Abstract
We investigated a family with an autosomal recessive syndrome of café-au-lait patches and childhood malignancy, notably supratentorial primitive neuroectodermal tumor. There was no cancer predisposition in heterozygotes; nor was there bowel cancer in any individual. However, autozygosity mapping indicated linkage to a region of 7p22 surrounding the PMS2 mismatch-repair gene. Sequencing of genomic PCR products initially failed to identify a PMS2 mutation. Genome searches then revealed a previously unrecognized PMS2 pseudogene, corresponding to exons 9–15, within a 100-kb inverted duplication situated 600 kb centromeric from PMS2 itself. This information allowed a redesigned sequence analysis, identifying a homozygous mutation (R802X) in PMS2 exon 14. Furthermore, in the family with Turcot syndrome, in which the first inherited PMS2 mutation (R134X) was described, a further truncating mutation was identified on the other allele, in exon 13. Further whole-genome analysis shows that the complexity of PMS2 pseudogenes is greater than appreciated and may have hindered previous mutation studies. Several previously reported PMS2 polymorphisms are, in fact, pseudogene sequence variants. Although PMS2 mutations may be rare in colorectal cancer, they appear, for the most part, to behave as recessive traits. For technical reasons, their involvement in childhood cancer, particularly in primitive neuroectodermal tumor, may have been underestimated.
Introduction
Embryonal tumors of the CNS are a heterogeneous group for which a precise diagnosis by morphological criteria is problematic. Most common among them is medulloblastoma, classically arising in the posterior fossa, probably from cerebellar granule cells (Pomeroy et al. 2002). Supratentorial primitive neuroectodermal tumor (SPNET) is a rarer, aggressive embryonal tumor, most likely derived from primitive neuroepithelial cells. It has features that suggest that it is biologically and genetically distinct from the histologically similar medulloblastoma (Russo et al. 1999; Pomeroy et al. 2002). SPNET has a poor prognosis, with median survival of <2 years (Dirks et al. 1996), although there are indications that this may be improved with aggressive therapy (Jakacki 1999; Yang et al. 1999; Strother et al. 2001).
Rather little is known about the molecular basis for SPNET. It is usually sporadic and has not been commonly reported in the major hereditary cancer syndromes. There are, however, occasional exceptions. Pineal or suprasellar primitive neuroectodermal tumor (PNET) can occur in patients with germline RB1 mutations (so-called “trilateral” retinoblastoma [MIM 180200]) (Paulino 1999). Also, childhood PNET can occur in patients with germline TP53 mutations (Li-Fraumeni syndrome [MIM 151623]) (Orellana et al. 1998), whereas somatic TP53 mutations have been found in adult SPNETs (Ho et al. 1996). Central PNET with features of rhabdoid differentiation can result from germline INI1 (hSNF5) mutations (SMARCB1 [MIM 601607]) (Sevenet et al. 1999; Taylor et al. 2000).
There are at least two reports of PNET in neurofibromatosis type 1 (NF1 [MIM 162200]). One of these tumors was peripheral, developing in a preexisting plexiform neurofibroma (Chan et al. 1996). The other was a large SPNET that developed after chemo- and radiotherapy for a brain stem astrocytoma (Raffel et al. 1989). Most intracranial tumors in NF1, however, are glial in origin, which raises the question of whether atypical tumors, including medulloblastomas or SPNETs, may be a pointer to a genetic disorder distinct from NF1. Although most children with café-au-lait spots (CALS) do indeed have NF1 (Korf 1992), the precise spectrum of tumors attributable to NF1 remains unclear (Korf 2000), and there are recent reports of an NF1-like phenotype in children with unexpected tumors (Ricciardone et al. 1999; Wang et al. 1999; Vilkki et al. 2001; Whiteside et al. 2002).
We report here on a heavily consanguineous family showing cosegregation of cutaneous CALS and early-onset brain tumors. This presentation led to an initial diagnosis of NF1 (although the consensus criteria were not met). Later, the clearly recessive inheritance pattern suggested the likelihood of an alternative, unrecognized cancer syndrome. Unexpectedly, the causative gene ultimately proved to be PMS2, prompting us to reconsider the accepted role of this gene in tumor predisposition.
PMS2 is one of the mammalian genes similar to the mutL DNA mismatch-repair (MMR) gene of Escherichia coli (Marti et al. 2002). The hMutLα heterodimer of PMS2 with MLH1 (another mutL homologue) is the major species providing mutL-like MMR activity in human cells (Li and Modrich 1995; Raschle et al. 1999; Harfe and Jinks-Robertson 2000). PMS2-mutated human cells accordingly show a mutation rate equal to or higher than that of MLH1-mutated cells (Kato et al. 1998). Despite this, although germline MLH1 mutations are the most common cause of hereditary nonpolyposis colon cancer (HNPCC) (Peltomaki 2001), PMS2 mutations are, in contrast, very rare, having been reported in only five families with cancer (Nicolaides et al. 1994; Hamilton et al. 1995; Miyaki et al. 1997; De Rosa et al. 2000; Trimbath et al. 2001).
The existence of several pseudogenes corresponding to the first five or so exons of PMS2 has been recognized; many of them are transcribed and could interfere with both genomic and cDNA-based mutation analyses (Horii et al. 1994; Nicolaides et al. 1995; Kondo et al. 1999). Reagents that allow discrimination between PMS2 and these pseudogenes have been described (Nicolaides et al. 1995). The experience we report here, however, suggests that even these may not permit a comprehensive genomic-based PMS2 mutation analysis and that new studies of PMS2 are warranted to further address its contribution to cancer predisposition.
