Abstract
The phylogeography of Y-chromosome haplogroups E (Hg E) and J (Hg J) was investigated in >2,400 subjects from 29 populations, mainly from Europe and the Mediterranean area but also from Africa and Asia. The observed 501 Hg E and 445 Hg J samples were subtyped using 36 binary markers and eight microsatellite loci. Spatial patterns reveal that (1) the two sister clades, J-M267 and J-M172, are distributed differentially within the Near East, North Africa, and Europe; (2) J-M267 was spread by two temporally distinct migratory episodes, the most recent one probably associated with the diffusion of Arab people; (3) E-M81 is typical of Berbers, and its presence in Iberia and Sicily is due to recent gene flow from North Africa; (4) J-M172(xM12) distribution is consistent with a Levantine/Anatolian dispersal route to southeastern Europe and may reflect the spread of Anatolian farmers; and (5) E-M78 (for which microsatellite data suggest an eastern African origin) and, to a lesser extent, J-M12(M102) lineages would trace the subsequent diffusion of people from the southern Balkans to the west. A 7%–22% contribution of Y chromosomes from Greece to southern Italy was estimated by admixture analysis.
It has been proposed that the observed decreasing frequency gradients of Y-chromosome superhaplogroups E (Hg E) (defined by the SRY4064 mutation) and J (Hg J) (characterized by the 12f2a-8kb allele) (Semino et al. 1996; Hammer et al. 1998; Rosser et al. 2000) reached southwestern Europe as a result of demic expansions of Neolithic agriculturalists from the Middle East (Semino et al. 1996; Hammer et al. 1998). The spatial frequency patterns of Hg E and Hg J, at this level of molecular resolution, accommodate both infiltrations of Neolithic agriculturalists into southwestern Europe and cultural adaptations in western and northern Europe by indigenous Mesolithic peoples. This is consistent with the Neolithic migration hypothesis (Ammerman and Cavalli-Sforza 1984; Cavalli-Sforza 2002). However, this first-order level of molecular resolution does not readily reflect apparent complexities in regional and local archaeological sequences. The archaeological records suggest that the large-scale clinal patterns of Hg E and Hg J reflect a mosaic of numerous small-scale, more regional population movements, replacements, and subsequent expansions overlying previous ranges. The recent findings of many biallelic markers, which subdivide these two haplogroups, give us the opportunity to investigate the contribution of different population movements that have spread Hg E and Hg J. Through analysis of the Alu insertion (YAP), the M174 and SRY4064 mutations, and the 12f2a deletion, we identified haplogroups D (YAP/M174), E (YAP/SRY4064), and J (12f2a) Y chromosomes in >2,400 males from 29 populations, mainly from Europe and the Mediterranean area but also from Africa and Asia. No subject belonged to the recently reported paragroup DE* (Weale at al. 2003), and only 6 belonged to the Asian-specific Hg D, whereas 501 were members of Hg E and 445 of Hg J. The survey of 36 biallelic markers in the Hg E and Hg J Y chromosomes allowed us to define the phylogenetic relationships of their numerous subclades (figs. 1 and 2) and to analyze their distributions in the various geographic areas (tables 1 and 2). In addition, the survey of eight microsatellites (figs. 3 and 4) in a subset of these samples allowed investigation of the relative dating of different subclades.
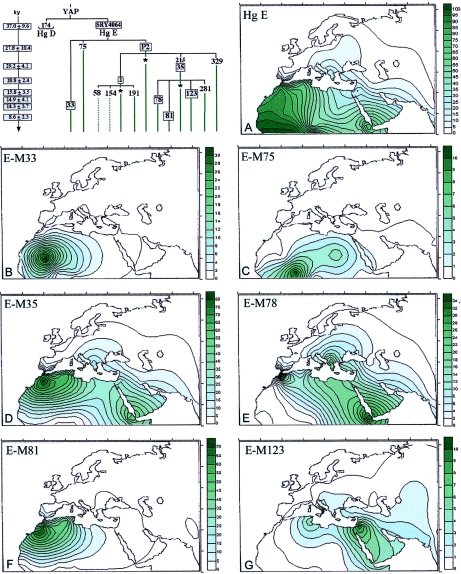
Figure 1.
Phylogeny and frequency distributions of Hg E and its main subclades (panels A–G). The numbering of mutations is according to the Y Chromosome Consortium (YCC) (YCC 2002; Jobling and Tyler-Smith 2003). To the left of the phylogeny, the ages (in 1,000 years) of the boxed mutations are reported, with their SEs (Zhivotovsky et al. 2004). Because the procedure used is based on STR data, it actually estimates the ages of STR variation observed within the corresponding haplogroup in the studied populations. With the exception of the value relative to SRY4064 mutation, which as been calculated as TD (with V0=0) between the sister clades Hg E-P2 and Hg E-M33, the other values were estimated as the average squared difference (ASD) in the number of repeats between all current chromosomes of a sample and the founder haplotype, which has an expected value μt for single-step mutations (Thomas et al. 1998) and wt for a general mutation scheme, where w is an average effective mutation rate at the loci, taken as 6.9×10-4 per 25 years (Zhivotovsky et al. 2004) (microsatellite data available on request). In some cases, because of small sample sizes or long time passed since the occurrence of the mutation, the founder haplotype could not be reliably estimated as a modal haplotype. Therefore, we constructed it from modal alleles at single loci, although this can underestimate the age if the candidate founder haplotype differs from the real one. To make the computation of the P2 and M35 ages independent from those of their most-represented subclades, the STR variation observed at only the “asterisk” lineages (e.g., E-P2*) has been used. The M35 estimate is in agreement with those of Bosch et al. (2001) and Cruciani et al. (2004 [in this issue]), obtained with different methods. The YAP insertion was studied as an amplified fragment-length polymorphism (Hammer and Horai 1995). The other mutations were investigated in a hierarchical order by use of the denaturing high-performance liquid chromatography (DHPLC) methodology (Underhill et al. 2001). Subhaplogroups observed in this study are illustrated by continuous lines, whereas subhaplogroups discussed elsewhere are indicated by dotted lines. For simplicity, the prefix “M” was omitted from the name of the marker mutations. Haplogroup-frequency surfaces were graphically computer reconstructed following the Kringing procedure (Delfiner 1976) by use of the Surfer System (Golden Software) and the data reported in table 1.
