Abstract
Previously, we have reported linkage of markers from chromosome 1q22 to schizophrenia, a finding supported by several independent studies. We have now examined the region of strongest linkage for evidence of linkage disequilibrium (LD) in a sample of 24 Canadian familial-schizophrenia pedigrees. Analysis of 14 microsatellites and 15 single-nucleotide polymorphisms (SNPs) from the 5.4-Mb region between D1S1653 and D1S1677 produced significant evidence (nominal P<.05) of LD between schizophrenia and 2 microsatellites and 6 SNPs. All of the markers exhibiting significant LD to schizophrenia fall within the genomic extent of the gene for carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase (CAPON), making it a prime positional candidate for the schizophrenia-susceptibility locus on 1q22, although initial mutation analysis of this gene has not identified any schizophrenia-associated changes within exons. Consistent with several recently identified candidate genes for schizophrenia, CAPON is involved in signal transduction in the NMDA receptor system, highlighting the potential importance of this pathway in the etiology of schizophrenia.
Schizophrenia (SCZD [MIM 181500]) is a serious neuropsychiatric illness estimated to affect ∼1% of the general population. Family, twin, and adoption studies have demonstrated that schizophrenia is predominantly a genetic disorder with a high heritability (reviewed in McGuffin et al. 1994). Multiple genetic and nongenetic factors are likely to be involved (Bassett et al. 2001b). As part of a genomewide search for loci contributing to risk for schizophrenia, we previously reported linkage to chromosome 1q21-22 (SCZD9 [MIM 604906]) in a group of 22 medium-sized Canadian families, selected for study because multiple relatives were clinically diagnosed with schizophrenia or with schizoaffective disorder (Brzustowicz et al. 2000). Parametric multipoint linkage analysis, under a recessive model of inheritance and using a narrow definition of illness (schizophrenia and schizoaffective disorder), produced a maximum heterogeneity LOD score of 6.50 between the markers D1S1653 and D1S1679. Simulation studies confirmed the genomewide significance of this LOD score, even when accounting for multiple tests, with P<.0002. Most recently, we have reported the results of fine linkage mapping of this 1q21-22 region in the same sample of individuals, narrowing the region most likely to harbor this susceptibility locus to ∼1 Mb between APOA2 and D1S2675 (Brzustowicz et al. 2002). Again, results showed a maximum multipoint heterogeneity LOD score of 6.50, under the same recessive-narrow genetic and diagnostic model (Brzustowicz et al. 2002). Here, we report the results of linkage disequilibrium (LD) analysis of this region.
The subjects for this study included the individuals that were used for our previous genome scan and fine linkage mapping studies (Brzustowicz et al. 2000, 2002). Two new families (n=17 participating subjects) and additional members of the previously studied families (n=25) were also included. Written informed consent was obtained from all subjects, after an explanation of possible consequences. Protocols were approved by the institutional review boards of Rutgers University, University of Toronto, and by the Centre for Addiction and Mental Health. The ascertainment procedures and composition of the original sample have been previously described in greater detail (Bassett et al. 1993; Bassett and Honer 1994; Brzustowicz et al. 2000). The current sample consisted of 24 Canadian families of Celtic (n=23) or German (n=1) descent, recruited for study because schizophrenic illness appeared to be segregating in a unilineal (one side of the family) autosomal–dominant-like manner.
