Abstract
Hereditary nonpolyposis colorectal cancer (HNPCC) accounts for ∼2% of all colorectal cancer (CRC) cases and is the most common hereditary CRC syndrome. We have previously reported a high incidence of microsatellite instability (MSI) and germline mismatch repair (MMR) gene mutations in young Hong Kong Chinese with CRC. Ongoing studies at the Hereditary Gastrointestinal Cancer Registry in Hong Kong have revealed a unique germline MSH2 c.1452–1455delAATG mutation that has not been reported in other ethnic groups. Detailed analysis showed that this specific MSH2 mutation constituted 21% of all germline MMR gene mutations and 36% of all MSH2 germline mutations identified. We designed a specific PCR-based diagnostic test on paraffin-embedded tissues and identified this germline mutation in 2 (1.5%) of 138 consecutive patients with early-onset CRC (<46 years of age at diagnosis). Haplotype analysis was performed using 11 microsatellite markers located between D2S391 and D2S123. All 10 families had the same disease haplotype, suggesting a founder effect. These 10 families all originated from the Chinese province of Guangdong, which historically included Hong Kong. It is the most populous of the Chinese provinces, with a population of >93 million. Further analysis suggested that this founder mutation may date back to between 22 and 103 generations ago. The identification of this MSH2 founder mutation has important implications for the design of mutation-detection strategies for the southern Chinese population. Since there were major emigrations from Hong Kong and Guangdong province during the 19th and 20th centuries, this finding is also significant for Chinese communities worldwide.
Hereditary nonpolyposis colorectal cancer (HNPCC) (MIM #114500) accounts for ∼2% of all colorectal cancer (CRC) cases (Aaltonen et al. 1998) and is the most common hereditary CRC syndrome. Germline mutations in the six DNA mismatch repair (MMR) genes, including MSH2, MLH1, MSH6, PMS1, PMS2, and MLH3, have been identified in kindreds with HNPCC, and inactivation of the MMR mechanism contributes to the microsatellite instability (MSI) phenotype detected in HNPCC tumors. Mutations in two of these MMR genes, MSH2 and MLH1, account for the majority of the kindreds with HNPCC (International Collaborative Group on HNPCC [ICG-HNPCC]) (Peltomaki 2001).
CRC is the second most common cancer in Hong Kong. We recently drew attention to the distinct epidemiology in the Hong Kong Chinese population, in which there is an incidence rate of CRC in the young population (<46 years of age at diagnosis) that is four times the rate in other countries, such as the United States, Scotland, and Japan (Yuen et al. 1997). To further clarify the origin of this apparent population susceptibility, we have studied the incidence of MSI and MMR gene mutations in relation to age at onset of patients with CRC. The results showed that a significant proportion of tumors from young patients exhibited high-level MSI and that many of them had a germline mutation in one of the MMR genes; thus, these patients did, in fact, exhibit HNPCC (Chan et al. 1999; Ho et al. 2000; Yuen et al. 2002). In addition, we have identified a founder mutation characterized by a 1.8-kb deletion that includes exon 11 in MLH1 in two unrelated Chinese families with HNPCC (Chan et al. 2001). The identification of founder mutations is not just of interest to researchers but has important implications in the design of mutation-detection strategies for ethnic populations. Previous identification of two founder MLH1 mutations in the Finnish population and three founder BRCA1/2 mutations in the Ashkenazi Jewish population have facilitated the genetic diagnosis in these populations (Nystrom-Lahti et al. 1995; Moisio et al. 1996; Shiri-Sverdlov et al. 2000).
