan populations (31% and 52% vs. 65%–80% in six Berber speaking groups from Morocco and Algeria [Bosch et al. 2001; Cruciani et al. 2002; present study]); in Egypt, where Berbers are restricted to a few villages, E-M81 is rare (1.9%), and the southernmost finding of E-M81 chromosomes on the continent is that here reported in the Tuareg from Niger (9.1%), who also speak a Berber language. Outside of Africa, E-M81 has been observed in all the six Iberian populations surveyed, with frequencies in the range of 1.6%–4.0% in northern Portuguese, southern Spaniards, Asturians, and Basques; 12.2% in southern Portuguese; and 41.1% in the Pasiegos from Cantabria. It has been suggested (Bosch et al. 2001) that recent gene flow may have brought E3b chromosomes from northwestern Africa into Iberia, as a consequence of the Islamic occupation of the peninsula, and that such gene flow left only a minor contribution to the current Iberian Y-chromosome pool. The relatively young TMRCA of 5.6 ky (95% CI 4.6–6.3 ky) that we estimated for haplogroup E-M81 and the lack of differentiation between European and African haplotypes in the network of E-M81 (fig. 2C) support the hypothesis of recent gene flow between northwestern Africa and Iberia. In this context, our data refine the conclusions of Bosch et al. (2001) in two ways. First, not all of the E3b chromosomes in Iberia can be regarded as a signature of African gene flow into the peninsula: in our data set, 8 of 15 E-M78 chromosomes belong to cluster α, denoting gene flow from mainland Europe (see above). Second, and more importantly, the degree of the African contribution is highly variable across different Iberian populations: the proportion of haplogroup E chromosomes of African origin (E[xE3b], E-M35*, and E-M81) was <5% in three Spanish locations; 10.0% and 14.2% in northern and southern Portugal, respectively; and >40% in the Pasiegos (table 1). A relatively high frequency of E-M81 in a different sample of Pasiegos (18%) and non-Pasiegos Cantabrians (17%) has also recently been reported (Maca-Meyer et al. 2003). Such differences in the relative African contribution to the male gene pool of different Iberian populations may reflect, at least in part, the different durations of Islamic influence and introgression in different parts of the peninsula, as well as drift/founder effects for the small Pasiegos group.
The E-M123 clade was found in Ethiopia (11.2%), the Near East (3.7%), Europe (1.7%), and northern Africa (0.9%). In our data set, all the E-M123 chromosomes also carry the M34 mutation (E-M34), with the exception of one E-M123* subject from Bulgaria. This paragroup has been previously reported only in one individual from Central Asia (Underhill et al. 2000). Although the frequency distribution of E-M34 could suggest that eastern Africa was the place in which the haplogroup arose, two observations point to a Near Eastern origin: (1) Within eastern Africa, the haplogroup appears to be restricted to Ethiopia, since it has not been observed in either neighboring Somalia or Kenya (present study) or Sudan (Underhill et al. 2000). By contrast, E-M34 chromosomes have been found in a large majority of the populations from the Near East so far analyzed (Underhill et al. 2000; Cinnioğlu et al. 2004; Semino et al. 2004 [in this issue]; present study). (2) E-M34 chromosomes from Ethiopia show lower variances than those from the Near East and appear closely related in the E-M34 network (fig. 2D). If our interpretation is correct, E-M34 chromosomes could have been introduced into Ethiopia from the Near East. The high frequency of E-M34 observed for some of the Ethiopian populations could be the consequence of subsequent genetic drift, which can also explain the lower frequencies (2.3% [Underhill et al. 2000] and 4.0% [Semino et al. 2002]) reported for two large independent samples of Ethiopians. From the Near East, E-M34 chromosomes could also have been introduced into Europe, possibly by Neolithic farmers, but the paucity of E-M34 chromosomes in southeastern Europe (Semino et al. 2004 [in this issue]; present study) weakens this hypothesis. Indeed, as for E-M78δ chromosomes, introduction of E-M34 from Africa directly to southern-central Europe cannot be excluded at the present.
Haplogroup E-V6 was observed only in eastern Africa (8.9% in Ethiopia, with a single occurrence in both Somalia and Kenya), further testifying to the richness of E3b lineages in this region. Although no clear inferences can be drawn on the basis of the current E-V6 frequency distribution data, the V6 polymorphism may prove to be a useful marker for future microevolutionary studies in eastern Africa.