For this study, we used a narrow definition of schizophrenia; only individuals with schizophrenia and chronic schizoaffective disorder were coded as affected. Direct interviews were conducted, using the Structured Clinical Interview for DSM-III-R (SCID-I) for major disorders and the SCID-II for personality disorders. The interviews, collateral information, and medical records were used to make consensus diagnoses for 319 subjects based on DSM-III-R criteria. For 11 additional subjects—2 alive, but unavailable for full phenotypic assessment, and 9 deceased—only medical records and collateral information were reviewed, through the consensus diagnosis procedure. Details of the diagnostic and ascertainment methods have been described elsewhere (Bassett et al. 1993; Bassett and Honer 1994). Consensus diagnosis (by E.W.C.C. and A.S.B.) of narrow-definition schizophrenia was not obtained for four subjects, possibly with affective psychotic illness, who were directly assessed, or for the two living subjects, who were not directly assessed; therefore, these individuals were coded as “diagnosis unknown.” Three additional subjects, each from a separate family, were coded as diagnosis unknown, because mental retardation complicated their assessment. Of the nine deceased subjects, four were coded as unknown, because conclusive diagnostic elements could not be obtained, and five were determined to be affected. In total, 85 subjects, or an average of 3.5 subjects per family, were considered affected with narrowly defined schizophrenia. There were 13 subjects with phenotypic data who were coded as unknown and 232 subjects considered unaffected, including 40 individuals with the schizophrenia spectrum diagnoses of nonaffective psychotic disorder, schizotypal personality disorder, or paranoid personality disorder. DNA samples were available on 11 additional subjects without phenotypic assessments, who were coded as unknown. DNA was unavailable on the nine deceased subjects. DNA was extracted from blood samples or lymphoblastoid cell lines using the GenePure system (Gentra Systems). Overall, 330 subjects were phenotypically evaluated and 332 subjects had DNA samples available for this study.
Microsatellite marker genotypes generated for our fine-mapping study, described elsewhere (Brzustowicz et al. 2002), were used for this study. SNP genotypes were generated by a primer-extension strategy (Pyrosequencing) and were typed as simplex assays using the automated PSQ HS96A platform (Ronaghi et al. 1998; Ahmadian et al. 2000). SNPs were identified either from dbSNP or by semiautomated fluorescent sequencing (Beckman CEQ 8000), using DNA from 16 members of these families. PCR primers surrounding the SNPs were designed using Primer3 (Whitehead Institute), and the sequencing primer used for the Pyrosequencing assay was designed using the Pyrosequencing SNP Primer Design Software v 1.0. Web Supplement. Table A1 (appendix A [online only]) lists the primers used for the Pyrosequencing assays. PCR reactions contained 40 ng of template DNA, 0.5 U Taq polymerase, 0.03 μM of each primer, 0.1 mM of dNTP, 3.0 mM of MgCl2, 50mM of KCl, 10mM of Tris—HCl (pH 9.0), and 0.1% Triton X-100 in a 20 μl volume. A touchdown amplification program was used. After 8 min at 95°C, 15 cycles were done at 94°C for 30 s, annealing for 20 s, and at 72°C for 20 s, with the annealing temperature starting at 60°C and decreasing by 0.5°C for each cycle. This was followed by 35 cycles at 94°C for 20 s, at 52°C for 20 s, at 72°C for 30 s, and then a final extension step at 72°C for 15 min.
Genotypes were checked for likely errors by use of the mistyping analysis of SIMWALK2 v2.82 (Sobel and Lange 1996; Sobel et al. 2001). Because different starting conditions can generate different results when SIMWALK2 is used, mistyping analyses were conducted at least four consecutive times, with varying starting conditions, using all markers simultaneously. Genotypes identified with a probability of mistyping ⩾0.25 were regenotyped. The repeated microsatellite genotypes still having a probability of mistyping ⩾0.25 were set to zero for further analyses, whereas flagged SNP genotypes were further verified by direct sequencing. Ultimately, only three SNP genotypes (0.06%) identified as potential errors could not be resolved; these all occurred in unaffected individuals, and the omission of those SNP genotypes from the study did not change the results of the LD analysis. Overall, 99.6% of all SNP genotypes initially attempted were available for further analysis, with 100% of SNP genotypes available for affected individuals. Allele frequencies (table 1) were calculated using FBAT v1.4.2 (Laird et al. 2000; Horvath et al. 2001).
Table 1.