In our previous studies, we observed a recurrent MSH2 c.1452–1455delAATG germline mutation that had not been reported in other ethnic groups (Chan et al. 1999; Yuen et al. 2002). Therefore, we examined the incidence of this germline MMR gene mutation in our Hereditary Gastrointestinal Cancer Registry, which is the only registry in Hong Kong that provides comprehensive service, including genetic diagnosis, for families with hereditary CRC syndromes (Hereditary Gastrointestinal Registry Hong Kong). The genetic diagnostic laboratory of the registry has established standard protocols for genetic diagnosis. The MSI status of the tumor was determined using a panel of 6–10 microsatellite markers. MSH2 and MLH1 protein expression was studied using immunohistochemical staining. Mutations were detected using a combination of in vitro synthesized protein (IVSP) assay, RT-PCR, and direct DNA sequencing methods. Details of these methodologies have been described in our previous studies (Leung et al. 1998; Chan et al. 1999; Chan et al. 2001; Yuen et al. 2002). Up to the end of 2002, the Registry had recorded 13 MLH1, 22 MSH2, and 3 MSH6 germline mutations. Of the 22 MSH2 germline mutations, 8 (36.4%) were c.1452–1455delAATG mutations. In all of these cancers, there was high-level MSI, and immunohistochemical staining showed loss of MSH2 protein. A truncated MSH2 protein product could be detected by IVSP, and the specific germline MSH2 mutation was confirmed by direct DNA sequencing.
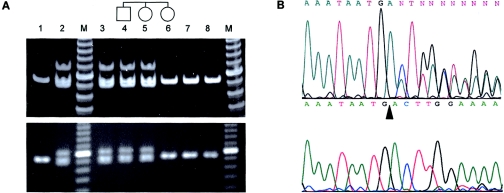
To explore the frequency of this germline MSH2 mutation in our population, 143 consecutive archival formalin-fixed paraffin-embedded CRC resection specimens, from patients whose age at diagnosis was <46 years, were retrieved from the pathology departments of three regional hospitals. DNA from tumor tissue and normal tissue was extracted by microdissection from paraffin sections. A diagnostic PCR test was designed by amplification of a 90-bp fragment covering the deleted region. Cases with the MSH2 c.1452–1455delAATG mutation showed a specific banding pattern by gel electrophoresis, with the presence of three bands that corresponded to heteroduplex, homozygous wild-type (90 bp in length), and homozygous mutant fragments (86 bp in length), respectively. DNA was successfully amplified by this PCR-based method from 138 cases. Two patients (1.5%) showed an abnormal banding pattern, as seen in the positive control (fig. 1A). Direct DNA sequencing confirmed the presence of MSH2 c.1452–1455delAATG in both the tumors and the normal tissues of these two patients (fig. 1B). In addition, one of the tumors had lost the wild-type allele.
Figure 1.
A, Diagnostic PCR test for the MSH2 c.1452–1455delAATG mutation. The paraffin sections, prepared from stored blocks, were assessed by light microscopy. Areas with the tumor or normal colonic mucosa were microdissected under a light microscope for DNA extraction by use of standard protocol. The primer sequences were as follows: forward primer CATTTGATCCTAATCTCAGTGAA; reverse primer CAAGATCTCTGGCTGCACTT. Forty cycles of PCR were performed in 20 μl with 2 μl–3 μl of DNA lysate from paraffin-embedded tissue, 1 × PCR buffer, 2 mM MgCl2, 4 pmol each of forward and reverse primer, 50 μM of each dNTP, and 0.1 U Platinum Taq polymerase (Invitrogen). The annealing temperature was 60°C. The PCR products were electrophoresed in 6% polyacrylamide (upper panel) or 3% agarose gels (lower panel). Lane 1 shows the negative control from an individual without the specific MSH2 mutation. Lane 2 indicates the positive control from an individual with the specific MSH2 mutation, showing an upper heteroduplex band, a middle wild-type band (90 bp), and a lower deletion mutant band (86 bp); M represents the 10-bp ladder. Lanes 3 and 4 show PCR amplicons from the paraffin-embedded nontumor colonic tissues of the two young patients who screened positive for the specific MSH2 mutation. Lanes 5 and 6 indicate PCR amplicons from genomic DNA of two family members of the patient in lane 4. Lanes 7 and 8 illustrate PCR amplicons from paraffin-embedded tissues of other young patients with CRC but without the specific mutation. B, Direct DNA sequencing of the PCR products amplified from DNA extracted from paraffin-embedded tissue of patient in lane 4 of figure 1A. The top panel illustrates the overlapping sequence after the point of deletion in microdissected normal tissue. The lower panel indicates the complete loss of the wild-type sequence in the microdissected tumor sample. The arrowhead denotes the site of the AATG deletion.