The paragroup E-M35* has been observed at high frequencies in both eastern (10.5%) and southern (15.2%) Africa, with rare occurrences in northern Africa and Europe (0.4% and 0.5%, respectively). The paragroup has a high microsatellite allele variance (0.63), comparable to that of the whole set of E3b(xE3b1*) chromosomes (0.53), suggesting that E-M35* is a collection of several lineages whose relationships to other E3b haplogroups remain to be established. Nevertheless, the observed distribution of E-M35* can shed light on the history of peopling of Africa. For example, we found E-M35* and E-M78 chromosomes in Bantu-speaking populations from Kenya (14.3%) but not in those living in central Africa (Cruciani et al. 2002), the area in which the Bantu expansion originated (Vansina 1984). In agreement with mtDNA data (Salas et al. 2002), this finding suggests a relevant contribution of eastern African peoples to the gene pool of the eastern Bantu. Also, the extensive interpopulation E-M35* microsatellite diversity (fig. 2A) between Ethiopians and Khoisan indicates that eastern Africans and Khoisan have been separated for a considerable period of time, as has been suggested elsewhere (Scozzari et al. 1999; Cruciani et al. 2002; Semino et al. 2002).
In conclusion, we detected the signatures of several distinct processes of migration and/or recurrent gene flow associated with the dispersal of haplogroup E3b lineages. Early events involved the dispersal of E-M78δ chromosomes from eastern Africa into and out of Africa, as well as the introduction of the E-M34 subclade into Africa from the Near East. Later events involved short-range migrations within Africa (E-M78γ and E-V6) and from northern Africa into Europe (E-M81 and E-M78β), as well as an important range expansion from the Balkans to western and southern-central Europe (E-M78α). This latter expansion was the main contributor to the present distribution of E3b chromosomes in Europe.
Acknowledgments
We would like to express our gratitude to all blood donors for their helpful collaboration, which made this study possible. We gratefully acknowledge Emanuele Guida, Jadwiga Jaruzelska, Kenneth K. Kidd, Judith R. Kidd, Damian Labuda, Jean-Paul Moisan, Valentino Romano, Laurent Varesi, Richard Villems, and the National Laboratory for the Genetics of Israeli Populations for DNA samples. We also thank two anonymous reviewers for their helpful comments. This research received support from Grandi Progetti Ateneo Università di Roma “La Sapienza” (to R.S.) and the Italian Ministry of the University (Progetti Ricerca Interesse Nazionale 2002 and 2003) (to R.S., A.N., and A.T.).
Electronic-Database Information
The URL for data presented herein is as follows:
- Y-STR Haplotype Reference Database, http://www.ystr.org/
References
- Ayub Q, Mohyuddin A, Qamar R, Mazhar K, Zerjal T, Mehdi SQ, Tyler-Smith C (2000) Identification and characterisation of novel human Y-chromosomal microsatellites from sequence database information. Nucleic Acids Res 28:e8 10.1093/nar/28.2.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between north-western Africa and the Iberian Peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, Bodmer J, et al (2002) A human genome diversity cell line panel. Science 296:261–262 10.1126/science.296.5566.261b [DOI] [PubMed] [Google Scholar]
- Cinnioğlu C, King R, Kivisild T, Kalfoğlu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA (2004) Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 114:127–148 10.1007/s00439-003-1031-4 [DOI] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW (1995) An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 15:427–441 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Karafet TM, Redd AJ, Jarjanazi H, Santachiara-Benerecetti S, Soodyall H, Zegura SL (2001) Hierarchical patterns of global human Y-chromosome diversity. Mol Biol Evol 18:1189–1203 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Redd AJ, Wood ET, Bonner MR, Jarjanazi H, Karafet T, Santachiara-Benerecetti S, Oppenheim A, Jobling MA, Jenkins T, Ostrer H, Bonne-Tamir B (2000) Jewish and Middle Eastern non-Jewish populations share a common pool of Y-chromosome biallelic haplotypes. Proc Natl Acad Sci USA 97:6769–6774 10.1073/pnas.100115997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Krüger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer N, Sánchez-Velasco P, Flores C, Larruga J-M, González A-M, Oterino A, Leyva-Cobián F (2003) Y chromosome and mitochondrial DNA characterization of Pasiegos, a human isolate from Cantabria (Spain). Ann Hum Genet 67:329–339 10.1046/j.1469-1809.2003.00045.x [DOI] [PubMed] [Google Scholar]
- Malaspina P, Ciminelli BM, Viggiano L, Jodice C, Cruciani F, Santolamazza P, Sellitto D, Scozzari R, Terrenato L, Rocchi M, Novelletto A (1997) Characterization of a small family (CAIII) of microsatellite-containing sequences with X-Y homology. J Mol Evol 44:652–659 [DOI] [PubMed] [Google Scholar]