LD between Schizophrenia and Markers within CAPON
|
Evidence of LD, given linkage |
||||||
| SNP Markersa | Distanceb(bp) | Location | Allelesc | Allele Frequencyd | Nominal Pe | Empiric Pf |
| ss16342088 | 0 | 5′ | A/G | .31 | .44 | .49 |
| 336H14-tcta1 | 3,997 | Intron 1 | (8) | (.70) | .74 | … |
| rs1572495 | 61,445 | Intron 1 | C/T | .12 | .018 | .021 |
| rs1538018 | 92,625 | Intron 2 | G/T | .30 | .032 | .047 |
| rs945713 | 97,810 | Intron 2 | A/G | .41 | .011 | .016 |
| rs1415263 | 128,187 | Intron 2 | C/T | .38 | .0011 | .0016 |
| 336H14-ca1 | 131,530 | Intron 2 | (12) | (.78) | .044 | … |
| rs4306106 | 134,138 | Intron 2 | C/T | .14 | .050 | .079 |
| rs3934467 | 144,821 | Intron 2 | C/T | .18 | .12 | .17 |
| rs3924139 | 154,256 | Intron 2 | A/G | .30 | .059 | .068 |
| D1S2675 | 172,105 | Intron 2 | (7) | (.65) | .036 | … |
| rs4145621 | 180,832 | Intron 2 | A/G | .41 | .0017 | .0016 |
| ss16342089 | 219,177 | Intron 2 | G/C | .22 | .087 | .10 |
| rs2661818 | 226,958 | Intron 4 | C/G | .43 | .0022 | .0032 |
| rs1508263 | 241,653 | Intron 4 | A/G | .45 | .53 | .55 |
| rs3751284 | 275,879 | Exon 6 | G/A | .30 | .84 | .84 |
| rs7521206 | 293,071 | Intron 8 | C/G | .43 | .54 | .58 |
| rs348624 | 297,400 | Exon 9 | G/A | .05 | .11 | .15 |
| D1S1679 | 323,908 | 3′ | (9) | (.83) | .92 | … |
Markers are listed from centromeric to telomeric; microsatellite markers are in italics.
Distance is measured from the first marker, ss16342088, on the basis of NCBI build 34 assembly.
The more common allele is listed first (for microsatellites, the number of observed alleles is in parentheses).
Frequency of the rare allele is listed (for microsatellites, the heterozygosity is listed in parentheses).
Nominal P values were calculated by PSEUDOMARKER.
Empiric P values meeting studywide significance level of P<.05 are in boldface italics.
SIMWALK2 v2.82 was also used to generate haplotypes for analysis of marker-to-marker LD. Ten SIMWALK2 runs were performed on the data after error checking, with a variety of initial analysis parameters, to identify the most likely haplotypes. Genotypes from all 15 SNPs, as well as from the seven microsatellite markers APOA2, B195G14.ttg1, B366H14.tcta1, B366H14.ca1, D1S2675, D1S1679, and D1S2768, were used simultaneously for haplotype generation. Marker order was set according to build 34 of the human genome assembly (UCSC). To identify and remove any haplotypes with one or more markers that could not be definitively phased, the resulting haplotypes were compared across runs. Thirteen founder haplotypes were removed because they could not be fully phased, and 178 fully phased founder haplotypes were defined in this sample. Founder haplotypes were then collected into input files for GOLD to assess marker-to-marker LD (Abecasis and Cookson 2000).
In testing for evidence of LD to schizophrenia, we wanted to use an analysis method that could take full advantage of the extended pedigree structure of the families in our study, but also one that would clearly separate the evidence for LD from the strong evidence for linkage in this sample. On this basis, we elected to use the program PSEUDOMARKER v0.9.7 to conduct analyses of LD (Goring and Terwilliger 2000). PSEUDOMARKER, unlike many popular TDT-based programs, explicitly separates the evidence for linkage and LD in families with multiple affected individuals. Simulation studies also indicate that PSEUDOMARKER has greater power than FBAT (Laird et al. 2000; Horvath et al. 2001) and PDT (Martin et al. 2000; Martin et al. 2001) to detect LD in this sample (Brzustowicz et al. 2003). For each marker, PSEUDOMARKER provides separate evaluations for evidence of linkage, of LD given linkage, of LD given no linkage, of linkage given LD, and for a joint test of linkage and LD. On the basis of the most favored model from our prior linkage results for the 1q22 locus, analyses were conducted using a recessive pseudomarker model and a narrow definition of “affected” (schizophrenia or schizoaffective disorder) (Brzustowicz et al. 2000).