Subjects and Methods
Clinical Features
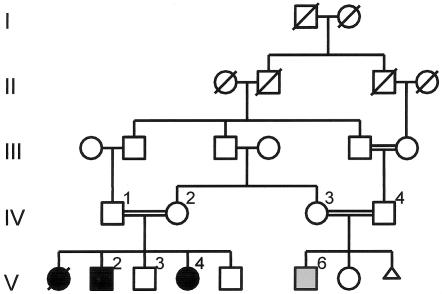
Three siblings developed brain tumors in childhood (fig. 1). Patient V-1 presented with left hemiparesis in 1991 at the age of 10 years. A multifocal right cerebral lesion with small-round-cell histology was classified as a high-grade B-cell non-Hodgkin lymphoma. She had had multiple CALS but was not personally examined, since she died in 1992. Patient V-2 also had multiple CALS. At 8 years of age, he presented with headaches and diplopia. A right temporoparietal mass was subtotally resected and found to be a PNET. Patient V-4 had multiple CALS and mild learning difficulties. At 14 years of age, she presented with impaired consciousness. A right frontal lobe lesion was resected and was again found to be a SPNET. (Its karyotype was near-tetraploid, with many cells also showing loss of one chromosome 13 and an unbalanced rearrangement resulting in 10q loss.) A fourth family member (V-6) also has CALS and mild learning difficulties; he is a double first cousin (aged 10 years) of the proband and so far has no history of cancer. No Lisch nodules or additional features of NF1 were present in any of the affected individuals. The parents of both sibships were first cousins. They were carefully examined and had no ocular or cutaneous signs of NF1. Thus, neither the proband nor any other family member met the diagnostic criteria for NF1 (National Institutes of Health 1988). The CALS in affected individuals had a ragged-edged, slightly diffuse appearance, not typical of the more sharply delineated CALS in NF1. There was no family history of colorectal or endometrial adenocarcinoma. Colonoscopy in patients IV-1 and IV-2 was normal. Full informed consent for genetic investigations was obtained from all subjects or their parents, and the studies were conducted under a protocol approved by the local ethics committee.
Figure 1.
Pedigree diagram. Linkage analysis was performed by use of DNA from the eight numbered individuals (IV-1–IV-4, V-2–V-4, V-6). Gray shading of the square representing patient V-6 indicates that he has CALS but no tumor.
Linkage Analysis
The Weber human genome screening set, version 8 (Research Genetics), comprising 395 autosomal microsatellite markers at ∼10 cM density, was used for an autozygosity search. PCR was performed on a RoboSeq 4200 automated biosystem (MWG Biotech), using 10-μl reactions containing 60 ng genomic DNA, 1 μM primers, 250 μM each dNTP, 5 U Taq polymerase (Promega), 1.5–3.0 mM MgCl2, 10 mM Tris-HCl (pH 8.0), 50 mM KCl, and 0.1% Triton X-100. Cycling conditions were 95°C for 120 s; 30 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 60 s; and 72°C for 7 min. Products were electrophoresed in multiplexed pools on an ABI 377 and analyzed with GeneScan and Genotyper 1.1.1. Additional individual markers were analyzed by similar methods.
Sequence Analysis of PMS2
Initially, we used primers (although with different buffer conditions) that were developed to allow the specific amplification of the PMS2 exons while avoiding coamplification of known pseudogene sequences (Nicolaides et al. 1994, 1995). We obtained gene-specific amplicons only for exons 1, 2, 6, 7, 8, and 9. New primers used for other exons (table 1) were designed as directed by multiple BLAST alignments (see below). Thermal cycling conditions (on a PTC-200 [MJ Research]) were as follows:
-
a.
94°C for 180 s; 35 cycles of 94°C for 30 s, 49°C for 30 s, and 72°C for 60 s;
-
b.
94°C for 180 s; 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 60 s;
-
c.
94°C for 180 s; 8 cycles of 94°C for 30 s, 58°C (−1°C/cycle) for 45 s, and 72°C for 60s; 32 cycles of 94°C for 30 s, 50°C for 45 s, and 72°C for 60 s; and 72°C for 420 s; and
-
d.
94°C for 180 s; 8 cycles of 94°C for 30 s, 63°C (−1°C/cycle) for 30 s, and 72°C for 60s; and 34 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s.
Table 1.
New PMS2-Specific Primers Used for Mutation Analysis
| Exon | PCR Primers | Protocol(Mg2+/mM) | Sequencing Primer |
| 3 | F: ACTGATAGCATGGGTCCG, R: CAAAATTCTGAGACATGTGA | c (1.5) | … |
| 4 | F: ACACTGTCTTGGGAAATG, R: ATTAATTTTCAGAGAGGTTTC | b (1.5) | … |
| 5 | F: CTCAACCATTTAGATCTTGA, R: AATAAAGCATTTCTCAATAAT | b (2.5) | … |
| 10 | F: ATTAAGCCCTTCCGTATTT, R: GGAAACACATTAGCTAAAAGC | b (1.5) | F: CCTGTAATCCTAGCTACT |
| 11 | F: GACCAGTCCTGAACTCCTAGCC, R: GCTGCAGTGAGCCAAGATCA | d (1.5) | F: CAGTCCACATCTGAAAAA, R: CTCGCAGGAACATGTGGA |
| 12 | F: GTGTACAGGTCTGAAAACTTG, R: CGCCTGGCCAACTAGATA | b (1.5) | … |
| 13 | F: CACTTAGCTGAGTAGTGTTGTTATTT, R: TGAACACCTGAAAGAGAGGAAAC | b (1.5) | … |
| 14 | F: TCCAAAAAGCATTTTGTGAGTT, R: GAGTTCAAGGTCACAGAGAACG | b (1.5) | … |
| 15 | F: AACTACTAAAACGTTGAACC, R: TTTTTGAGACACAGTCTTGT | a (2.0) | … |
PCR products were directly sequenced on both strands by use of the BigDye (Applied Biosystems) or ET (Amersham) dye-terminator cycle sequencing kits and ABI 377 or MegaBace 500 sequencers, respectively. Exon 13 products heterozygous for the 2184delTC mutation were cloned to allow resolution of the two alleles.
Analysis of PMS2-Associated Repeat Elements
Following MegaBLAST genome searches (MegaBLAST Web site), pairwise comparisons between regions containing PMS2-related sequences were performed by use of PipMaker (Schwartz et al. 2000) and RepeatMasker (RepeatMasker Web site). As described below, all PMS2 pseudogenes are associated with a copy of a ∼17-kb sequence that we refer to as “PPJ” (pseudo-PMS2-juxtaposed). A total of 20 PPJ copies exist on chromosome 7. Two regions of the PPJ sequence, free from high-copy repeat, were identified, and all 20 copies of each were aligned by use of ClustalW (Thompson et al. 1994). By use of PHYLIP (PHYLIP Web site), two phylogenetic trees were constructed for each of these two alignments: one by a parsimony method, one by a distance method, and both by bootstrapping. The groupings that were suggested from both aligned regions were concordant. The distance trees demonstrated the existence of two main subgroups of PPJ repeats, each containing nine members, and two outliers (at 4.78 Mb and 6.47 Mb).