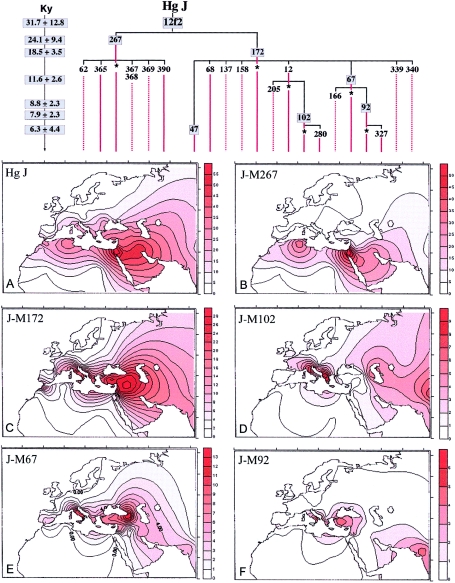
Figure 2.
Phylogeny and frequency distributions of Hg J and its main subclades (panels A–F). The numbering of mutations is according to the YCC (YCC 2002; Jobling and Tyler-Smith 2003). To the left of the phylogeny, the ages (in 1,000 years) of the boxed mutations are reported, with their SEs (Zhivotovsky et al. 2004). With the exception of the age relative to the 12f2 mutation, which has been estimated as TD (with V0=0) between the combined data of the two sister clades Hg J-M267 and Hg J-M172, the other values have been determined as ASD, as described in figure 1. The 12f2a marker was examined as an RFLP by Southern blotting (Passarino et al. 1998); the other mutations were investigated in hierarchical order by use of DHPLC methodology (Underhill et al. 2001). Three new mutations, M327, M280, and M390, were found in this study. M327 is a T→C transition at np 404 within the STS containing mutation M92, M280 is a G→A transition at np 330 within the STS containing the mutation M67, and M390 is an A insertion after nt 175 in the STS containing the M365 mutation. Conventions used are the same as for figure 1. The frequency surfaces were drawn using the data reported in table 2 and, for Hg J (panel A), also the data from Rosser et al. (2000), Quintana-Murci et al. (2001), and Scozzari et al. (2001).
Table 1.
Population Frequencies of Hg E and Its Subclades
|
Hg E |
Frequency of E Subhaplogroupb |
Hg D |
|||||||||||||||
| Population/Regiona | No. | % | 2*c | 58 | 191 | 154 | P2* | 329 | 35* | 123 | 78 | 81 | 281 | 33 | 75 | No. | % |
| Arab (Morocco)d (49) | 37 | 75.5 | 42.9 | 32.6 | |||||||||||||
| Arab (Morocco)e (44) | 32 | 72.7 | 6.8 | 2.3 | 11.4 | 52.3 | |||||||||||
| Berber (Morocco)d (64) | 55 | 85.9 | 4.7 | 10.9 | 68.7 | 1.6 | |||||||||||
| Berber (north-central Morocco)e (63) | 55 | 87.3 | 9.5 | 7.9 | 1.6 | 65.1 | 3.2 | ||||||||||
| Berber (southern Morocco)e (40) | 35 | 87.5 | 2.5 | 7.5 | 12.5 | 65.0 | |||||||||||
| Saharawish (North Africa)e (29) | 24 | 82.7 | 3.4 | 75.9 | 3.4 | ||||||||||||
| Algerian (32) | 21 | 65.6 | 3.1 | 3.1 | 6.3 | 53.1 | |||||||||||
| Tunisian (58) | 32 | 55.2 | 3.4 | 3.4 | 5.2 | 15.5 | 27.6 | ||||||||||
| Malif (44) | 37 | 84.1 | 20.5 | 29.5 | 34.1 | ||||||||||||
| Burkina Fasod (106) | 105 | 99.1 | 67.9 | 1.9 | 13.2 | .9 | 3.8 | 11.3 | |||||||||
| North Cameroond (152) | 69 | 45.4 | 20.3 | 12.5 | 1.3 | 7.9 | 3.3 | ||||||||||
| South Cameroond (89) | 83 | 93.3 | 43.8 | 40.4 | 9.0 | ||||||||||||
| Senegaleseg (139) | 136 | 97.8 | 80.6 | .7 | 2.9 | 5.0 | .7 | .7 | 5.0 | 2.9 | |||||||
| Bantu (South Africa)f (53) | 44 | 83.0 | 54.7 | 5.7 | 3.8 | 1.9 | 1.9 | 15.1 | |||||||||
| Khoisan (South Africa)d (90) | 59 | 65.6 | 31.1 | 11.1 | 1.1 | 16.7 | 5.6 | ||||||||||
| Sudanf (40) | 12 | 30.0 | 17.5 | 5.0 | 2.5 | 5.0 | |||||||||||
| Ethiopian (Oromo)g (78) | 62 | 79.5 | 12.8 | 2.6 | 19.2 | 5.1 | 35.9 | 2.6 | 1.3 | ||||||||
| Ethiopian (Amhara)g (48) | 22 | 45.8 | 10.4 | 10.4 | 2.1 | 22.9 | |||||||||||
| Iraqi (218) | 20 | 9.2 | .9 | 2.8 | 5.5 | ||||||||||||
| Lebanese (42) | 8 | 19.0 | 4.8 | 11.9 | 2.4 | ||||||||||||
| Ashkenazim Jewish (77) | 14 | 18.2 | 1.3 | 11.7 | 5.2 | ||||||||||||
| Sephardim Jewish (40) | 12 | 30.0 | 2.5 | 10.0 | 12.5 | 5.0 | |||||||||||
| Turkish (Istanbul) (46) | 6 | 13.0 | 2.2 | 8.7 | 2.2 | ||||||||||||
| Turkish (Konya) (117) | 17 | 14.5 | 1.7 | 12.8 | 1 | .9 | |||||||||||
| Georgian (41) | 0 | .0 | |||||||||||||||
| Balkarian (southern Caucasus) (39) | 1 | 2.6 | 2.6 | ||||||||||||||
| Northern Greek (Macedonia) (59) | 12 | 20.3 | 1.7 | 18.6 | |||||||||||||
| Greek (84) | 20 | 23.8 | 2.4 | 21.4 | |||||||||||||
| Albanian (44) | 11 | 25.0 | 25.0 | ||||||||||||||