Simulation studies were conducted to assess the significance of our findings. The analysis of multiple markers for LD raises the possibility of false positive findings due to multiple testing. However, as marker-to-marker disequilibrium is likely to exist over small physical distances, these tests are not independent, and a Bonferroni correction for multiple testing would be overly conservative. In addition, we were concerned about the possibility that the strong linkage signal in this sample could bias the analysis of LD. Our goal was to provide an appropriate correction of type I error in the evidence of LD, under the situation of strong linkage and marker-to-marker LD. We therefore devised a simulation strategy to address this issue. First, the general segregation of haplotypes within the pedigrees was defined using SIMWALK2 with the haplotype label option. The population of fully phased founder haplotypes was then shuffled and randomly reassigned to the segregating haplotype labels. On average, this procedure is expected to preserve the marker-to-marker LD (since the original real founder haplotypes are used) and to preserve the marker-to-disease linkage signal of the original sample (since the same pattern of allele sharing among relatives within each pedigree is maintained), but it also should generate a random marker-to-disease LD signal. We analyzed 2,500 replicates. Owing to the predominance of SNP markers within the region of interest and to the computational burden of conducting PSEUDOMARKER analysis with highly polymorphic microsatellite markers (since computation time increases exponentially with the number of alleles), simulations were restricted to the set of 15 SNPs. Each simulated replicate was analyzed by the same analysis method as the real data (recessive pseudomarker analysis with a narrow disease definition), and a distribution of results was assembled to allow empiric evaluation of the results of the real-data analysis.
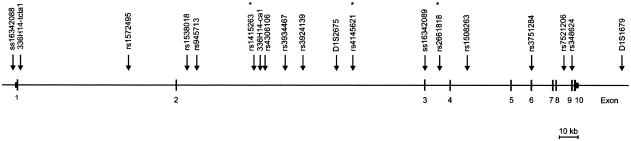
Evidence of LD between schizophrenia susceptibility and markers from chromosome 1q22 was assessed in two stages. For the first stage of analysis, the 14 microsatellites from D1S1653 to D1S1677, that were previously used for the fine genetic linkage mapping of this region (Brzustowicz et al. 2002), were screened for LD, using PSEUDOMARKER, under a recessive pseudomarker model. Consistent with prior results, all these markers produce significant evidence (nominal P<.05) of linkage to schizophrenia, but only two—336H14-ca1 (nominal P=.044) and D1S2675 (nominal P=.036)—produced significant evidence (nominal P<.05) of LD to schizophrenia, given linkage. As both of these markers were located within the genomic extent of CAPON, or carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase, additional markers within this gene were selected for further study.
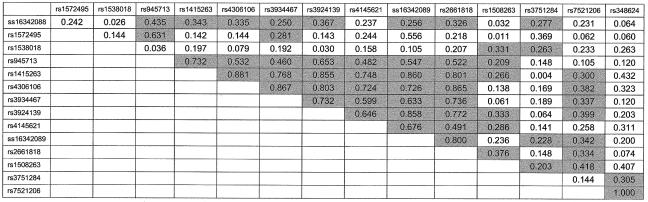
For the second stage of analysis, 15 SNPs from CAPON were evaluated for LD (fig. 1). Twelve of these were from intronic sequences, one 5′ to the gene (ss16342088), one from exon 6 (rs3751284), and one from exon 9 (rs348624). SNPs rs3751284 and rs348624 represent the only SNPs in the CAPON coding sequence that were observed during resequencing of 16 individuals from our linkage sample or that are listed in dbSNP, build 118. Both of these SNPs represent synonymous substitutions. Marker-to-marker LD was calculated for all marker pairs, with significant pairwise LD detected through much of the region (fig. 2).
Figure 1.
Map of genomic structure of CAPON with genetic markers indicated. Asterisks (*) indicate SNPs that reach studywide empirical significance for LD at the P<.05 level.
Figure 2.
LD (D′) between pairs of SNP markers. Pairs with significant evidence (P<.05) of LD are shaded.
Results of LD analysis of markers from CAPON are presented in table 1. Of the 15 SNPs from CAPON, 6 exhibited significant LD to schizophrenia (nominal P<.05). We conducted simulation studies to determine empiric P values for these results, both for each marker individually and for the entire set of SNP markers. Empiric P values for individual markers indicated that, for this collection of pedigrees and for this pattern of haplotype segregation, the nominal P values produced by PSEUDOMARKER were slightly anticonservative (table 1), although results for all six SNPs remained significant. For the full set of 15 SNPs used in this set of pedigrees, simulations indicated that a nominal P value of <.0025 was required to reach studywide empirical significance at the P<.05 level. Markers rs1415263, rs4145621, and rs2661818 attained this level of significance, with studywide empirical P values of .019, .036, and .045, respectively. Not surprisingly, these three markers exhibit significant marker-to-marker LD (fig. 2). We also used simulation results to calculate the overall probability of observing, by chance, 6 of 15 SNPs showing nominally significant LD to schizophrenia, given the existing pattern of marker-to-marker LD. An empiric P value of .015 was obtained.