Results
Linkage Mapping
The pedigree structure is shown in figure 1. Cytogenetic analysis (G-banding), including chromosome breakage studies, had shown a normal karyotype in all affected individuals. For purposes of linkage, patient V-6, who has CALS but no tumor, was assigned as affected. The three living affected individuals, one unaffected sibling, and their four parents were analyzed for autozygosity by use of 395 autosomal markers. Two regions were concordantly homozygous in the three affected individuals: 9q34.3 (D9S158) and 7p22.1 (GATA24F03 [D7S1819 and D7S517], 4.14 Mb). The 9q region was eliminated by demonstrating heterozygosity for closely flanking markers D9S1818 and D9S1838. In contrast, two additional 7p22.1 markers, D7S481 (5.78 Mb) and D7S511 (4.35 Mb), were found to be concordantly homozygous. Multipoint analysis of the entire genome-search data set confirmed that this was the only genomic region with a LOD score >3. It is significant that this homozygous region includes the location of the PMS2 MMR gene (5.7 Mb).
Mutation Analysis
Sequencing of PMS2 was initially performed by use of published PCR primers (Nicolaides et al. 1995). Amplification of exons 10 and 11 failed, and no sequence variants unique to the affected individuals were found in the remaining exons. The strong genetic evidence implicating this locus, however, prompted close examination of the patient sequences. Compared with the published PMS2 cDNA sequence, a number of apparently heterozygous or homozygous base changes were observed, in the amplicons for exons 3, 4, 5, 13, and 14. These were consistently present in both unaffected and affected individuals. The appearance of heterozygous changes within an autozygous region suggested that there might still be coamplification of two or more targets. This is probably attributable at least in part to the fact that we had not employed the unusual PCR buffer conditions of the original report (Nicolaides et al. 1995); these conditions are necessary to obtain PMS2-specific products (B. Vogelstein, personal communication). Even so, the previously reported partial PMS2 pseudogenes (Horii et al. 1994; Nicolaides et al. 1995; Kondo et al. 1999) did not explain the anomalous results for exons 13–14. We therefore performed a search for all PMS2-related sequences in the draft genome sequence. As detailed below, this identified 14 pseudogenes, each corresponding to most or all of the PMS2 exon 1–5 region. (Some of these correspond to the pseudogenes previously identified by others.) However, in addition, we identified a novel 100-kb genomic duplicon containing copies of exons 9 and 11–15. All of these pseudogenes are located on chromosome 7.
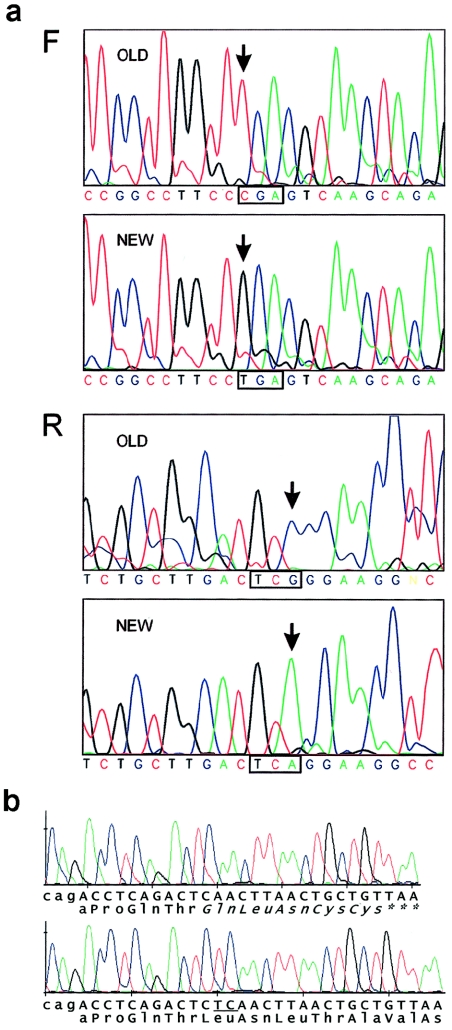
Alignment of the sequences of PMS2 and all of these pseudogenes allowed the design of completely specific PCR primer pairs for exons (3–5, 10–15) that had previously failed or had coamplified with pseudogene sequences. After sequencing the new amplicons, a homozygous 2428C→T mutation was found in exon 14 of PMS2 in all three affected individuals. This is a nonsense mutation (R802X). Figure 2 shows a comparison between the exon 14 sequences obtained by use of the new specific primers and the published pair. All four parents and the unaffected sibling were heterozygous for this mutation. The original mutation analysis failed to detect the R802X mutation because, under the conditions we used, the original primers actually preferentially amplify the previously unrecognized pseudogene (which we refer to below as “ψ0”).
Figure 2.
a, Mutation analysis of PMS2 exon 14 (ABI377). Forward (F) and reverse (R) strand sequences are shown of uncloned PCR products generated by use of the published primer pair (upper panels) or the redesigned PMS2-specific primers (lower panels). The mutated codon is boxed, and the arrows indicate the mutated nucleotide. b, Exon 13 mutation in the patient with Turcot syndrome (Hamilton et al. 1995) (MegaBace). PCR products were cloned to show both deleted (top) and normal alleles. The lowercase nucleotides are the end of intron 12. The deleted dinucleotide (within a 2-nt repeat) is underlined, and the resulting predicted novel C-terminus is italicized.
Recessive Inheritance of Turcot Syndrome
The first report of an inherited PMS2 mutation was in siblings with Turcot syndrome (MIM 276300) (Hamilton et al. 1995). Both had inherited a heterozygous R134X mutation from their unaffected father. We wondered whether this might indicate that R134X, like R802X, is a recessive mutation, and we looked for a second mutation by genomic analysis, as described above. A heterozygous 2-bp deletion was found in exon 13, within a repeated dinucleotide (CTCT) at codon 728–729 (2184delTC). This brings a stop codon in frame after five novel amino acid residues. The mutation was confirmed by sequencing cloned PCR products (fig. 2b), and its specificity for PMS2 exon 13 (as opposed to ψ0) was confirmed by the presence of TTT, rather than TTC, at codon 751 in these clones. Both siblings were compound heterozygotes (R134X/2184delTC), and 2184delTC was confirmed to be maternal in origin by analysis of both parental DNAs.