- Malaspina P, Cruciani F, Santolamazza P, Torroni A, Pangrazio A, Akar N, Bakalli V, Brdicka R, Jaruzelska J, Kozlov A, Malyarchuk B, Mehdi SQ, Michalodimitrakis E, Varesi L, Memmi MM, Vona G, Villems R, Parik J, Romano V, Stefan M, Stenico M, Terrenato L, Novelletto A, Scozzari R (2000) Patterns of male-specific inter-population divergence in Europe, West Asia and North Africa. Ann Hum Genet 64:395–412 10.1046/j.1469-1809.2000.6450395.x [DOI] [PubMed] [Google Scholar]
- Malaspina P, Tsopanomichalou M, Duman T, Stefan M, Silvestri A, Rinaldi B, Garcia O, Giparaki M, Plata E, Kozlov AI, Barbujani G, Vernesi C, Papola F, Ciavarella G, Kovatchev D, Kerimova MG, Anagnou N, Gavrila L, Veneziano L, Akar N, Loutradis A, Michalodimitrakis EN, Terrenato L, Novelletto A (2001) A multistep process for the dispersal of a Y chromosomal lineage in the Mediterranean area. Ann Hum Genet 65:339–349 10.1046/j.1469-1809.2001.6540339.x [DOI] [PubMed] [Google Scholar]
- Mathias N, Bayés M, Tyler-Smith C (1994) Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet 3:115–123 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Torroni A, Bandelt H-J (2002) In search of geographical patterns in European mitochondrial DNA. Am J Hum Genet 71:1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roewer L, Arnemann J, Spurr NK, Grzeschik K-H, Epplen JT (1992) Simple repeat sequences on the human Y chromosome are equally polymorphic as their autosomal counterparts. Hum Genet 89:389–394 [DOI] [PubMed] [Google Scholar]
- Roewer L, Kayser M, Dieltjes P, Nagy M, Bakker E, Krawczak M, de Knijff P (1996) Analysis of molecular variance (AMOVA) of Y-chromosome-specific microsatellites in two closely related human populations. Hum Mol Genet 5:1029–1033 10.1093/hmg/5.7.1029 [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Richards M, De la Fe T, Lareu MV, Sobrino B, Sanchez-Diz P, Macaulay V, Carracedo A (2002) The making of the African mtDNA landscape. Am J Hum Genet 71:1082–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Malaspina P, Santolamazza P, Ciminelli BM, Torroni A, Modiano D, Wallace DC, Kidd KK, Olckers A, Moral P, Terrenato L, Akar N, Qamar R, Mansoor A, Mehdi SQ, Meloni G, Vona G, Cole DEC, Cai W, Novelletto A (1997) Differential structuring of human populations for homologous X and Y microsatellite loci. Am J Hum Genet 61:719–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Pangrazio A, Santolamazza P, Vona G, Moral P, Latini V, Varesi L, Memmi MM, Romano V, De Leo G, Gennarelli M, Jaruzelska J, Villems R, Parik J, Macaulay V, Torroni A (2001) Human Y-chromosome variation in the western Mediterranean area: implications for the peopling of the region. Hum Immunol 62:871–884 10.1016/S0198-8859(01)00286-5 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, Malaspina P, Torroni A, Sellitto D, Arredi B, Destro-Bisol G, De Stefano G, Rickards O, Martinez-Labarga C, Modiano D, Biondi G, Moral P, Olckers A, Wallace DC, Novelletto A (1999) Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 65:829–846 (erratum 66:346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, Lin AA , Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner JP, Zhivotovsky LA, Roy K, Torroni A, Cavalli-Sforza LL, Underhill PA, Santachiara-Benerecetti AS (2004) Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the Neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 74:1023–1034 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiæ M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA (2002) Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 70:265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origins of Old Testament priests. Nature 394:138–140 10.1038/28083 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Lahr MM, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 10.1046/j.1469-1809.2001.6510043.x [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Vansina J (1984) Western Bantu expansion. J African History 25:129–145 [Google Scholar]
- Weale ME, Shah T, Jones AL, Greenhalgh J, Wilson JF, Nymadawa P, Zeitlin D, Connell BA, Bradman N, Thomas MG (2003) Rare deep-rooting Y chromosome lineages in humans: lessons for phylogeography. Genetics 165:229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Tatum OL, Deaven LL, Longmire JL (1999) New, male-specific microsatellite markers from the human Y chromosome. Genomics 57:433–437 10.1006/geno.1999.5782 [DOI] [PubMed] [Google Scholar]
- Whitfield LS, Sulston JE, Goodfellow PN (1995) Sequence variation of the human Y chromosome. Nature 378:379–380 10.1038/378379a0 [DOI] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 74:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]