Although powerful for linkage analysis, this sample of 24 independent pedigrees would not be expected to have much power to detect LD. Overall, a total of 178 founder haplotypes could be reconstructed for the region investigated, with 90 of these observed to be transmitted to individuals with schizophrenia. Thus, for purposes of LD analysis, this set of 330 subjects would be expected to behave, at best, like a collection of 45 trios, a modest-size sample for a complex disease-association study. This makes the detection of significant LD in this sample all the more striking.
Our evidence for LD is centered within the CAPON gene. The only previous study of LD involving this region investigated six microsatellite markers spanning 39 cM from 1p21.1-q23.3 in 80 Spanish nuclear families, and the authors reported significant LD to schizophrenia only for D1S1679 (Rosa et al. 2002), which is located within 25 kb of the 3′ end of CAPON. The next nearest markers tested were D1S1653 and D1S1677, which map 3.8 Mb 5′ and 1.2 Mb 3′ to CAPON, respectively. Linkage studies also support a 1q22 positional candidate for schizophrenia. Several independent studies, including our own, have shown strong or suggestive linkage to this region (Shaw et al. 1998; Brzustowicz et al. 2000, 2002; Gurling et al. 2001; Hwu et al. 2003). A large meta-analysis of genome scans also supports a schizophrenia susceptibility locus in the 1q region (Lewis et al. 2003). It is important to note that, previously, we found that an estimated 75% of these families were linked to 1q22 and also that there was significant evidence for linkage to 13q32 in an estimated 65% of families (Brzustowicz et al. 2000), indicating that several schizophrenia susceptibility genes, including CAPON, may be active in the same sample.
The expression profile of CAPON further supports this gene as an appealing candidate for schizophrenia susceptibility. Human transcripts have been isolated from the brain and testes of adults and infants (UniGene database, NCBI). Rat studies reveal expression in the adult brain, but not in the adrenal, bladder, heart, kidney, lung, or skeletal muscle; the highest expression was in the cerebral cortex and medulla oblongata (Jaffrey et al. 1998). Currently, nothing is known about the fetal expression pattern of CAPON. However, CAPON is known to participate in neuronal signal transduction and other pathways that are essential for both neuronal proliferation and neuronal differentiation in development and for synaptic plasticity throughout life (Hata and Takai 1999; Boehning and Snyder 2003).
CAPON primarily functions as an adaptor protein targeting neuronal nitric oxide synthase (nNOS). The C-terminal amino acids of CAPON bind to the PDZ domain of the neuronal—but not the other—forms of NOS, and CAPON and nNOS are colocalized in rat brains (Jaffrey et al. 1998). CAPON competes with PSD95 for binding to nNOS. PSD95 is a postsynaptic density scaffolding protein that binds to both nNOS and NMDA receptor subunits in neurons, thereby allowing activation of nNOS by Ca++ influx through the NMDA receptor. This produces NMDA receptor-mediated NO release into the synaptic structures. CAPON, by competitively binding to nNOS, disrupts this coupling and, thus, inhibits this local NO release (Jaffrey et al. 1998). This places CAPON at the scene of NMDA glutamate neurotransmission, long proposed to be involved in schizophrenia. Abnormal expression of PSD95 and of related post-synaptic density proteins has also implicated NMDA receptor-associated intracellular pathways and signal-transduction abnormalities in schizophrenia (Clinton et al. 2003).
CAPON also targets nNOS to Dexras1 (Fang et al. 2000), thereby facilitating the NO-dependent activation of this small G-protein (Jaffrey et al. 2002b; Boehning and Snyder 2003), which is involved in modulating signal transduction (Graham et al. 2002). Recently, the N-terminus of CAPON has also been found to interact with synapsins I, II, and III through a phosphotyrosine-binding domain, thereby changing the subcellular localization of nNOS (Jaffrey et al. 2002a). Synapsins are a family of phosphoproteins involved in synaptogenesis and in regulating neurotransmitter release (Ferreira and Rapoport 2002).