Distribution and Evolution of PMS2 Pseudogenes
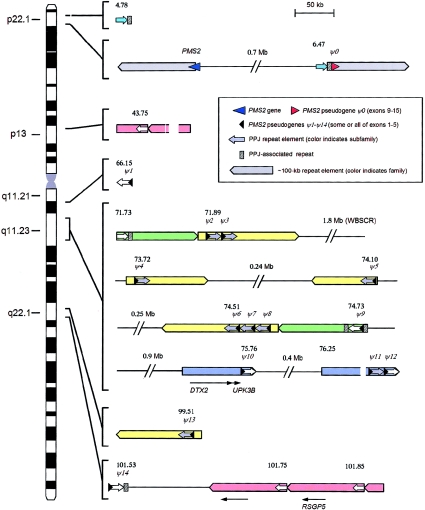
A number of studies have demonstrated the existence of a family of transcripts similar to PMS2 and their corresponding genomic sequences (related to PMS2 exons 1–5) on chromosome 7q (Horii et al. 1994; Nicolaides et al. 1995; Osborne et al. 1997; Chadwick et al. 2000; Valero et al. 2000). However, the full extent of this family has not been systematically examined by use of recent whole-genome sequence data. To ensure the specificity of our mutation analysis, we first performed a BLAST search of the human genome with the PMS2 reference cDNA NM_000535. This revealed a total of 15 pseudogene loci on chromosome 7, none of them processed. In addition to PMS2 itself (15 exons), there is one locus (ψ0) containing exons 9 and 11–15 and 14 loci (ψ1–ψ14) that each contain some or all of exons 1–5. There are no recognizable pseudogenes on other chromosomes. A search of the mouse genome with the reference mouse cDNA showed no evidence for comparable intrachromosomal duplications.
Some of the ψ1–ψ14 loci lie in close proximity, in a pattern that suggested that the ∼7-kb PMS2 genomic fragment containing exons 1–5 was part of a larger duplicated structure at the pseudogene loci. In fact, further analysis showed that all of these pseudogenes (ψ1–ψ14) are part of a larger sequence of ∼23 kb (including the PMS2 section), with the PMS2 section located at one end of the 23-kb block. It is curious that, despite being a feature common to all of the pseudogenes (ψ1–ψ14), the ∼16 kb of genomic sequence 3′ to the (ψ1–ψ14) exons is not present in the PMS2 gene itself. We refer to this ∼16-kb element as a PPJ element. Further BLAST searching revealed eight additional PPJ copies that are not associated with a 7-kb PMS2 exon 1–5 pseudogene segment. Two of these (comparatively poorly conserved) are on chromosome 17, and the others are on chromosome 7. One is in close proximity to the 5′ end of the ψ0 pseudogene (see below).
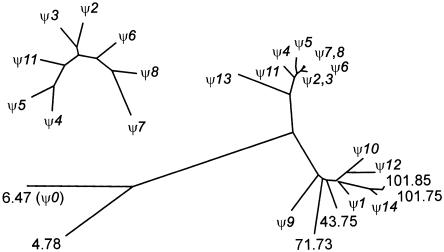
The distribution of the PPJ repeats (indicated by block arrows) is represented in figure 3. For insight into the mechanism of dispersal of the PPJ elements and their associated pseudogenes, two nonredundant segments of sequence were used for a phylogenetic comparison between all 20 of the chromosome 7 PPJ copies. A distance-based tree (fig. 4) shows that they fall into three groups (indicated in fig. 3 as arrows of different colors). Their mechanism of dispersal appears complex. In some cases, there is evidence for local endoreduplication (e.g., PPJ elements associated with ψ2-ψ3 and ψ6-ψ7-ψ8), but, in another case (ψ11-ψ12), PPJ elements that are evolutionarily divergent are juxtaposed, suggesting transposition or recombinational joining of different PPJ arrays.
Figure 3.
Distribution of PMS2 pseudogenes and related sequences on chromosome 7. Symbols are as defined in the key. The members of the pseudogene-associated repeat family (PPJ repeats) can be classified by sequence similarity into two main subgroups (indicated by white and gray block arrows) and two outlying members (turquoise). The type 1 members (gray) appear to be the younger of the two main PPJ subgroups, having (with the exception of the ψ13-associated PPJ element) shorter distance branches on the phylogeny tree. Larger blocks of different colors indicate different higher-order repeat elements. Those shown in yellow and green correspond, respectively, to the type A and type C repeat elements identified elsewhere in studies of the Williams syndrome deletion region (Valero et al. 2000). (Other large repeat blocks in this region that are not associated with PPJ elements are not shown.) Numbers indicate the approximate positions (in Mb) of each PPJ element on the draft genome sequence (build 33).
Figure 4.
Phylogenetic tree illustrating the relationships between the different members of the family with the 23-kb PPJ element. The tree was generated by using the DNADIST algorithm of the PHYLIP package to analyze a ClustalW-generated alignment of all 20 copies of a ∼1.5-kb segment of the PPJ element. (Details are available on request.) Branch lengths, therefore, give an indication of degree of divergence. The pseudogene-associated PPJ elements are referred to by the corresponding identity (e.g., ψ1). Those not associated with a pseudogene are given a number representing their sequence coordinate on chromosome 7 (National Center for Biotechnology Information [NCBI] release 33). The inset shows a tree generated from a subset of these PPJ elements by use of the same algorithm, to show more clearly the interrelationships of the closely similar ψ2–ψ8 PPJ elements. The structure of this tree implies that the local duplications leading to these PPJ clusters (and, hence, associated pseudogene clusters) occurred after the large-scale (∼100-kb) duplications around the Williams syndrome deletion region.