We then analyzed the detailed family histories of patients carrying this specific germline mutation. These were obtained using both in-person and telephone interviews with the patients and their relatives, as described previously (Chan et al. 1999; Ho et al. 2000). The majority of these families have strong family or personal history of cancer. A total of 43 cancers were noted in these families: 29 CRCs, 4 uterine cancers, 4 brain tumors, 2 breast cancers, 1 ovarian cancer, 1 stomach cancer, and 2 disseminated cancers of unknown origin. The majority of these cancers were diagnosed before the age of 50 years. Although these families did have multiple individuals with a history of cancer, only three families (H041, H043, and H046) satisfied the Amsterdam criteria/Bethesda criteria I. Family H004 satisfied the Amsterdam II criteria. Family H004 and four others (H032, H037, H061, and H073) satisfied Bethesda criteria II and/or III (Rodriguez-Bigas et al. 1997). For the remaining three individuals (from families H037, H072, and W043), the family history could not be traced.
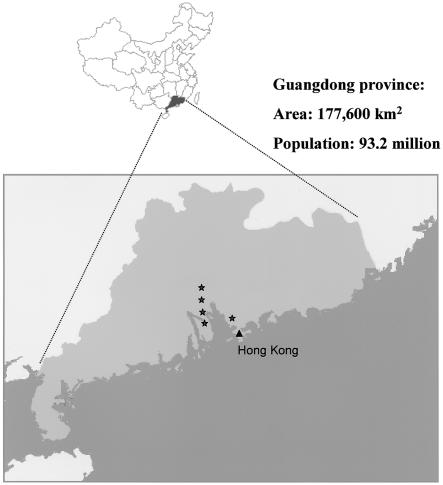
These individuals and their families were interviewed further to explore the possibility of the existence of kinship among them. Documented family histories indicated that the most ancient and traceable ancestors of these families did not have any blood relationship to one another. They all originated from the Guangdong province of southern China, which is the origin of most Hong Kong inhabitants. Further inquiries revealed that the ancestors of these patients originally resided in at least five different counties/regions within Guangdong province (fig. 2).
Figure 2.
Map of China, with Guangdong province highlighted. The stars denote the locations of the counties/cities from which six of the families with the MSH2 c.1452–1455delAATG mutation in our study originated. In the four remaining families, the exact counties/cities of origin in Guangdong province are not known.
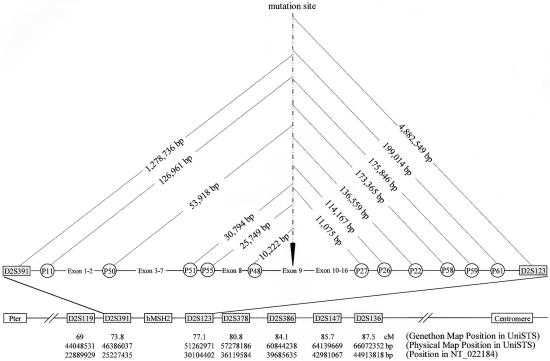
Whenever possible, members from two generations were invited for haplotype analysis. Venous blood was sampled, with informed consent, for DNA extraction by use of standard protocols. Of the 10 families with this specific MSH2 germline mutation, many are small families, and, for cultural reasons, the older generations were reluctant to give consent for genotype analysis. Therefore, there were only two families (H046 and H073) with DNA available from members of two consecutive generations. To estimate the frequency of the disease haplotype in the general population, 51 genomic DNA samples from two consecutive generations of 23 families (5 families with both parents and one descendant; 18 families with either the father or mother and one descendant) without this MSH2 mutation were used as the control population. Initially, seven microsatellite markers (Tel-D2S119-D2S391-MSH2-D2S123-D2S378-D2S386-D2S147-D2S136-Cen) were selected from the Genethon genetic map on the basis of information from UniSTS database of the National Center for Biotechnology Information (NCBI). These markers have also been recorded in a single human genome contig (GenBank accession number NT_022184). Their order and their physical position in relation to the location of the specific 4-bp deletion in exon 9 of MSH2 are listed in figure 3. By use of these seven markers, we determined that the disease haplotype for family H046 was 2-2-10-2-2-1-2, whereas for family H073 the haplotype was 1-2-9-1-2-1-1. Comparison of the two disease haplotypes showed a low level of linkage disequilibrium, which might be due to the fact that these markers were far away from the mutation site.