CAPON has not previously been proposed as a candidate gene for schizophrenia. The genetic evidence, expression pattern, and known functions of this gene, however, are consistent with prevailing models of schizophrenia pathogenesis, including anomalous neurodevelopment (Bassett et al. 2001a) and abnormal synaptic connectivity (Harrison 1999). Two recent reviews of the most plausible candidate susceptibility genes for schizophrenia (Harrison and Owen 2003; Sawa and Snyder 2003) propose that most or all are involved in schizophrenia susceptibility through interlinked processes in glutamatergic synapses, particularly those processes involving NMDA receptors. CAPON’s role in mediating NMDA receptor-associated signaling would be consistent with this model of schizophrenia pathogenesis. In addition, the specific binding of CAPON to nNOS implicates NO, which can have both neuroprotective and neurotoxic functions. The NO system is directly involved in mediating neuronal response to hypoxia (Boehning and Snyder 2003) and could fit an integrated model, including nongenetic factors such as in utero hypoxia (Zornberg et al. 2000) and childhood head injury (AbdelMalik et al. 2003).
Screening of the coding region of CAPON in individuals from our linkage sample has failed to identify any mutations that would be predicted to be associated with altered protein function. This is consistent with current results for other candidate genes for schizophrenia (Harrison and Owen 2003). Given the possibility of trinucleotide repeat expansions in schizophrenia (Bassett and Husted 1997; Margolis et al. 1999), we examined a small imperfect CAG repeat within exon 7 ([CAG]2[TCT]2[CAG]6) coding for polyglutamine with interspersed leucines, for evidence of expansion. We found no evidence of instability at that site.
CAPON has a large, nearly 300-kb, genomic extent, only 1.5 kb of which represents coding sequence. Therefore, there are many potential sites for regulatory sequences that could be disrupted and that could lead to altered protein expression. The markers that exhibit empirically significant evidence of LD to schizophrenia span 99 kb and define a region that includes most of the very large (133-kb) intron 2 of CAPON. The existence of an additional gene or genes within this intron cannot be excluded, providing the possibility of other positional candidates within the region of LD.
There are now two reports of significant LD between schizophrenia and markers in the CAPON region: this sample and that of Rosa et al. (2002). It will be very interesting to see if this association can be detected in other schizophrenia samples.
Acknowledgments
This work was supported by grant R01 MH62440 from the National Institutes of Mental Health (L.M.B.), the Canadian Institutes of Health Research (A.S.B.), and The Bill Jefferies Schizophrenia Endowment Fund (A.S.B.). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through contract N01-HG-65403 from the National Institutes of Health to the Johns Hopkins University. Thanks to Christina Lucey and Vivian Wong for assistance with figure preparation and to Tero Hiekkalinna for assistance with PSEUDOMARKER.
Appendix A: Supplemental Data
Table A1.