| Croatian (57) | 5 | 8.8 | 1.8 | 7.0 | |||||||||||||
| Hungarian (53) | 5 | 9.4 | 1.9 | 7.5 | |||||||||||||
| Ukrainian (93) | 8 | 8.6 | 1.1 | 7.5 | |||||||||||||
| Polish (99) | 4 | 4.0 | 4.0 | ||||||||||||||
| Italian (north-central Italy) (56) | 6 | 10.7 | 10.7 | ||||||||||||||
| Italian (Calabria 1) (80) | 18 | 22.5 | 1.3 | 2.5 | 16.3 | 1.3 | 1.3 | ||||||||||
| Italian (Calabria 2)h (68) | 16 | 23.5 | 1.5 | 13.2 | 5.9 | 2.9 | |||||||||||
| Italian (Apulia) (86) | 12 | 13.9 | 2.3 | 11.6 | |||||||||||||
| Italian (Sicily) (55) | 15 | 27.3 | 5.5 | 3.6 | 12.7 | 5.5 | |||||||||||
| Italian (Sardinia) (139) | 7 | 5.0 | .7 | 1.4 | 2.9 | ||||||||||||
| Dutch (34) | 0 | .0 | |||||||||||||||
| Bearnais (27) | 1 | 3.7 | 3.7 | ||||||||||||||
| French Basque (45) | 0 | .0 | |||||||||||||||
| Spanish Basque (48) | 1 | 2.1 | 2.1 | ||||||||||||||
| Catalan (33) | 2 | 6.1 | 3.0 | 3.0 | |||||||||||||
| Andalusian (76) | 7 | 9.2 | 3.9 | 5.3 | |||||||||||||
| Andalusiane (37) | 4 | 10.8 | 2.7 | 2.7 | 5.4 | ||||||||||||
| Hindu (India) (47) | 0 | .0 | |||||||||||||||
| Tharu (Nepal) (98) | 0 | .0 | 4 | 4.1 | |||||||||||||
| Chinese (65) | 0 | .0 | 1 | 1.5 | |||||||||||||
Numbers in parentheses indicate the number of Y chromosomes analyzed. The population samples include those reported by Semino et al. (2000, 2002), Passarino et al. (1998), and Al-Zahery et al. (2003).
An asterisk (*) indicates chromosomes that belong to a clade but not its subclades.
The clade 2* also includes the subhaplogroups classified elsewhere as M116.1, M155 (Underhill et al. 2000), M10, and M149 (Cruciani et al. 2002).
Data from Cruciani et al. (2002) and F. Cruciani, personal communication.
Data from Bosch et al. (2001).
Data from Underhill et al. (2000).
Data from Semino et al. (2002).
The sample “Calabria 2” refers to the Albanian community of the Cosenza province (Torroni et al. 1990).
Table 2.
Population Frequencies of Hg J and Its Subclades
|
Frequency of J subhaplogroupb |
|||||||||||||||||
|
Hg J |
M172 |
M267c |
|||||||||||||||
| Population/Regiona | No. | % | 172* | 158 | 12* | 102* | 280 | 47 | 67* | 92* | 327 | 68 | Total % | 267* | 62 | 365 | 390 |
| Arab (Morocco)d (49) | 20 | 20.4 | 10.2 | 10.2 | 10.2 | ||||||||||||
| Arab (Morocco)e (44) | 7 | 15.9 | 2.3 | 13.6 | |||||||||||||
| Berber (Morocco)d (64) | 4 | 6.3 | 6.3 | ||||||||||||||
| Berber (Morocco)e (103) | 11 | 10.7 | 2.9 | 7.8 | |||||||||||||
| Saharawish (North Africa)e (29) | 5 | 17.2 | 17.2 | ||||||||||||||
| Algerian (20) | 7 | 35.0 | 35.0 | ||||||||||||||
| Tunisian (73) | 25 | 34.2 | 1.4 | 1.4 | 1.4 | 4.1 | 30.1 | ||||||||||
| Sudanf (40) | 0 | .0 | |||||||||||||||
| Ethiopian (Amhara) (48) | 17 | 35.4 | 2.1 | 2.1 | 33.3 | ||||||||||||
| Ethiopian (Oromo) (78) | 3 | 3.8 | 1.3 | 1.3 | 2.6 | ||||||||||||
| Iraqi (156) | 79 | 50.6 | 10.2 | 2.6 | 2.6 | 4.5 | 1.3 | 1.3 | 22.4 | 28.2 | |||||||
| Lebanese (40) | 15 | 37.5 | 20.0 | 2.5 | 2.5 | 25.0 | 10.0 | 2.5 | |||||||||
| Muslim Kurdg (95) | 38 | 40.0 | 28.4 | 11.6 | |||||||||||||
| Palestinian Arabg (143) | 79 | 55.2 | 16.8 | 38.4 | |||||||||||||
| Bedouing (32) | 21 | 65.6 | 3.1 | 62.5 | |||||||||||||
| Ashkenazim Jewish (82) | 31 | 37.8 | 12.2 | 1.2 | 4.9 | 4.9 | 23.2 | 14.6 | |||||||||
| Sephardim Jewish (42) | 17 | 40.5 | 23.8 | 2.4 | 2.4 | 28.6 | 11.9 | ||||||||||
| Turkish (Istanbul) (73) | 18 | 24.7 | 11.0 | 2.7 | 4.1 | 17.8 | 5.5 | 1.4 | |||||||||
| Turkish (Konya) (129) | 41 | 31.8 | 17.8 | .8 | .8 | 3.1 | 4.6 | .8 | 27.9 | 3.1 | .8 | ||||||
| Georgian (45) | 15 | 33.3 | 8.9 | 2.2 | 13.3 | 2.2 | 26.7 | 4.4 | 2.2 | ||||||||
| Balkarian (southern Caucasus) (16) | 4 | 25.0 | 12.5 | 6.3 | 6.3 | 25.0 | |||||||||||