Figure 3.
Schematic representation of MSH2 and the surrounding regions of chromosome 2. This map is based on the information provided by UniSTS, NCBI. The location and order of the loci used in the present study, as well as their physical and genetic positions with reference to the MSH2 c.1452–1455delAATG mutation site, are shown. These data are extracted from the Genethon map, the Marshfield genetic map (Marshfield Center for Medical Genetics), and the contig NT_022184.
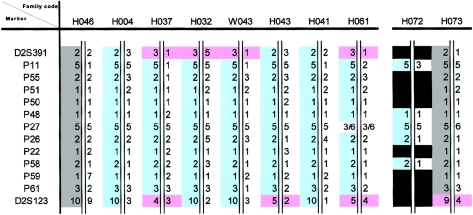
Eleven microsatellite markers located between D2S391 and D2S123 were further selected. These included three tetranucleotide repeats (P50, P51, and P55) and three dinucleotide repeats (P22, P26, and P27), which had been used elsewhere for MSH2 haplotype analysis (Desai et al. 2000), as well as five novel dinucleotide repeats (P11, P48, P58, P59, and P61). The physical locations of these microsatellite markers in relation to MSH2, as defined by the human genome contig NT_022184, are also shown in figure 3. By use of this second set of 11 microsatellite markers spanning a region of 3.3 cM, flanked by D2S391 and D2S123, the same disease haplotype (5-2-1-1-1-5-2-1-2-1-3) (fig. 4) was identified for both family H046 and family H073.
Figure 4.
Disease haplotypes of the 10 families with the specific MSH2 mutation. Haplotype analysis was performed as described elsewhere (Chan et al. 2001). The regions covering these microsatellite markers were amplified by PCR by use of fluorescent dye-labeled M13(−21)-tagged forward primers. The PCR amplicons were analyzed using the automated 377 DNA Sequencer (Applied Biosystems) with the GeneScan program. Alleles were numbered consecutively according to increasing size. The initial eight families with the specific MSH2 mutation are shown on the left; the two families screened positive by the PCR-based diagnostic method are shown on the right. Also shown are the phased disease haplotype (gray), the reconstructed disease haplotype (blue), loci not amplifiable from paraffin tissues (black), and the observable recombination events (pink).
Genotype analysis was performed using both sets of markers for the remaining eight families, and the haplotypes were reconstructed by PHASE (Stephens et al. 2001), which used Bayesian methods to predict the haplotype distribution, conditional on observed genotypes. Again, linkage disequilibrium was not significant when the first set of seven Genethon markers was used. With the second set of 11 microsatellite markers, seven of the eight families revealed an identical disease haplotype to that found in families H046 and H073 (fig. 4). For the last family (H061), a mutation in the marker P27 was present, resulting in an unmatched haplotype. When this locus was ignored, an identical haplotype could be reconstructed as the consensus disease haplotype (fig. 4).
The allele frequencies of the 11 microsatellite loci on the mutation-bearing (n=10) and control (n=74) chromosomes are listed in table 1. Compared with the control chromosomes, allele frequencies of all of these loci on the disease haplotype were in excess of the frequencies on the mutation-bearing chromosomes. Moreover, the disease haplotype was not found in any of the 74 haplotypes from the 23 parent/child-pair control population. These data suggest that this germline MSH2 c.1452–1455delAATG mutation is a founder mutation in the southern Chinese population.