SNP Primer Sequences
|
Primer (5′→3′) |
|||
| SNP | Biotinylated PCR | Unlabeled PCR | Pyrosequencing |
| ss16342088 | ctctaaaacctggcacagtaactca | cgtttggagtcagaggacatt | gaatatgagatgtaaatgaa |
| rs1572495 | gtgttttcctacctctcacgttttc | gggttctaaagggactgtacgatt | agatttgagctgagcc |
| rs1538018 | gtgttacaacttcagctgttctgag | ctttgtccaggagtcactttatcc | tcagcaccatttactcc |
| rs945713 | gagataaggaaatatgaaaaacaagtg | gccattatttgattttatgaaactctt | catttggatatcccaaag |
| rs1415263 | tggtatcaaccaaactagcaaagaa | tgtggtaaaatggaaatctgtcaa | aggcaacatacttgatga |
| rs4306106 | tggaagctttatggagagaaattat | taagaatatatcgctttcagggtca | aaactcagtaaagctaccc |
| rs3934467 | aggctttcgaggaacagtctc | gtgtagcttctgaactctctgtgga | gtgccccggcacac |
| rs3924139 | gttattgagcatggcagtatacaagg | tcaagtaaaaggtctctgaaacaactg | agaaatttatgatactagcc |
| rs4145621 | gggacccatctcaatatgttttt | ctgttgctatcatctattgtccatc | tgtggtttgatggagaa |
| ss16342089 | agacatccacctgcttttacatc | tgatgagatcagtgcatctatacag | gaaaaaaggtcccca |
| rs2661818 | atttagcgtattggcctcacc | aaaacagcccagggaacc | ttcaacctccttccact |
| rs1508263 | aaaggcattactagctcctcttttt | cagagcagtttcagtagggagttg | ttacttggtagtgaaagaaa |
| rs3751284 | ccaagctatgagaatcgttcgg | agtgcctacctgggtctcctga | ctgttgctgttcctctc |
| rs7521206 | tcaaagaattaagccccttt | gccgtagtgtcacatcactct | ctgtgccacatcactg |
| rs348624 | acacttgctgaaggaccagttg | tgacagcttcagttccagctctt | gcaaaagctgatgcac |
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Center for Inherited Disease Research (CIDR), http://www.cidr.jhmi.edu/
- National Center for Biotechnology Information, dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=OMIM
- Pyrosequencing Technical Support, http://techsupport.pyrosequencing.com
- UCSC Genome Browser, http://genome.cse.ucsc.edu/
- UniGene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene
- Whitehead Institute, http://www.genome.wi.mit.edu/genome-software/other/primer3.html
References
- AbdelMalik P, Husted J, Chow EW, Bassett AS (2003) Childhood head injury and expression of schizophrenia in multiply affected families. Arch Gen Psychiatry 60:231–236 10.1001/archpsyc.60.3.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J (2000) Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem 280:103–110 10.1006/abio.2000.4493 [DOI] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, O’Neill S, Brzustowicz LM (2001a) Genetic insights into the neurodevelopmental hypothesis of schizophrenia. Schizophr Bull 27:417–430 [DOI] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, Waterworth DM, Brzustowicz L (2001b) Genetic insights into schizophrenia. Can J Psychiatry 46:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Collins EJ, Nuttall SE, Honer WG (1993) Positive and negative symptoms in families with schizophrenia. Schizophr Res 11:9–19 10.1016/0920-9964(93)90033-F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Honer WG (1994) Evidence for anticipation in schizophrenia. Am J Hum Genet 54:864–870 [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Husted J (1997) Anticipation or ascertainment bias in schizophrenia? Penrose’s familial mental illness sample. Am J Hum Genet 60:630–637 [PMC free article] [PubMed] [Google Scholar]
- Boehning D, Snyder SH (2003) Novel neural modulators. Ann Rev Neurosci 26:105–131 10.1146/annurev.neuro.26.041002.131047 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS (2002) Fine mapping of the schizophrenia susceptibility locus on chromosome 1q22. Hum Hered 54:199–209 10.1159/000070665 [DOI] [PubMed] [Google Scholar]
- Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS (2000) Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 288:678–682 10.1126/science.288.5466.678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Simone J, Mohseni P, Hayter JE, Chow EWC, Bassett AS (2003) Linkage disequilibrium mapping of a schizophrenia susceptibility locus on 1q22. Am J Hum Genet suppl 73:183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Haroutunian V, Davis KL, Meador-Woodruff JH (2003) Altered transcript expression of NMDA receptor-associated postsynaptic proteins in the thalamus of subjects with schizophrenia. Am J Psychiatry 160:1100–1109 10.1176/appi.ajp.160.6.1100 [DOI] [PubMed] [Google Scholar]
- Fang M, Jaffrey SR, Sawa A, Ye K, Luo X, Snyder SH (2000) Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron 28:183–193 10.1016/S0896-6273(00)00095-7 [DOI] [PubMed] [Google Scholar]