| Northern Greek (Macedonia) (56) | 8 | 14.3 | 3.6 | 5.4 | 3.6 | 12.5 | 1.8 | ||||||||||
| Greek (92) | 21 | 22.8 | 4.3 | 6.5 | 2.2 | 4.3 | 3.3 | 20.6 | 2.2 | ||||||||
| Albanian (56) | 13 | 23.2 | 14.3 | 3.6 | 1.8 | 19.6 | 3.6 | ||||||||||
| Croatian (48) | 3 | 6.2 | 6.2 | 6.2 | |||||||||||||
| Hungarian (49) | 1 | 2.0 | 2.0 | 2.0 | |||||||||||||
| Ukrainian (82) | 6 | 7.3 | 2.4 | 2.4 | 1.2 | 1.2 | 7.3 | ||||||||||
| Polish (97) | 1 | 1.0 | 1.0 | 1.0 | |||||||||||||
| Italian (north-central Italy) (52) | 14 | 26.9 | 5.8 | 9.6 | 9.6 | 1.9 | 26.9 | ||||||||||
| Italian (Calabria 1) (57) | 14 | 24.6 | 14.0 | 1.8 | 3.5 | 3.5 | 22.8 | 1.8 | |||||||||
| Italian (Calabria 2)h (45) | 9 | 20.0 | 4.4 | 8.9 | 6.6 | 20.0 | |||||||||||
| Italian (Apulia) (86) | 27 | 31.4 | 16.3 | 3.5 | 2.3 | 7.0 | 29.1 | 2.3 | |||||||||
| Italian (Sicily) (42) | 10 | 23.8 | 11.9 | 2.4 | 2.4 | 16.7 | 7.1 | ||||||||||
| Italian (Sardinia) (144) | 18 | 12.5 | 2.8 | 2.1 | 2.8 | 2.1 | 9.7 | 2.8 | |||||||||
| Dutch (34) | 0 | .0 | |||||||||||||||
| Bearnais (26) | 2 | 7.7 | 3.8 | 3.8 | 7.7 | ||||||||||||
| French Basque (44) | 6 | 13.6 | 13.6 | 13.6 | |||||||||||||
| Spanish Basque (48) | 0 | .0 | |||||||||||||||
| Catalan (28) | 1 | 3.6 | 3.6 | 3.6 | |||||||||||||
| Andalusian (93) | 8 | 8.6 | 2.2 | 1.1 | 3.2 | 1.1 | 7.5 | 1.1 | |||||||||
| Hunza (Pakistan)f (38) | 5 | 13.2 | 2.6 | 7.9 | 10.5 | 2.6 | |||||||||||
| Pakistan-Indiaf (88) | 21 | 23.9 | 3.4 | 1.1 | 2.3 | 3.4 | 1.1 | 4.5 | 15.9 | 7.9 | |||||||
| Hindu (India) (76) | 4 | 5.3 | 2.6 | 1.3 | 1.3 | 5.3 | |||||||||||
| Tharu (Nepal) (50) | 7 | 14.0 | 8.0 | 6.0 | 14.0 | ||||||||||||
| Central Asiaf (184) | 40 | 21.7 | 6.5 | .5 | 2.2 | .5 | 1.1 | .5 | .5 | 11.9 | 9.2 | .5 | |||||
| Chinese (65) | 0 | .0 | |||||||||||||||
Numbers in parentheses indicate the number of Y chromosomes analyzed. The population samples include those reported by Santachiara-Benerecetti et al. (1993), Semino et al. (1996, 2000, 2002), Passarino et al. (1998), and Al-Zahery et al. (2003).
An asterisk (*) indicates chromosomes that belong to a clade but not its subclades.
All chromosomes classified as J* (because of not belonging to J-M172) by Cruciani et al. (2002), Nebel et al. (2001), and Bosch et al. (2001) were considered members of J-M267*.
Data from Cruciani et al. (2002).
Data from Bosch et al. (2001). These samples were not subclassified and are reported only in the “M172 Total” column.
Data from Underhill et al. (2000).
Data from Nebel et al. (2001).
The sample “Calabria 2” refers to the Albanian community of the Cosenza province (Torroni et al. 1990).
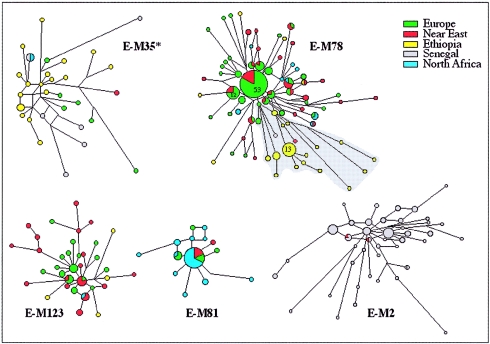
Figure 3.
Networks of the STR haplotypes of the main subhaplogroups of Hg E. These networks were obtained by the analysis of a subset of the samples for the following microsatellites: YCAIIa, YCAIIb (Mathias et al. 1994), DYS19, DYS389, DYS390, DYS391, and DYS392 (Roewer et al. 1996). The phylogenetic relationships between the microsatellite haplotypes were determined using the program NETWORK 2.0b (Fluxus Engineering). Networks were calculated by the median-joining method (ɛ=0) (Bandelt et al. 1995), weighting the STR loci according to their relative variability in Hg E and, with the exception of E-M81, after having processed the data with the reduced-median method. Circles represent the microsatellite haplotypes. Unless otherwise indicated by a number on the pie chart, the area of the circles and the area of the sectors are proportional to the haplotype frequency in the haplogroup and in the geographic area indicated by the color. The smallest circle of each network corresponds to one Y chromosome. The shaded area in E-M78 indicates the branch characterized by the DYS392-12 allele.