Table 1.
Linkage Disequilibrium between the D2S391 and D2S123 Markers and Age Estimation of the Founder MSH2 c.1452–1455delAATG Mutation [Note]
|
Frequency forMarker Allele on |
||||||||
| Marker | Allele Size(bp) | Mutation-BearingChromosomes | NormalChromosomes | LD(δ) | PhysicalDistance(kb) | GeneticDistance(cM) | RecombinationFraction(θ) | Estimated Age(generations) |
| D2S391 | 183 | .556 (5/9) | .284 (21/74) | .379 | 1,278.587 | .935 | .009352 | 103 |
| P11 | 190 | 1.000 (10/10) | .405 (30/74) | 1.000 | 124.911 | .091 | .000914 | … |
| P50 | 302 | 1.000 (9/9) | .761 (54/71) | 1.000 | 53.895 | .039 | .000394 | … |
| P51 | 270 | 1.000 (9/9) | .257 (19/74) | 1.000 | 30.635 | .022 | .000224 | … |
| P55 | 205 | 1.000 (9/9) | .357 (25/70) | 1.000 | 25.726 | .019 | .000188 | … |
| P48 | 144 | 1.000 (10/10) | .716 (53/74) | 1.000 | 10.084 | .007 | .000074 | … |
| P27 | 160 | .900 (9/10) | .459 (34/74) | … | 110.929 | .081 | .000811 | … |
| P26 | 166 | 1.000 (10/10) | .378 (28/74) | 1.000 | 114.423 | .084 | .000837 | … |
| P22 | 263 | 1.000 (9/9) | .620 (44/71) | 1.000 | 136.940 | .100 | .001002 | … |
| P58 | 156 | 1.000 (10/10) | .284 (21/74) | 1.000 | 175.654 | .128 | .001285 | … |
| P59 | 210 | 1.000 (8/8) | .391 (27/69) | 1.000 | 178.198 | .130 | .001303 | … |
| P61 | 160 | 1.000 (8/8) | .324 (24/74) | 1.000 | 201.307 | .147 | .001472 | … |
| D2S123 | 160 | .556 (5/9) | .000 (0/74) | .556 | 3,598.226 | 2.632 | .026317 | 22 |
Note.— The linkage disequilibrium (LD) index was calculated according to Bengtsson and Thomson (1981): δ=(Pd−Pn)/(1−Pn), where Pd and Pn are the frequencies for the marker allele on mutation-bearing and normal chromosomes, respectively. The physical and genetic distances are expressed with reference to the specific MSH2 mutation site. The appropriate physical-to-genetic distance conversion factor was calculated by linear regression analysis of genetic versus physical map position by use of seven markers in proximity to MSH2, with known centimorgan (cM) (Genethon and Marshfield maps) and megabase (Mb) (UniSTS, NCBI) information, as highlighted in figure 3. The conversion factor was calculated to be 0.7314 cM/Mb (using data from Genethon), which compared well with that from the Marshfield genetic map (0.7342) (using data from the Marshfield Center for Medical Genetics). The Kosambi’s function (Colombo et al. 2000) was applied to convert the genetic map distance into the recombination fraction (θ). Estimation of the age of the mutation was based on the following formula (Risch et al. 1995): generations = logδ/log(1−θ).
For age estimation of this founder mutation, allele-frequency distributions of the individual markers in disease and control chromosomes were compared using the Mantel-Haenszel common odds ratio estimate. The linkage disequilibrium for the 13 microsatellite loci was calculated according to the method employed by Bengtsson and Thomson (1981). On the basis of the recombination fraction and the linkage disequilibrium of loci D2S391 and D2S123, the age of the disease haplotype was calculated to be between 22 and 103 generations (Risch et al. 1995) (table 1).