- Ferreira A, Rapoport M (2002) The synapsins: beyond the regulation of neurotransmitter release. Cell Mol Life Sci 59:589–595 10.1007/s00018-002-8451-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring HH, Terwilliger JD (2000) Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet 66:1310–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TE, Prossnitz ER, Dorin RI (2002) Dexras1/AGS-1 inhibits signal transduction from the Gi-coupled formyl peptide receptor to Erk-1/2 MAP kinases. J Biol Chem 277:10876–10882 10.1074/jbc.M110397200 [DOI] [PubMed] [Google Scholar]
- Gurling HM, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, Petursson H, Curtis D (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ (1999) The neuropathology of schizophrenia: a critical review of the data and their interpretation. Brain 122:593–624 10.1093/brain/122.4.593 [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Owen MJ (2003) Genes for schizophrenia? Recent findings and their pathophysiological implications. Lancet 361:417–419 10.1016/S0140-6736(03)12379-3 [DOI] [PubMed] [Google Scholar]
- Hata Y, Takai Y (1999) Roles of postsynaptic density-95/synapse-associated protein 90 and its interacting proteins in the organization of synapses. Cell Mol Life Sci 56:461–472 10.1007/s000180050445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM (2001) The family based association test method: strategies for studying general genotype—phenotype associations. Eur J Hum Genet 9:301–306 10.1038/sj.ejhg.5200625 [DOI] [PubMed] [Google Scholar]
- Hwu HG, Liu CM, Fann CS, Ou-Yang WC, Lee SF (2003) Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry 8:445–452 10.1038/sj.mp.4001235 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Benfenati F, Snowman AM, Czernik AJ, Snyder SH (2002a) Neuronal nitric-oxide synthase localization mediated by a ternary complex with synapsin and CAPON. Proc Natl Acad Sci USA 99:3199–3204 10.1073/pnas.261705799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey SR, Fang M, Snyder SH (2002b) Nitrosopeptide mapping: a novel methodology reveals s-nitrosylation of dexras1 on a single cysteine residue. Chem Biol 9:1329–1335 10.1016/S1074-5521(02)00293-4 [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snowman AM, Eliasson MJ, Cohen NA, Snyder SH (1998) CAPON: a protein associated with neuronal nitric oxide synthase that regulates its interactions with PSD95. Neuron 20:115–124 10.1016/S0896-6273(00)80439-0 [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X (2000) Implementing a unified approach to family-based tests of association. Genet Epidemiol 19:S36–S42 [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 73:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, McInnis MG, Rosenblatt A, Ross CA (1999) Trinucleotide repeat expansion and neuropsychiatric disease. Arch Gen Psychiatry 56:1019–1031 10.1001/archpsyc.56.11.1019 [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Kaplan NL (2001) Correcting for a potential bias in the pedigree disequilibrium test. Am J Hum Genet 68:1065–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL (2000) A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet 67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Asherson P, Owen M, Farmer A (1994) The strength of the genetic effect: is there room for an environmental influence in the aetiology of schizophrenia? Br J Psychiatry 164:593–599 [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P (1998) A sequencing method based on real-time pyrophosphate. Science 281:363, 365 10.1126/science.281.5375.363 [DOI] [PubMed] [Google Scholar]
- Rosa A, Fañanás L, Cuesta MJ, Peralta V, Sham P (2002) 1q21-q22 locus is associated with susceptibility to the reality-distortion syndrome of schizophrenia spectrum disorders. Am J Med Genet 114:516–518 10.1002/ajmg.10526 [DOI] [PubMed] [Google Scholar]
- Sawa A, Snyder SH (2003) Schizophrenia: neural mechanisms for novel therapies. Mol Med 9:3–9 [PMC free article] [PubMed] [Google Scholar]
- Shaw SH, Kelly M, Smith AB, Shields G, Hopkins PJ, Loftus J, Laval SH, Vita A, De Hert M, Cardon LR, Crow TJ, Sherrington R, DeLisi LE (1998) A genomewide search for schizophrenia susceptibility genes. Am J Med Genet 81:364–376 [DOI] [PubMed] [Google Scholar]
- Sobel E, Lange K (1996) Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet 58:1323–1337 [PMC free article] [PubMed] [Google Scholar]
- Sobel E, Sengul H, Weeks DE (2001) Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered 52:121–131 10.1159/000053366 [DOI] [PubMed] [Google Scholar]
- Zornberg GL, Buka SL, Tsuang MT (2000) Hypoxic-ischemia-related fetal/neonatal complications and risk of schizophrenia and other nonaffective psychoses: a 19-year longitudinal study. Am J Psychiatry 157:196–202 10.1176/appi.ajp.157.2.196 [DOI] [PubMed] [Google Scholar]