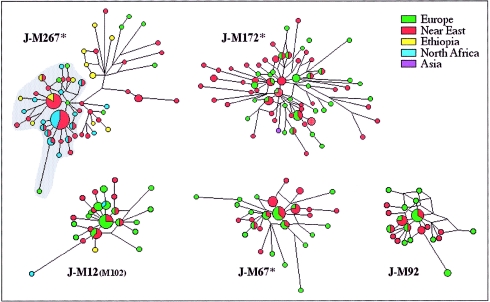
Figure 4.
Network of the STR haplotypes of the main subhaplogroups of Hg J. These networks were obtained by the analysis of a subset of the samples for the following microsatellites: YCAIIa, YCAIIb (Mathias et al. 1994), DYS388 (Thomas et al. 1999), DYS19, DYS389, DYS390, DYS391, and DYS392 (Roewer et al. 1996), by the same procedures used for Hg E (fig. 3). Apart from the YCAII system in Hg J-M267, which was considered as a stable marker in this haplogroup (see text), the STR loci were weighted according to their relative variability in Hg J. The most complex networks, J-M267* and J-M172*, were calculated by the median-joining method (ɛ=0) on the preprocessed data with the reduced-median method; the other networks were calculated by using only the reduced-median algorithm. The shaded area in J-M267* indicates the branch characterized by the YCAIIa-22/YCAIIb-22 motif. For the areas of the circles and the sectors, see figure 3. The expansion time of this branch was calculated using TD (Zhivotovsky 2001), which gives 8.7 and 4.3 ky, respectively, for the earliest and the latest bounds of the expansion time. The former estimate was calculated by using the variance in the number of repeats of the remaining six loci, assuming a variance at the beginning of population separation (V0) equal to zero, and thus gives an upper bound for the TD (Zhivotovsky 2001). The latter assumes a linear approximation of the within-population variance in repeat scores as a function of time and takes a predicted value of V0 prior to population split; because the linearity can be achieved in a case of infinite population size only and because each survived haplogroup started from one individual and could maintain small size for a long time, the linear approximation overestimates V0 and thus might be considered as a lower bound for divergence times (L.A.Z., unpublished method).
Hg E (fig. 1A) is observed in Africa, Europe, and the Near East and includes the subhaplogroups E-M33, E-M75, and the most widespread subclade, E-P2. The latter includes three clusters, two of which, E-M2 and E-M35, are the most widespread. Haplogroups E-M33 (fig. 1B), E-M75 (fig. 1C), and the not-shown E-P2* and E-M2 are virtually absent in European populations and appear to be geographically restricted to sub-Saharan Africa. The E-P2* lineages were observed mainly in Ethiopians, whereas E-M2, which is considered a signature of the Bantu expansion (Hammer et al. 1998; Passarino et al. 1998; Scozzari et al. 1999), shows its highest frequency (>80%) in Senegal and has been sporadically observed in North Africa and Iraq. E-M35 (fig. 1D) has been found in Africa, the Near East, and Europe, where it is believed to have arrived in Neolithic times (Hammer et al. 1998; Semino et al. 2000). In particular, from among its subgroups, E-M78 (fig. 1E) is present in Europe, the Middle East, and North and East Africa. However, whereas no preferential YCAII microsatellite motif is observed in the Middle East, prevalent associations with YCAIIa21-YCAIIb19 in Europe and YCAIIa22-YCAIIb19 in Africa are found. E-M81 (fig. 1F) is almost absent in Europe (with the exception of Sicily and Iberia) and the Middle East but characterizes the majority of the Y chromosomes of populations from northwestern Africa. E-M123 (fig. 1G) is spread in the Near East and is also observed in North Africa and Europe but does not reach the western European regions. E-M281 and E-M329 are geographically restricted, having been seen only in Ethiopians (two subjects each). The remaining 37 E-M35* Y chromosomes were found mainly in Africa, with a high frequency in the Ethiopians and the Khoisan.
Both phylogeography and microsatellite variance suggest that E-P2 and its derivative, E-M35, probably originated in eastern Africa. This inference is further supported by the presence of additional Hg E lineal diversification and by the highest frequency of E-P2* and E-M35* in the same region. The distribution of E-P2* appears limited to eastern African peoples. The E-M35* lineage shows its highest frequency (19.2%) in the Ethiopian Oromo but with a wider distribution range than E-P2*. Indeed, it is also found at high frequency (16.7%) in the Khoisan of South Africa (Underhill et al. 2000; Cruciani et al. 2002) (suggesting, once again, their ancient relationship with Ethiopians) and observed in southern Europe (present study). It is interesting that both E-P2* and E-M35* and their derivatives, E-M78 and E-M123, exhibit in Ethiopians the 12-repeat allele at the DYS392 microsatellite locus, an allele scarcely seen (Y-Chromosome STR Database), especially in other haplogroups and other populations (A.S.S.-B., unpublished data). In addition, the Ethiopian DYS392-12 allele is usually associated with the unusually short DYS19-11 allele, which is typical of this area. These findings are not easily explained. One possible scenario is that an ancient differentiation of the E-P2 haplogroup occurred in loco (East Africa). However, this also implies a low mutability of the associated microsatellite motif (DYS392-12/DYS19-11). Alternatively, the microsatellite motif may be due to homoplasy.
The first scenario is more likely, since this unique microsatellite haplotype occurs in E-P2*, E-M35*, and E-M78 but is almost absent in all other haplogroups and populations. In addition, the high stability of the DYS392 locus (Brinkmann et al. 1998; Nebel et al. 2001) and of the shorter alleles of DYS19 (Carvalho-Silva et al. 1999) has been reported elsewhere. Moreover, the observation that the derivative E-M78 displays the DYS392-12/DYS19-11 haplotype suggests that it also arose in East Africa. This is illustrated by the microsatellite network (fig. 3, shaded area), which reveals that the Ethiopian branch harboring DYS392-12 is not shared with either Near Eastern or European populations. The very low frequency of E-M123 in Ethiopia does not allow any inferences about the origin of this clade. The network of E-M78 and that of E-M123 are in agreement with the hypothesis of their ancient presence in the Near East and their subsequent expansion into the southern Balkans. The divergence time (TD) (Zhivotovsky 2001) between the Near East and European lineages has been estimated to a range of 7–14 thousand years (ky) ago. Cinnioğlu et al. (2004) found a high degree of variance of E-M123 in Turkey, which has been interpreted as being due to multiple founders rather than a single early dispersal event that has remained geographically circumscribed. E-M81 has the lowest variance and a compact network (fig. 3), indicating either its relatively recent origin followed by expansion or its recent expansion after a bottleneck. In Europe, this clade is restricted to the southernmost regions, such as Iberia and Sicily, and the absence of microsatellite variation suggests a very recent arrival from North Africa, consistent with previous observations (Bosch et al. 2001). The frequency pattern and the microsatellite network of E-M2(xM191) (fig. 3) indicate a West African origin followed by expansion, a result that is in agreement with the findings of Cruciani et al. (2002).