Up to the end of 2003, 695 germline mutations were recorded in the ICG-HNPCC database. Mutations of MSH2, MLH1, and MSH6 account for 38.7%, 56.5% and 4.7% of all germline mutations, respectively. Our present study reported a frequent germline MSH2 c.1452–1455delAATG mutation in the southern Chinese population. This particular mutation was not found among the 269 cases of MSH2 mutations in the ICG-HNPCC database. Haplotype analysis suggested that it was a founder mutation that occurred between 22 and 103 generations ago.
Founder mutations in MMR genes explain a substantial fraction of instances of HNPCC in some ethnic groups. In Finland, two mutations in MLH1 (a 3.5-kb genomic deletion that includes exon 16 and a splice-acceptor site mutation of exon 6) account for 63% of all disease-causing mutations in families with HNPCC (Nystrom-Lahti et al. 1995). Hutter et al. described a third founder MLH1 mutation (MLH1 c.2141G→A) in several apparently unrelated families in the Valais region of Switzerland (Hutter et al. 1996). We also identified a founder mutation of MLH1 (a 1.8-kb genomic deletion that includes exon 11) in two apparently unrelated southern Chinese families (Chan et al. 2001). The MSH2 IVS5+3A→T mutation has been observed in families with HNPCC of many different populations and accounts for 14% of all known pathogenic MSH2 mutations. Whereas families in Newfoundland, Canada, have been found to share a common disease haplotype, those from other countries, such as the United States, England, Italy, Japan, and Hong Kong, do not, and the mutation appears to arise de novo (Froggatt et al. 1999; Desai et al. 2000). More recently, two MSH2 founder mutations (MSH2 c.1906G→C and MSH2 deletion of exons 1–6) were recorded to account for one-third of families with HNPCC in Ashkenazi Jews and one-tenth of American families with HNPCC, respectively (Foulkes et al. 2002; Wagner et al. 2003). Our current findings have provided evidence for another founder mutation of MSH2 that is important in the Chinese population. From our own Registry database, we determined that this mutation accounts for 21% of all pathogenic germline MMR mutations and 36% of MSH2 mutations detected so far. The identification of founder mutations in MMR genes has practical implications, in that ethnic-specific mutation-screening strategies should be employed before a more general search for pathogenic mutations is undertaken. Indeed, by use of this approach, another three unrelated young patients with CRC who harbored this MSH2 founder mutation were recently identified during the preparation of this manuscript.
The existence of these ethnic-specific founder mutations may contribute variably to the general overall incidence of CRC. For example, MSH2 c.1906G→C was found in 2%–3% of early-onset CRCs in the Ashkenazi Jewish population (Foulkes et al. 2002). In Hong Kong, our previous epidemiology study identified a high incidence of early-onset CRC, as compared with other countries (Yuen et al. 1997). To date, 10%–12% of all CRCs in Hong Kong occur in individuals <46 years of age (Hong Kong Cancer Registry). We show here that 1.5% of these early-onset CRCs are related to this particular founder mutation. It remains to be seen whether a cumulative effect of founder mutations of various cancer-susceptibility genes with different penetrances may contribute to the observed high incidence of early-onset CRC in our population.
The significance of our finding is not limited to Hong Kong locally. All 10 families carrying the founder mutation originated from the Guangdong province in China, which is in the immediate vicinity of Hong Kong. Guangdong (also known as Canton) province has an area of 177,600 km2 and a total population size of 86.42 million (National Bureau of Statistics of China). Historically, Hong Kong was part of Guangdong province, and the ancestors of the majority of current Hong Kong residents were from Guangdong. Thus, together with the 6.8 million inhabitants in Hong Kong, Guangdong has the largest provincial population size, with >93 million people. Several major emigration waves from Guangdong and Hong Kong in the 19th and 20th centuries led to the establishment of large Chinese communities in the United States, Canada, Australia, and the United Kingdom (Chinn 1969; Hoexter 1976) (Canada’s Digital Collections; Chinese Museum [Australia]; The Chinese in New Zealand; Library of Congress [United States]). Since we have noticed this founder effect in Hong Kong, we believe its effects may be seen in other Chinese communities of Guangdong descent as well. Our local experience in the Registry shows that many of these emigrants have lost contact with their relatives and extended families in mainland China. At times, it is difficult, if not impossible, to have a comprehensive family medical history, thus making a firm clinical diagnosis of HNPCC challenging. Similar to our previous reports (Chan et al. 1999; Yuen et al. 2002), most of the families studied here with this germline mutation do not have a family history that satisfies the Amsterdam criteria. Thus, our findings have important implications for the design of mutation-detection strategies for HNPCC in the southern Chinese population worldwide.
