Abstract
Nicotine is the major addictive substance in cigarettes, and genes involved in sensing nicotine are logical candidates for vulnerability to nicotine addiction. We studied six single-nucleotide polymorphisms (SNPs) in the CHRNA4 gene and four SNPs in the CHRNB2 gene with respect to nicotine dependence in a collection of 901 subjects (815 siblings and 86 parents) from 222 nuclear families with multiple nicotine-addicted siblings. The subjects were assessed for addiction by both the Fagerstrom Test for Nicotine Dependence (FTND) and the Revised Tolerance Questionnaire (RTQ). Because only 5.8% of female offspring were smokers, only male subjects were included in the final analyses (621 men from 206 families). Univariate (single-marker) family-based association tests (FBATs) demonstrated that variant alleles at two SNPs, rs1044396 and rs1044397, in exon 5 of the CHRNA4 gene were significantly associated with a protective effect against nicotine addiction as either a dichotomized trait or a quantitative phenotype (i.e., age-adjusted FTND and RTQ scores), which was consistent with the results of the global haplotype FBAT. Furthermore, the haplotype-specific FBAT showed a common (22.5%) CHRNA4 haplotype, GCTATA, which was significantly associated with both a protective effect against nicotine addiction as a dichotomized trait (Z=-3.04, P<.005) and significant decreases of age-adjusted FTND (Z=-3.31, P<.005) or RTQ scores (Z=-2.73, P=.006). Our findings provide strong evidence suggesting a common CHRNA4 haplotype might be protective against vulnerability to nicotine addiction in men.
Nearly one-third of adults worldwide are smokers, and the majority started the habit as adolescents (World Health Organization 1997). In 2000, smoking caused ∼2.43 million deaths in industrialized countries (∼19% of total adult mortality) and ∼2.41 million deaths in developing countries (∼9% of total adult mortality) (Ezzati and Lopez 2003).
Smoking cigarettes is both psychologically and physiologically addictive (Nair and Brandt 2000). A vast body of literature indicates that nicotine is the component of tobacco smoke that leads to addiction (Stolerman and Jarvis 1995). Nicotine addiction (i.e., tobacco addiction [MIM #188890]), like many other drug dependencies, is believed to be a complex, multifactorial behavior with both genetic and environmental determinants. Twin and adoption studies have shown that heritabilities of vulnerabilities for both smoking initiation (SI) and smoking persistence (SP) are at least 50% (Li 2003). A familial aggregation study among siblings of nuclear families (Niu et al. 2000) also suggests that genetic influences may be an important determinant in vulnerability to nicotine addiction. Furthermore, studies using animal models found that genetic factors play a critical role in both behavioral and physiological effects of nicotine (Batra et al. 2003). However, the identification of nicotine-addiction genes has lagged far behind environmental risk-factor classification (Straub et al. 1999; Batra et al. 2003).
Nicotine functions by binding to nicotinic acetylcholine receptors (nAChRs), which, in turn, modulate the release of dopamine in the mesolimbic system (Pidoplichko et al. 1997). To date, nine α (α2–α10) and three β (β2–β4) subunit genes have been identified (Mansvelder and McGehee 2002). Among them, α4 and β2 subunits are the most widely and concurrently expressed high-affinity nAChR subunits in the brain (Mathieu-Kia et al. 2002) and are both upregulated under chronic nicotine exposure (Whiteaker et al. 1998). The gene that encodes the human nAChR α4 subunit (CHRNA4 [MIM 118504] [National Center for Biotechnology Information {NCBI} locus ID 1137]) was mapped by FISH to 20q13.2-13.3 (Steinlein et al. 1994). The gene is ∼17 kb in size and comprises six exons (Steinlein et al. 1996). The nAChR β2 subunit gene (CHRNB2 [MIM 118507] [NCBI locus ID 1141]) was first mapped by FISH to chromosome 1q21 (Rempel et al. 1998) and was further narrowed to chromosome 1q21.3 (Lueders et al. 1999). The genomic sequence of the CHRNB2 gene is ∼12 kb and comprises six exons (Lueders et al. 2002). Genetic polymorphisms in the CHRNA4 gene have been reported to be associated with autosomal dominant nocturnal frontal lobe epilepsy (Hirose et al. 1999; Chioza et al. 2000; Steinlein et al. 2000; Rozycka et al. 2003), attention-deficit/hyperactivity disorder (ADHD) (Kent et al. 2001; Todd et al. 2003), Alzheimer disease (Kawamata and Shimohama 2002), and febrile convulsions (Chou et al. 2003), but there has been no prior study of the association of this gene with addictive behaviors. Studies on CHRNB2 gene variants are also quite limited, with two previous studies showing no associations with nicotine dependence (Silverman et al. 2000; Lueders et al. 2002).
To identify families affected by nicotine addiction, we screened a population of ∼25,000 siblings, aged 20–60 years, from November 2000 to July 2001, by use of the Fagerstrom Test for Nicotine Dependence (FTND) (Heatherton et al. 1991; Niu et al. 2000). The siblings resided in a relatively remote region (Huoqiu County) of Anhui Province, China. A total of 901 subjects (815 siblings [age 47.0 years ± 10.1 years, men 78.8%]; 86 parents) from 222 qualified nuclear families (22 [9.9%] families had both parents, 42 [18.9%] families had one single parent, and 158 [71.2%] families had no available parents) were finally recruited. The criteria used for subject selection were as follows: (1) at least two siblings, aged 20–60 years, with FTND scores ⩾8 and (2) at least one parent or one additional sibling. In addition to the FTND, the Revised Tolerance Questionnaire (RTQ) (Tate and Schmitz 1993) was also administered to eligible individuals. All subjects gave their written informed consent, and the study protocol was approved by the institutional review boards of the Harvard School of Public Health and the Anhui Medical University. Because only 10 women offspring were smokers (5.8%), all women and nonsmoking male offspring were excluded in the final analyses. A total of 621 male subjects from 206 nuclear families were finally used in the analysis. Among the 577 smoking male offspring, FTND scores (8.1 ± 2.1) and RTQ scores (35.1 ± 7.0) were found to be moderately but significantly correlated (Pearson correlation coefficient r=0.52, P<.0001), which was in accord with results of other independent studies (Tate and Schmitz 1993; Niu et al. 2000).