The 12f2a mutation, which characterizes haplogroup J, was observed in 445 subjects. Hg J harbors two main clades (see phylogeny in fig. 2), J-M267 (Cinnioğlu et al. 2004) and J-M172. J-M172 is the more frequent and currently differentiates into eight subhaplogroups defined by mutations M12/M102, M47, M67/M92, M68, M137, M158, M339, and M340, four of which occur at informative frequencies. The less-heterogeneous clade J-M267 includes all of the other 12f2a Y chromosomes that were reported elsewhere as Eu10 (Semino et al. 2000). Its current level of subdivision includes five scarcely represented subclusters defined by mutations M62, M365, M367/M368, and M369 (Cinnioğlu et al. 2004) and by the new mutation M390. Similar to Hg E, different geographic distributions are displayed by the various subhaplogroups of J (fig. 2). J-M172 (fig. 2C), which occurs as frequently as J-M267 (fig. 2B) in some Middle Eastern populations, is the more prevalent in Europe. Among its subclades, J-M137, J-M158, J-M339, and J-M340 were reported elsewhere as single observations (Underhill et al. 2000; Cinnioğlu et al. 2004) and have not been observed in this study. Likewise, J-M47 and J-M68 characterize very few Near Eastern and Asian samples. However, J-M12 and J-M67 and their derivatives are informative, being diffused in Europe and observed also in Asia. J-M12 is almost totally represented by its sublineage J-M102, which shows frequency peaks in both the southern Balkans and north-central Italy (fig. 2D). The history underlying this apparent affinity remains uncertain. J-M67 (fig. 2E) includes J-M67* lineages (not shown), which are most frequent in the Caucasus, and J-M92, which indicates affinity between Anatolia and southern Italy (fig. 2F). Finally, the J-M172* lineages display a decreasing frequency gradient from the Near East toward western Europe and strongly contribute to the overall gradient of Hg J. J-M267 is notable, since this haplogroup shows its highest frequencies in the Middle East, North Africa, and Ethiopia (fig. 2B) and its lowest in Europe, having been observed only in the Mediterranean area. Of its five subhaplogroups, only two have been observed: the J-M365 (in two Turks and one Georgian) and the new subclade J-M390 (in one Lebanese).
The extent of differentiation of Hg J, observed both with the biallelic and microsatellite markers, points to the Middle East as its likely homeland. In this area, J-M172 and J-M267 are equally represented and show the highest degree of internal variation, indicating that it is most likely that these subclades also arose in the Middle East. However, their different frequencies in different Middle Eastern countries and in Europe suggest distinct demography processes, possibly in population groups that underwent different temporal expansions. This is especially true for J-M172. The majority of its lineages are undifferentiated and thus potentially paraphyletic (fig. 4). Although J-M172* encompasses most of the M172 Y chromosomes in continental Europe and India (Kivisild et al. 2003; present study), their degree of affinity and shared history remain uncertain. The J-M67*, J-M92, and J-M102 representatives reflect more distinctive origins and dispersal patterns. Whereas J-M67* and J-M92 show higher frequencies and variances in Europe (0.40 and 0.32, respectively) and in Turkey (0.32 and 0.30, respectively [Cinnioğlu et al. 2004]) than in the Middle East (0.17 and 0.09, respectively), J-M12(M102) shows its maximum frequency in the Balkans. In spite of the relative high value of variance of this haplogroup in Turkey (Cinnioğlu et al. 2004)—which, however, could be due to multiple arrivals—the pattern of distribution and the network of J-M12(M102) (figs. 2 and 4) are consistent with its diffusion in Europe from the southern Balkans. On the contrary, J-M67* and J-M92 could have arrived in Europe from Anatolia via the Bosphorus isthmus, as well as by seafaring Neolithic populations who reached southern Italy. J-M67* and J-M92 could represent, at least in part, the Y-chromosome component that King and Underhill (2002) found to correlate with the distribution, from Anatolia toward Europe, of archaeological painted pottery and anthropomorphic figurines. On the other hand, J-M67– and J-M12–related lineages have been observed in Pakistan and India; thus, they probably have marked other migratory events, but the small number of J subclades in these regions (Underhill et al. 2000; Kivisild et al. 2003; present study) does not allow an evaluation of the mode and time of their arrival.
Southern Italy (Apulia and Calabria) contains sites of the early Neolithic period (Whitehouse 1968), but we know from history that these regions were subsequently colonized by the Greeks (Peloponnesians). To test the relative contribution of Greek colonists versus putative earlier Neolithic settlers, an admixture analysis (Bertorelle and Excoffier 1998) was performed, using E-M78 and J-M172(xM12) as signatures of Greek and Anatolian lineages, respectively. The Anatolian source population was based on 523 Turks, of whom 118 were J-M172(xM12) and 25 were E-M78 (Cinnioğlu et al. 2004). The Greek population comprised 36 Peloponnesian samples, 5 of which were J-M172(xM12) and 17 of which were E-M78 (R.K., unpublished data). In spite of the small Peloponnesian sample size, the high E-M78 frequency (47%) observed here is consistent with that (44%) independently found in the same region (Di Giacomo et al. 2003) for the YAP chromosomes harboring microsatellite haplotypes (A. Novelletto, personal communication) typical of Hg E-M78 (Cruciani et al. 2004 [in this issue]; present study). The admixture analysis yielded an admixture proportion from Greece of 0.07±0.15 for the Calabrian samples and of 0.22±0.15 for the Apulian samples. SD was determined by bootstrapping 1,000 replicates.