There are also several different examples of PPJ dispersal via intrachromosomal duplication of larger segments containing them. Some of these were previously described (as “type A” and “type C” repeats) in an analysis of the repeated sequences that flank the Williams syndrome deletion region (Valero et al. 2000). However, there are also three novel sets of large (∼100-kb) duplicons. One of these contains the exon 9–15 pseudogene (ψ0) and 3′ flanking region, in inverted orientation ∼740 kb upstream of PMS2. Another duplicon (associated with ψ10 and ψ11-ψ12) contains the DTX2 and UPK3B genes. A third (containing the non-PMS2-associated PPJ elements at 43.70 Mb, 100.69 Mb, and 100.79 Mb) contains the calcium-promoted Ras inactivator gene (RSGP5, CAPRI). Thus, there are five different large duplicons associated with PPJ elements on chromosome 7. Only 3 of the 20 PPJ elements are not part of one of these larger duplicons. The close similarity of the ψ2 and ψ3 PPJ elements and of the ψ6, ψ7, and ψ8 PPJ elements to each other further suggests that each of these clusters originated subsequent to the larger-scale genomic duplications around the Williams syndrome deletion region (fig. 4).
Discussion
Phenotype
Although CALS are the hallmark lesion of NF1, their presence alone does not establish the diagnosis (National Institutes of Health 1988). Nonetheless, this is the diagnostic label applied to most children with CALS (Burwell et al. 1982; Korf 1992), including, initially, the children in the family described here. It is accepted that patients with NF1 have an increased risk of malignancy (Korf 2000)—most commonly, peripheral malignant nerve sheath tumors and glial tumors, including astrocytomas (Baptiste et al. 1989). CNS neoplasms (other than neurofibromas and optic gliomas) occurred in 2% of NF1 probands in a large national series (Friedman and Birch 1997), and there have also been other suggestions of an association between medulloblastoma and NF1 (Perilongo et al. 1993; Martinez-Lage et al. 2002). In the family described here, however, none of the affected individuals met the National Institutes of Health consensus criteria. Furthermore, the pedigree structure strongly suggested a recessive disorder. This experience shows that diagnostic caution is needed when patients present with the constellation of brain tumors and CALS.
Five recent reports demonstrate the association of homozygous loss of MMR function with the combination of CALS and early-onset neoplasia (particularly hematological). This phenotype has been observed with mutation of MLH1 (Ricciardone et al. 1999; Wang et al. 1999; Vilkki et al. 2001), MSH2 (Whiteside et al. 2002), and PMS2 (Trimbath et al. 2001). In the families with MLH1 mutations (Ricciardone et al. 1999; Wang et al. 1999; Vilkki et al. 2001), there were phenotypes consistent with HNPCC in heterozygotes. The family reported here, in contrast, has no features suggesting HNPCC, nor any other clear heterozygote effect. The phenotype is one of CALS with brain tumors, typical neither of NF1 nor of HNPCC. This presentation appears highly characteristic, and we propose that it should be considered indicative of a disorder caused by recessive mutations in MMR genes, not of NF1. In isolated cases, however, the recognition of this phenotype could be problematic, especially since up to half of NF1 cases result from new mutations (Ruggieri and Huson 2001). It is thus very likely that other individuals with this disorder will have been misdiagnosed with NF1. SPNET, a rare tumor not usually found in the context of cancer-predisposition syndromes, is a particularly striking feature of the present family. It remains to be determined whether PMS2 mutations are a frequent underlying factor in nonfamilial PNET.
Genetic Behavior of PMS2 Mutations
Inherited mutations of PMS2 have been described in only five families, whereas mutations of its partner MLH1 are a frequent cause of HNPCC (Peltomaki 2001). This discrepancy has never been adequately explained. In particular, current biochemical knowledge does not support the idea that functional redundancy among the possible MLH1 partners (PMS1, PMS2, and MLH6) underlies the rarity of PMS2 mutations in cancer families (Li and Modrich 1995; Kato et al. 1998; Raschle et al. 1999; Harfe and Jinks-Robertson 2000).
Critical review of known families (table 2) suggests that PMS2 mutations behave as recessive traits. In four families (Hamilton et al. 1995; De Rosa et al. 2000; Trimbath et al. 2001), there are homozygous or compound heterozygous truncating mutations, with little or no evidence of cancer predisposition in heterozygotes. Mutation 2361delCTTC (De Rosa et al. 2000) truncates the protein at amino acid 788, and it is the most similar mutation to the R802X mutation in the present family. These mutations may remove the ability to interact with MLH1 through the PMS2 C-terminal domain (amino acids 675–850), defined elsewhere by in vitro studies (Guerrette et al. 1999). It is interesting to note that there is good existing evidence that R802X is a biochemically null mutation. This mutation was previously found in the HEC-1-A and HEC-1-B endometrial cancer cell lines (Kuramoto et al. 1972; Risinger et al. 1995). The R802X-hemizygous HEC-1-A line is almost completely lacking in MMR activity, unlike HEC-1-B, which also retains a normal PMS2 allele (Glaab et al. 1998). This, along with the correction of the severe microsatellite instability in HEC-1-A by reintroduction of functional PMS2 (Risinger et al. 1998), confirms that R802X is a null mutation.
Table 2.
Genetic and Clinical Summary of Reported Familial PMS2 Mutations
| Mutation | Exon | Inheritance | >1 SiblingAffected? | Tumor Type | Reference |
| R802X | 14 PMS2-specific | Recessive | Yes | PNET, non-Hodgkin lymphoma | Present study |
| 1145ins20a | 10/11 junction | Recessive | Yes | CALS, colon cancer, acute lymphoblastic leukemia, astrocytoma, ovarian neuroectodermal tumor, endometrial cancer, “brain tumor” | Trimbath et al. 2001 |
| 1221delG, 2361delCTTC | 11b, 14b | Recessive | Yes | Oligodendroglioma, colon cancer, neuroblastoma | De Rosa et al. 2000 |
| R134X, 2184delTC | 5, 13 | Recessivec | Yes | CALS, glioblastoma, colon cancer, non-Hodgkin lymphoma | Hamilton et al. 1995; present study |
| E705K | 12 | Recessived | No | Astrocytoma, lymphoma, colon cancer | Miyaki et al. 1997 |
Renumbered from original reference, according to 1 = A of initiator codon.
On the basis of presented data, one cannot absolutely exclude the possibility of this mutation being within the ψ0 pseudogene.
Dominant inheritance was initially suggested, but clinical data are more consistent with recessive inheritance (see text and also discussion in Liu et al. 2001).
Only one heterozygous mutation was found, but data are consistent with recessive inheritance (see text).