Acknowledgments
This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region (HKU 7330/00M), the Committee on Research and Conference grant (10203881) from the University of Hong Kong, and donations from the Hong Kong Cancer Fund and the Hong Kong Society of Gastroenterology. We thank Dr. Paul Yip and Dr. Jurg Ott for their useful advice and discussions. We also thank Dr. Neil Hanchard and Dr. M. S. Chan for their help with analysis using the PHASE program.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Canada’s Digital Collections, “Across the Generations: A History of the Chinese in Canada,” http://collections.ic.gc.ca/generations/index2.html
- Chinese Museum (Australia), http://home.vicnet.net.au/~mcah/cah.html
- GenBank, http://ncbi.nlm.nih.gov/Genbank/ (for human genome contig [accession number NT_022184])
- Hereditary Gastrointestinal Cancer Registry Hong Kong, http://www.generations.hk.com/index_e.php3
- Hong Kong Cancer Registry, http://www.ha.org.hk/cancereg
- International Collaborative Group on Hereditary Non-Polyposis Colorectal Cancer (ICG-HNPCC), http://www.nfdht.nl [DOI] [PubMed]
- Library of Congress (United States), “Hong Kong,” http://www.loc.gov/loc/lcib/9708/hongkong.html
- Marshfield Center for Medical Genetics, http://research.marshfieldclinic.org
- National Bureau of Statistics of China, http://www.stats.gov.cn/english/newrelease/statisticalreports
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HNPCC) [PubMed]
- The Chinese in New Zealand, http://www.stevenyoung.co.nz/chinesevoice/index.htm
References
- Aaltonen LA, Salovaara R, Kristo P, Canzian F, Hemminki A, Peltomaki P, Chadwick RB, Kaariainen H, Eskelinen M, Jarvinen H, Mecklin JP, de la Chapelle A (1998) Incidence of hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med 338:1481–1487 10.1056/NEJM199805213382101 [DOI] [PubMed] [Google Scholar]
- Bengtsson BO, Thomson G (1981) Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens 18:356–363 [DOI] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Chung LP, Ho JW, Kwan KY, Chan AS, Ho JC, Leung SY, Wyllie AH (1999) Frequent microsatellite instability and mismatch repair gene mutations in young Chinese patients with colorectal cancer. J Natl Cancer Inst 91:1221–1226 10.1093/jnci/91.14.1221 [DOI] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Ho JW, Chan AS, Kwan K, Chung LP, Lam PW, Tse CW, Leung SY (2001) A novel germline 1.8-kb deletion of hMLH1 mimicking alternative splicing: a founder mutation in the Chinese population. Oncogene 20:2976–2981 10.1038/sj.onc.1204376 [DOI] [PubMed] [Google Scholar]
- Chinn TW (1969) A history of the Chinese in California: a syllabus. Chinese Historical Society of America, San Francisco [Google Scholar]
- Colombo R, Bignamini AA, Carobene A, Sasaki J, Tachikawa M, Kobayashi K, Toda T (2000) Age and origin of the FCMD 3′-untranslated-region retrotransposal insertion mutation causing Fukuyama-type congenital muscular dystrophy in the Japanese population. Hum Genet 107:559–567 10.1007/s004390000421 [DOI] [PubMed] [Google Scholar]