The TD of the two sister clades J-M267 and J-M172 was estimated, with V0=0, and turned out to be 31.7 ky (see phylogeny in fig. 2). This estimate, however, is not easily interpretable, because such old haplogroups are differently represented in different regions where they probably underwent multiple bottlenecks. The lower internal variance of J-M267 in the Middle East and North Africa, relative to Europe and Ethiopia, is suggestive of two different migrations. In the absence of additional binary polymorphisms allowing further informative subdivision of J-M267, the YCAII microsatellite system provides important insights. The majority of J-M267 Y chromosomes harbor the single-banded motif YCAIIa22-YCAIIb22 in the Middle East (>70%) and in North Africa (>90%), whereas this association is much less frequent in Ethiopia and only sporadically found in southern Europe. Considering the distribution of this YCAII single-banded pattern—which, besides the usual stepwise mutational mechanism, could be due to a stable mutational event (one locus deletion or a single-nucleotide mutation in the primer sequence)—we suggest that the motif YCAIIa22-YCAIIb22 potentially characterizes a monophyletic clade of J-M267. A comparable situation is observed within Hg I-M170, in which the single-banded haplotype YCAIIa21-YCAIIb21 parallels a biallelic marker (O.S., unpublished data).
According to this interpretation, the first migration, probably in Neolithic times, brought J-M267 to Ethiopia and Europe, whereas a second, more-recent migration diffused the clade harboring the microsatellite motif YCAIIa22-YCAIIb22 in the southern part of the Middle East and in North Africa. In this regard, it is worth noting that the median expansion time of the J-M267-YCAIIa22-YCAIIb22 clade was estimated to be 8.7–4.3 ky, by use of the TD approach (see fig. 4 legend), and that this clade includes the modal haplotype DYS19-14/DYS388-17/DYS390-23/DYS391-11/DYS392-11 of the Galilee (Nebel et al. 2000) and of Moroccan Arabs (Bosch et al. 2001). These results are consistent with the proposal that this haplotype was diffused in recent time by Arabs who, mainly from the 7th century a.d., expanded to northern Africa (Nebel et al. 2002).
In conclusion, high-resolution Y-chromosome haplotyping and particular microsatellite associations reveal regional population differentiations, an East Africa homeland for E-M78, and recent gene-flow episodes consistent with the Neolithic in Europe. In particular, the spatial distributions of J-M172*, J-M267, E-M78, and E-M123 indicate expansions from the Middle East toward Europe that most likely occurred during and after the Neolithic, that of J-M102 illustrates population expansions from the southern Balkans, and that of E-M81 reveals recent gene flow from North Africa. Distinct histories of J-M267* lineages are suggested: an expansion from the Middle East toward East Africa and Europe and a more-recent diffusion (marked by the YCAIIa-22/YCAIIb-22 motif) of Arab people from the southern part of the Middle East toward North Africa.
Acknowledgments
We are grateful to all the donors for providing blood samples and to the people who contributed to their collection. In particular, we thank Ahmet Arslan, Agnese Brega, and B. Kindar (for samples from Turks); Jaume Bertranpetit and Anne Cambon-Thomsen (for samples from Catalans, Basques, and Bearnais); Aiping Liu (for samples from Chinese); J. Garcia-Puche (for samples from Andalusians); and Adriana Grasso and F. Pignatelli (for samples from Apulians). We warmly acknowledge two anonymous reviewers for their helpful and constructive criticism. This research was supported by Progetto Finalizzato CNR “Beni Culturali” (A.S.S.-B.), National Institutes of Health grants GM28428 and GM55273 (L.L.C.-S.), Progetto MIUR-CNR Genomica Funzionale-Legge 449/97 (A.T. and L.L.C.-S), Fondo d’Ateneo per la Ricerca dell’Università di Pavia (A.S.S.-B and A.T.), the Italian Ministry of the University's Progetti Ricerca Interesse Nazionale 2002 and 2003 (A.T.), and Fondo Investimenti Ricerca di Base 2001 (A.T.).
Electronic-Database Information
The URLs for data presented herein are as follows:
- Fluxus Engineering, http://www.fluxus-engineering.com (for NETWORK 2.0b)
- Y-Chromosome STR Database, http://www.cstl.nist.gov/biotech/strbase/y_strs.htm
References
- Al-Zahery N, Semino O, Benuzzi G, Magri C, Passarino G, Torroni A, Santachiara-Benerecetti AS (2003) Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol Phylogenet Evol 28:458–472 10.1016/S1055-7903(03)00039-3 [DOI] [PubMed] [Google Scholar]
- Ammerman AJ, Cavalli-Sforza LL (1984) Neolithic transition and the genetics of populations in Europe. Princeton University Press, Princeton, NJ [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle G, Excoffier L (1998) Inferring admixture proportions from molecular data. Mol Biol Evol 15:1298–1311 [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann B, Klintschar M, Neuhuber F, Hühne J, Rolf B (1998) Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet 62:1408–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Silva DR, Santos FR, Hutz MH, Salzano FM, Pena SD (1999) Divergent human Y-chromosome microsatellite evolution rates. J Mol Evol 49:204–214 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL (2002) Demic diffusion as the basic process of human expansions. In: Bellwood P, Renfrew C (eds) Examining the farming/language dispersal hypothesis. McDonald Institute for Archaeological Research, Cambridge, United Kingdom, pp 79–88 [Google Scholar]
- Cinnioğlu C, King R, Kivisild T, Kalfoğlu E, Atasoy S, Cavalleri GL, Lillie AS, Roseman CC, Lin AA, Prince K, Oefner PJ, Shen P, Semino O, Cavalli-Sforza LL, Underhill PA (2004) Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet 114:127–148 10.1007/s00439-003-1031-4 [DOI] [PubMed] [Google Scholar]