Evidence for dominant PMS2 mutations is less convincing. The R134X mutation (Hamilton et al. 1995) displayed a dominant negative effect on MMR in hamster fibroblasts, but not in human fibroblasts (Nicolaides et al. 1998; Yamada et al. 2003). Despite this, the clinical data were more suggestive of recessive inheritance, and we have now confirmed the presence of a second truncating mutation on the other allele in this patient. Heterozygous mutation E705K was reported in another patient with Turcot syndrome (Miyaki et al. 1997), but, as with R134X, this mutation was inherited from a clinically unaffected parent, who also lacked the constitutional MMR defect present in the patient. Miyaki et al. therefore suspected an undetected mutation on the patient’s other allele. A classical, “two-hit” (germline and somatic) loss of tumor-suppressor function was found in the first reported case of PMS2-related HNPCC (Nicolaides et al. 1994), but it is not clear if the germline mutation was inherited.
All in all, there remains no convincing evidence demonstrating segregation of the heterozygous PMS2 mutation with a cancer-predisposition syndrome. Mice heterozygous for a Pms2 disruption do not exhibit increased spontaneous mutagenesis, again suggesting that Pms2-related genetic instability may be recessive (Narayanan et al. 1997). In contrast, present evidence indicates that recessively inherited PMS2 pathology is a clearly defined entity and could be implicated in unusual types of neoplasm, particularly in childhood.
Pseudogenes Confounding Previous Studies of PMS2 in Cancer
Our experience suggests that some previous PMS2 mutation analyses may have suffered both from failure to detect pathogenic mutations and from misinterpretation of sequence changes within the pseudogene exons. The following specific points may be highlighted:
-
1.
The ψ0 exon 12 and 15 sequences are identical to those of PMS2. Previous analyses of these exons could not have distinguished which target was being analyzed.
-
2.
The ψ0 exons 13 and 14 each contain a single nucleotide substitution. Both of these (codon 751 TTT→TTC and codon 775 AAC→AGC) have been reported elsewhere as polymorphisms (Basil et al. 1999). (Observed heterozygosities >50% could have indicated the true nature of the sequence changes.) Sequence comparison predicts that neither the primers used by Basil et al. (1999) nor those of Nicolaides et al. (1995) would be selective for the real PMS2 exons 13–14. Our own experience, in fact, suggests that the published primers may, under some conditions, preferentially amplify pseudogene sequences (fig. 2). Again, therefore, some previous analyses of these exons in families with cancer may not have distinguished pseudogene and PMS2 targets.
-
3.
The large ψ0 exon 11 differs at 23 positions from PMS2. Again, two of these (codon 479 CAC→CAG, codon 496 CAC→CAT) were reported as PMS2 polymorphisms (Basil et al. 1999), whereas anomalous PCR results pointed others to the existence of an exon 11 pseudogene (Chadwick et al. 2000). The latter authors concluded that the exon 11 pseudogene was not transcribed since no corresponding RT-PCR product was obtainable (Chadwick et al. 2000), but, unfortunately, an RT-PCR primer located in exon 10 was used. Our finding, that exon 10 is missing from ψ0, explains the failure to detect pseudogene transcripts. EST databases show approximately equal numbers of spliced transcripts representing ψ0 and the corresponding region of PMS2. The ψ0 (9–15) pseudogene is therefore transcribed, and it will interfere with cDNA-based screens for PMS2 mutations, not just those based on genomic PCR.
-
4.
The location of the ∼100-kb inverted repeat associated with ψ0 raises the interesting possibility that an inversion could occur through intrachromosomal recombination between this repeat and its PMS2-associated copy. An inversion polymorphism of this kind is known to result from recombination between the inverted repeats flanking the Williams-Beuren syndrome critical region (WBSCR) (Osborne et al. 2001). Such an inversion would not physically disrupt PMS2 unless its breakpoint occurred within the much smaller ∼16-kb segment of the repeat that lies upstream of exon 15.
In conclusion, we suggest that the role of PMS2 as a familial cancer gene merits reexamination. In light of present evidence, PMS2 mutations are more likely to result in a recessively inherited childhood cancer syndrome than in a “classical” autosomal dominant HNPCC-type disease.
Acknowledgments
This work was partly supported by a Medical Research Council Training Fellowship to M.D.V. We thank Dr. Bert Vogelstein for comments on the manuscript and for kindly supplying DNA from the family with Turcot syndrome that was studied by Hamilton et al. (1995).
Electronic-Database Information
The URLs for data presented herein are as follows:
- MegaBLAST, http://www.ncbi.nlm.nih.gov/genome/seq
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for retinoblastoma, Li-Fraumeni syndrome, SMARCB1, NF1, and Turcot syndrome)
- PHYLIP, http://evolution.genetics.washington.edu/phylip.html
- RepeatMasker, http://woody.embl-heidelberg.de/repeatmask
References
- Baptiste M, Nasca P, Metzger B, Field N, MacCubbin P, Greenwald P, Armbrustmacher V, Waldman J, Carlton K (1989) Neurofibromatosis and other disorders among children with CNS tumors and their families. Neurology 39:487–492 [DOI] [PubMed] [Google Scholar]
- Basil JB, Swisher EM, Herzog TJ, Rader JS, Elbendary A, Mutch DG, Goodfellow PJ (1999) Mutational analysis of the PMS2 gene in sporadic endometrial cancers with microsatellite instability. Gynecol Oncol 74:395–399 10.1006/gyno.1999.5486 [DOI] [PubMed] [Google Scholar]