- Desai DC, Lockman JC, Chadwick RB, Gao X, Percesepe A, Evans DG, Miyaki M, Yuen ST, Radice P, Maher ER, Wright FA, de la Chapelle A (2000) Recurrent germline mutation in MSH2 arises frequently de novo. J Med Genet 37:646–652 10.1136/jmg.37.9.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Thiffault I, Gruber SB, Horwitz M, Hamel N, Lee C, Shia J, et al (2002) The founder mutation MSH2*1906G→C is an important cause of hereditary nonpolyposis colorectal cancer in the Ashkenazi Jewish population. Am J Hum Genet 71:1395–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froggatt NJ, Green J, Brassett C, Evans DG, Bishop DT, Kolodner R, Maher ER (1999) A common MSH2 mutation in English and North American HNPCC families: origin, phenotypic expression, and sex specific differences in colorectal cancer. J Med Genet 36:97–102 [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Yuen ST, Chung LP, Kwan KY, Chan TL, Leung SY, Chan AS, Tse C, Lam PW, Luk IS (2000) Distinct clinical features associated with microsatellite instability in colorectal cancers of young patients. Int J Cancer 89:356–360 [DOI] [PubMed] [Google Scholar]
- Hoexter CK (1976) From Canton to California. Four Winds, New York [Google Scholar]
- Hutter P, Couturier A, Scott RJ, Alday P, Delozier-Blanchet C, Cachat F, Antonarakis SE, Joris F, Gaudin M, D’Amato L, Buerstedde JM (1996) Complex genetic predisposition to cancer in an extended HNPCC family with an ancestral hMLH1 mutation. J Med Genet 33:636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung SY, Chan TL, Chung LP, Chan AS, Fan YW, Hung KN, Kwong WK, Ho JW, Yuen ST (1998) Microsatellite instability and mutation of DNA mismatch repair genes in gliomas. Am J Pathol 153:1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisio AL, Sistonen P, Weissenbach J, de la Chapelle A, Peltomaki P (1996) Age and origin of two common MLH1 mutations predisposing to hereditary colon cancer. Am J Hum Genet 59:1243–1251 [PMC free article] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Kristo P, Nicolaides NC, Chang SY, Aaltonen LA, Moisio AL, Jarvinen HJ, Mecklin JP, Kinzler KW, Vogelstein B (1995) Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med 1:1203–1206 [DOI] [PubMed] [Google Scholar]
- Peltomaki P (2001) Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet 10:735–740 10.1093/hmg/10.7.735 [DOI] [PubMed] [Google Scholar]
- Risch N, de Leon D, Ozelius L, Kramer P, Almasy L, Singer B, Fahn S, Breakefield X, Bressman S (1995) Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet 9:152–159 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, Srivastava S (1997) A National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer Inst 89:1758–1762 10.1093/jnci/89.23.1758 [DOI] [PubMed] [Google Scholar]
- Shiri-Sverdlov R, Oefner P, Green L, Baruch RG, Wagner T, Kruglikova A, Haitchick S, Hofstra RM, Papa MZ, Mulder I, Rizel S, Bar Sade RB, Dagan E, Abdeen Z, Goldman B, Friedman E (2000) Mutational analyses of BRCA1 and BRCA2 in Ashkenazi and non-Ashkenazi Jewish women with familial breast and ovarian cancer. Hum Mutat 16:491–501 [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, Nakagawa H, Geugien M, Jaghmohan-Changur S, Breukel C, Meijers-Heijboer H, Morreau H, van Puijenbroek M, Burn J, Coronel S, Kinarski Y, Okimoto R, Watson P, Lynch JF, de la Chapelle A, Lynch HT, Fodde R (2003) Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 72:1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen ST, Chan TL, Ho JW, Chan AS, Chung LP, Lam PW, Tse CW, Wyllie AH, Leung SY (2002) Germline, somatic and epigenetic events underlying mismatch repair deficiency in colorectal and HNPCC-related cancers. Oncogene 21:7585–7592 10.1038/sj.onc.1205968 [DOI] [PubMed] [Google Scholar]
- Yuen ST, Chung LP, Leung SY, Luk IS, Chan SY, Ho JC, Ho JW, Wyllie AH (1997) Colorectal carcinoma in Hong Kong: epidemiology and genetic mutations. Br J Cancer 76:1610–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]