- Cruciani F, La Fratta R, Santolamazza P, Sellitto D, Pascone R, Moral P, Watson E, Guida V, Colomb EB, Zaharova B, Lavinha J, Vona G, Aman R, Calì F, Akar N, Richards M, Torroni A, Novelletto A, Scozzari R (2004) Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveal multiple migratory events within and out of Africa. Am J Hum Genet 74:1014–1022 (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfiner P (1976) Linear estimation of non-stationary spatial phenomena. Guarasio M, David M, Haijbegts C (eds) Advanced geostatistics in the mining industry. Dordrecht, Reidel, pp 49–68 [Google Scholar]
- Di Giacomo F, Luca F, Anagnou N, Ciavarella G, Corbo RM, Cresta M, Cucci F, Di Stasi L, Agostiano V, Giparaki M, Loutradis A, Mammi C, Michalodimitrakis EN, Papola F, Pedicini G, Plata E, Terrenato L, Tofanelli S, Malaspina P, Novelletto A (2003) Clinal patterns of human Y chromosomal diversity in continental Italy and Greece are dominated by drift and founder effects. Mol Phylogenet Evol 28:387–395 10.1016/S1055-7903(03)00016-2 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Horai S (1995) Y chromosomal DNA variation and the peopling of Japan. Am J Hum Genet 56:951–962 (erratum 56:1512) 7717406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 15:427–441 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- King R, Underhill PA (2002) Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages. Antiquity 76:707–714 [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk H-V, Stepanov V, Gölge M, Usanga E, Papiha SS, Cinnioğlu C, King R, Cavalli-Sforza LL, Underhill PA, Villems R (2003) The genetic heritage of earliest settlers persists in both the Indian tribal and caste populations. Am J Hum Genet 72:313–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias N, Bayes M, Tyler-Smith C (1994) Highly informative compound haplotypes for the human Y chromosome. Hum Mol Genet 3:115–123 [DOI] [PubMed] [Google Scholar]
- Nebel A, Filon D, Hohoff C, Faerman M, Brinkmann B, Oppenheim A (2001) Haplogroup-specific deviation from the stepwise mutation model at the microsatellite loci DYS388 and DYS392. Eur J Hum Genet 9:22–26 10.1038/sj.ejhg.5200577 [DOI] [PubMed] [Google Scholar]
- Nebel A, Filon D, Weiss DA, Weale M, Faerman M, Oppenheim A, Thomas MG (2000) High-resolution Y chromosome haplotypes of Israeli and Palestinian Arabs reveal geographic substructure and substantial overlap with haplotypes of Jews. Hum Genet 107:630–641 10.1007/s004390000426 [DOI] [PubMed] [Google Scholar]
- Nebel A, Landau-Tasseron E, Filon D, Oppenheim A, Faerman M (2002) Genetic evidence for the expansion of Arabian tribes into the southern Levant and North Africa. Am J Hum Genet 70:1594–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passarino G, Semino O, Quintana-Murci L, Excoffier L, Hammer M, Santachiara-Benerecetti AS (1998) Different genetic components in the Ethiopian population, identified by mtDNA and Y-chromosome polymorphisms. Am J Hum Genet 62:420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Krausz C, Zerjal T, Sayar SH, Hammer MF, Mehdi SQ, Ayub Q, Qamar R, Mohyuddin A, Radhakrishna U, Jobling MA, Tyler-Smith C, McElreavey K (2001) Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am J Hum Genet 68:537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roewer L, Kayser M, Dieltjes P, Nagy M, Bakker E, Krawczak M, de Knijff P (1996) Analysis of molecular variance (AMOVA) of Y-chromosome-specific microsatellites in two closely related human populations. Hum Mol Genet 5:1029–1033 10.1093/hmg/5.7.1029 [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santachiara-Benerecetti AS, Semino O, Passarino G, Torroni A, Brdicka R, Fellous M, Modiano G (1993) The common Near-Eastern origin of Ashkenazi and Sephardi Jews supported by Y-chromosome similarity. Ann Hum Genet 57:55–64 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Pangrazio A, Santolamazza P, Vona G, Moral P, Latini V, Varesi L, Memmi MM, Romano V, De Leo G, Gennarelli M, Jaruzelska J, Villems R, Parik J, Macaulay V, Torroni A (2001) Human Y-chromosome variation in the western Mediterranean area: implications for the peopling of the region. Hum Immunol 62:871–884 10.1016/S0198-8859(01)00286-5 [DOI] [PubMed] [Google Scholar]
- Scozzari R, Cruciani F, Santolamazza P, Malaspina P, Torroni A, Sellitto D, Arredi B, Destro-Bisol G, De Stefano G, Rickards O, Martinez-Labarga C, Modiano D, Biondi G, Moral P, Olckers A, Wallace DC, Novelletto A (1999) Combined use of biallelic and microsatellite Y-chromosome polymorphisms to infer affinities among African populations. Am J Hum Genet 65:829–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Brega A, Fellous M, Santachiara-Benerecetti AS (1996) A view of the Neolithic demic diffusion in Europe through two Y chromosome-specific markers. Am J Hum Genet 59:964–968 [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA (2002) Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet 70:265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Bradman N, Flin HM (1999) High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 105:577–581 10.1007/s004390051148 [DOI] [PubMed] [Google Scholar]
- Thomas MG, Skorecki K, Ben-Ami H, Parfitt T, Bradman N, Goldstein DB (1998) Origins of Old Testament priests. Nature 394:138–140 10.1038/28083 [DOI] [PubMed] [Google Scholar]
- Torroni A, Semino O, Rose G, De Benedictis G, Brancati C, Santachiara Benerecetti AS (1990) Mitochondrial DNA polymorphisms in the Albanian population of Calabria (southern Italy). Int J Anthropol 5:97–104 [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 10.1046/j.1469-1809.2001.6510043.x [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Weale ME, Shah T, Jones AL, Greenhalgh J, Wilson JF, Nymadawa P, Zeitlin D, Connell BA, Bradman N, Thomas M (2003) Rare deep-rooting Y chromosome lineages in humans: lessons for phylogeography. Genetics 165:229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse R (1968) The early Neolithic of southern Italy. Antiquity 42:188–193 [Google Scholar]
- Y Chromosome Consortium (YCC) (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 11827954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA (2001) Estimating divergence time with use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol 18:700–709 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 74: 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]