- Burwell RG, James NJ, Johnston DI (1982) Cafe-au-lait spots in schoolchildren. Arch Dis Child 57:631–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick RB, Meek JE, Prior TW, Peltomaki P, de La Chapelle A (2000) Polymorphisms in a pseudogene highly homologous to PMS2. Hum Mutat 16:530 [DOI] [PubMed] [Google Scholar]
- Chan GC, Nicholls JM, Lee AC, Chan LC, Lau YL (1996) Malignant peripheral neuroectodermal tumor in an infant with neurofibromatosis type 1. Med Pediatr Oncol 26:215–219 [DOI] [PubMed] [Google Scholar]
- De Rosa M, Fasano C, Panariello L, Scarano MI, Belli G, Iannelli A, Ciciliano F, Izzo P (2000) Evidence for a recessive inheritance of Turcot’s syndrome caused by compound heterozygous mutations within the PMS2 gene. Oncogene 19:1719–1723 10.1038/sj.onc.1203447 [DOI] [PubMed] [Google Scholar]
- Dirks PB, Harris L, Hoffman HJ, Humphreys RP, Drake JM, Rutka JT (1996) Supratentorial primitive neuroectodermal tumors in children. J Neurooncol 29:75–84 [DOI] [PubMed] [Google Scholar]
- Friedman JM, Birch PH (1997) Type 1 neurofibromatosis: a descriptive analysis of the disorder in 1,728 patients. Am J Med Genet 70:138–143 [DOI] [PubMed] [Google Scholar]
- Glaab WE, Risinger JI, Umar A, Kunkel TA, Barrett JC, Tindall KR (1998) Characterization of distinct human endometrial carcinoma cell lines deficient in mismatch repair that originated from a single tumor. J Biol Chem 273:26662–26669 10.1074/jbc.273.41.26662 [DOI] [PubMed] [Google Scholar]
- Guerrette S, Acharya S, Fishel R (1999) The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem 274:6336–6341 10.1074/jbc.274.10.6336 [DOI] [PubMed] [Google Scholar]
- Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, Berk T, Cohen Z, Tetu B, Burger PC, Wood PA, Taqi F, Booker SV, Petersen GM, Offerhaus GJA, Tersmette AC, Giardiello FM, Vogelstein B, Kinzler KW (1995) The molecular basis of Turcot’s syndrome. N Engl J Med 332:839–847 10.1056/NEJM199503303321302 [DOI] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S (2000) DNA mismatch repair and genetic instability. Annu Rev Genet 34:359–399 10.1146/annurev.genet.34.1.359 [DOI] [PubMed] [Google Scholar]
- Ho YS, Hsieh LL, Chen JS, Chang CN, Lee ST, Chiu LL, Chin TY, Cheng SC (1996) p53 gene mutation in cerebral primitive neuroectodermal tumor in Taiwan. Cancer Lett 104:103–113 10.1016/0304-3835(96)04238-3 [DOI] [PubMed] [Google Scholar]
- Horii A, Han HJ, Sasaki S, Shimada M, Nakamura Y (1994) Cloning, characterization and chromosomal assignment of the human genes homologous to yeast PMS1, a member of mismatch repair genes. Biochem Biophys Res Commun 204:1257–1264 10.1006/bbrc.1994.2598 [DOI] [PubMed] [Google Scholar]
- Jakacki RI (1999) Pineal and nonpineal supratentorial primitive neuroectodermal tumors. Childs Nerv Syst 15:586–591 10.1007/s003810050547 [DOI] [PubMed] [Google Scholar]
- Kato T, Yatagai F, Glickman BW, Tachibana A, Ikenaga M (1998) Specificity of mutations in the PMS2-deficient human tumor cell line HEC-1-A. Mutat Res 422:279–283 [DOI] [PubMed] [Google Scholar]
- Kondo E, Horii A, Fukushige S (1999) The human PMS2L proteins do not interact with hMLH1, a major DNA mismatch repair protein. J Biochem (Tokyo) 125:818–825 [DOI] [PubMed] [Google Scholar]
- Korf BR (1992) Diagnostic outcome in children with multiple cafe au lait spots. Pediatrics 90:924–927 [PubMed] [Google Scholar]
- Korf BR (2000) Malignancy in neurofibromatosis type 1. Oncologist 5:477–485 [DOI] [PubMed] [Google Scholar]
- Kuramoto H, Tamura S, Notake Y (1972) Establishment of a cell line of human endometrial adenocarcinoma in vitro. Am J Obstet Gynecol 114:1012–1019 [DOI] [PubMed] [Google Scholar]
- Li GM, Modrich P (1995) Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc Natl Acad Sci USA 92:1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Yan H, Kuismanen S, Percesepe A, Bisgaard ML, Pedroni M, Benatti P, Kinzler KW, Vogelstein B, Ponz de Leon M, Peltomaki P, Lindblom A (2001) The role of hPMS1 and hPMS2 in predisposing to colorectal cancer. Cancer Res 61:7798–7802 [PubMed] [Google Scholar]
- Marti TM, Kunz C, Fleck O (2002) DNA mismatch repair and mutation avoidance pathways. J Cell Physiol 191:28–41 10.1002/jcp.10077 [DOI] [PubMed] [Google Scholar]
- Martinez-Lage JF, Salcedo C, Corral M, Poza M (2002) Medulloblastomas in neurofibromatosis type 1: case report and literature review. Neurocirugia (Astur) 13:128–131 [DOI] [PubMed] [Google Scholar]
- Miyaki M, Nishio J, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Nagato M, Chong JM, Koike M, Terada T, Kawahara Y, Fukutome A, Tomiyama J, Chuganji Y, Momoi M, Utsunomiya J (1997) Drastic genetic instability of tumors and normal tissues in Turcot syndrome. Oncogene 15:2877–2881 10.1038/sj.onc.1201668 [DOI] [PubMed] [Google Scholar]
- Narayanan L, Fritzell JA, Baker SM, Liskay RM, Glazer PM (1997) Elevated levels of mutation in multiple tissues of mice deficient in the DNA mismatch repair gene Pms2. Proc Natl Acad Sci USA 94:3122–3127 10.1073/pnas.94.7.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institutes of Health (1988) Neurofibromatosis: conference statement. National Institutes of Health Consensus Development Conference. Arch Neurol 45:575–578 [PubMed] [Google Scholar]
- Nicolaides NC, Carter KC, Shell BK, Papadopoulos N, Vogelstein B, Kinzler KW (1995) Genomic organization of the human PMS2 gene family. Genomics 30:195–206 10.1006/geno.1995.9885 [DOI] [PubMed] [Google Scholar]
- Nicolaides NC, Littman SJ, Modrich P, Kinzler KW, Vogelstein B (1998) A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol Cell Biol 18:1635–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, Venter JC, Dunlop MG, Hamilton SR, Petersen GM, de la Chapelle A, Vogelstein B, Kinzler KW (1994) Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371:75–80 10.1038/371075a0 [DOI] [PubMed] [Google Scholar]
- Orellana C, Martinez F, Hernandez-Marti M, Castel V, Canete A, Prieto F, Badia L (1998) A novel TP53 germ-line mutation identified in a girl with a primitive neuroectodermal tumor and her father. Cancer Genet Cytogenet 105:103–108 10.1016/S0165-4608(98)00015-6 [DOI] [PubMed] [Google Scholar]
- Osborne LR, Herbrick JA, Greavette T, Heng HH, Tsui LC, Scherer SW (1997) PMS2-related genes flank the rearrangement breakpoints associated with Williams syndrome and other diseases on human chromosome 7. Genomics 45:402–406 10.1006/geno.1997.4923 [DOI] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui LC, Scherer SW (2001) A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet 29:321–325 10.1038/ng753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino AC (1999) Trilateral retinoblastoma: is the location of the intracranial tumor important? Cancer 86:135–141 [DOI] [PubMed] [Google Scholar]
- Peltomaki P (2001) Deficient DNA mismatch repair: a common etiologic factor for colon cancer. Hum Mol Genet 10:735–740 10.1093/hmg/10.7.735 [DOI] [PubMed] [Google Scholar]
- Perilongo G, Felix CA, Meadows AT, Nowell P, Biegel J, Lange BJ (1993) Sequential development of Wilms tumor, T-cell acute lymphoblastic leukemia, medulloblastoma and myeloid leukemia in a child with type 1 neurofibromatosis: a clinical and cytogenetic case report. Leukemia 7:912–915 [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR (2002) Prediction of central nervous system embryonal tumor outcome based on gene expression. Nature 415:436–442 10.1038/415436a [DOI] [PubMed] [Google Scholar]
- Raffel C, McComb JG, Bodner S, Gilles FE (1989) Benign brain stem lesions in pediatric patients with neurofibromatosis: case reports. Neurosurgery 25:959–964 [DOI] [PubMed] [Google Scholar]
- Raschle M, Marra G, Nystrom-Lahti M, Schar P, Jiricny J (1999) Identification of hMutLβ, a heterodimer of hMLH1 and hPMS1. J Biol Chem 274:32368–32375 10.1074/jbc.274.45.32368 [DOI] [PubMed] [Google Scholar]
- Ricciardone MD, Ozcelik T, Cevher B, Ozdag H, Tuncer M, Gurgey A, Uzunalimoglu O, Cetinkaya H, Tanyeli A, Erken E, Ozturk M (1999) Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res 59:290–293 [PubMed] [Google Scholar]
- Risinger JI, Umar A, Barrett JC, Kunkel TA (1995) A hPMS2 mutant cell line is defective in strand-specific mismatch repair. J Biol Chem 270:18183–18186 10.1074/jbc.270.31.18183 [DOI] [PubMed] [Google Scholar]
- Risinger JI, Umar A, Glaab WE, Tindall KR, Kunkel TA, Barrett JC (1998) Single gene complementation of the hPMS2 defect in HEC-1-A endometrial carcinoma cells. Cancer Res 58:2978–2981 [PubMed] [Google Scholar]
- Ruggieri M, Huson SM (2001) The clinical and diagnostic implications of mosaicism in the neurofibromatoses. Neurology 56:1433–1443 [DOI] [PubMed] [Google Scholar]
- Russo C, Pellarin M, Tingby O, Bollen AW, Lamborn KR, Mohapatra G, Collins VP, Feuerstein BG (1999) Comparative genomic hybridization in patients with supratentorial and infratentorial primitive neuroectodermal tumors. Cancer 86:331–339 [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586 10.1101/gr.10.4.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevenet N, Lellouch-Tubiana A, Schofield D, Hoang-Xuan K, Gessler M, Birnbaum D, Jeanpierre C, Jouvet A, Delattre O (1999) Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet 8:2359–2368 10.1093/hmg/8.13.2359 [DOI] [PubMed] [Google Scholar]
- Strother D, Ashley D, Kellie SJ, Patel A, Jones-Wallace D, Thompson S, Heideman R, Benaim E, Krance R, Bowman L, Gajjar A (2001) Feasibility of four consecutive high-dose chemotherapy cycles with stem-cell rescue for patients with newly diagnosed medulloblastoma or supratentorial primitive neuroectodermal tumor after craniospinal radiotherapy: results of a collaborative study. J Clin Oncol 19:2696–2704 [DOI] [PubMed] [Google Scholar]
- Taylor MD, Gokgoz N, Andrulis IL, Mainprize TG, Drake JM, Rutka JT (2000) Familial posterior fossa brain tumors of infancy secondary to germline mutation of the hSNF5 gene. Am J Hum Genet 66:1403–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimbath JD, Petersen GM, Erdman SH, Ferre M, Luce MC, Giardiello FM (2001) Cafe-au-lait spots and early onset colorectal neoplasia: a variant of HNPCC? Fam Cancer 1:101–105 [DOI] [PubMed] [Google Scholar]
- Valero MC, de Luis O, Cruces J, Perez Jurado LA (2000) Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: the low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 69:1–13 10.1006/geno.2000.6312 [DOI] [PubMed] [Google Scholar]
- Vilkki S, Tsao JL, Loukola A, Poyhonen M, Vierimaa O, Herva R, Aaltonen LA, Shibata D (2001) Extensive somatic microsatellite mutations in normal human tissue. Cancer Res 61:4541–4544 [PubMed] [Google Scholar]
- Wang Q, Lasset C, Desseigne F, Frappaz D, Bergeron C, Navarro C, Ruano E, Puisieux A (1999) Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res 59:294–297 [PubMed] [Google Scholar]
- Whiteside D, McLeod R, Graham G, Steckley JL, Booth K, Somerville MJ, Andrew SE (2002) A homozygous germ-line mutation in the human MSH2 gene predisposes to hematological malignancy and multiple cafe-au-lait spots. Cancer Res 62:359–362 [PubMed] [Google Scholar]
- Yamada NA, Castro A, Farber RA (2003) Variation in the extent of microsatellite instability in human cell lines with defects in different mismatch repair genes. Mutagenesis 18:277–282 10.1093/mutage/18.3.277 [DOI] [PubMed] [Google Scholar]
- Yang HJ, Nam DH, Wang KC, Kim YM, Chi JG, Cho BK (1999) Supratentorial primitive neuroectodermal tumor in children: clinical features, treatment outcome and prognostic factors. Childs Nerv Syst 15:377–383 10.1007/s003810050418 [DOI] [PubMed] [Google Scholar]