A SNP set of 30 CHRNA4 SNPs and 24 CHRNB2 SNPs from NCBI dbSNP (dbSNP Home Page) or published literatures was evaluated by the PCR-RFLP method in a panel of 45 previously established B-lymphoblastoid cell line DNA samples from the same geographical region. Among the 54 SNPs evaluated, 18 (33.3%) were validated to be polymorphic (CHRNA4, 14 SNPs; CHRNB2, 4 SNPs). From the 14 CHRNA4 SNPs, we excluded 1 low-heterozygosity (HET) SNP (i.e., HET ⩽18%; minor allele frequency [MAF] ⩽10%), as well as an additional 7 SNPs that were in perfect linkage disequilibrium (LD) with at least one of the remaining six CHRNA4 SNPs (the extent of LD was measured by use of |D′|, which was calculated according to Lewontin [1964]). Finally, we handpicked six CHRNA4 SNPs and four CHRNB2 SNPs on the basis of both HET (HET>18%, which implies MAF>10%) and pairwise LD (0.5<|D′|⩽1.0) criteria. Large-scale genotyping for these 10 SNPs in the 901 DNA samples was performed by use of either PCR-RFLP or PCR-OLA (oligonucleotide ligation assay). The PCR primer sequences (designed by use of the Primer3 or SNPkit [Hao et al. 2002]), as well as genotyping methods, are summarized in table A1 (online only).
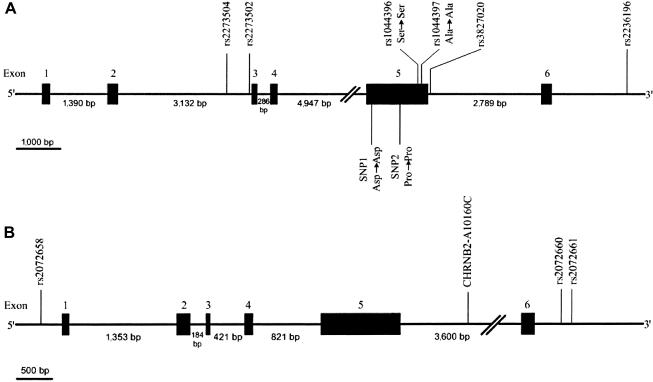
The distributions of the six CHRNA4 SNPs and the four CHRNB2 SNPs are depicted in fig. 1A and 1b, respectively. Among the six SNPs residing in the CHRNA4 gene, rs1044396 (C→T transition; Ser→Ser) and rs1044397 (G→A transition; Ala→Ala) are synonymous changes located in exon 5, whereas all others are located either in an intronic region (rs2273504 [G→A transition], rs2273502 [C→T transition], rs3827020 [T→C transition]) or in the 3′ UTR region (rs2236196 [A→G transition]). Among the four SNPs residing in the CHRNB2 gene, rs2072658 (G→A transition) is located in the 5′ UTR region, whereas A10160C is located at intron 5, and rs2072660 (C→T transition) and rs2072661 (G→A transition) are located in the 3′ UTR. In the DNA panel of 45 cell line samples, all 10 SNPs had a MAF>10% and all were in Hardy-Weinberg equilibrium (P>.05 in respective χ2 tests).
Figure 1.
A, Schematic representation of the six SNPs located on the CHRNA4 gene, showing their locations according to the information from the chromosome 20 genomic contig (GenBank accession number NT_011333). SNP1 and SNP2 in exon 5 are synonymous SNPs revealed by direct DNA sequencing. B, Schematic representation of the four SNPs located on the CHRNB2 gene, showing their locations according to the information from the chromosome 1 genomic contig (GenBank accession number NT_004668).
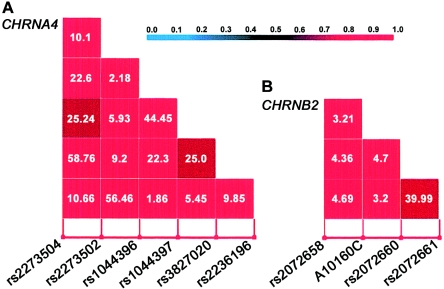
The pairwise |D′| values for the six CHRNA4 SNPs were found to be quite high (range 0.84–1.00, median 1.00) (fig. 2A), which indicates that these six SNPs can be considered to be one haplotype block. Similarly, the four CHRNB2 SNPs were also found in almost complete LD with respect to each other (range 0.86–1.00, median 1.00) and, therefore, could also be grouped in one block (fig. 2B). Thus, we either analyzed one SNP at a time by use of univariate (single-marker) family-based association tests (FBATs) or considered SNPs that were on the same gene as a single haplotype block and tested the associations by use of the haplotype FBAT for various nicotine-addiction phenotypes (both qualitative and quantitative). As mentioned above, all analyses were performed in men only. In the univariate FBAT, a significant association of CHRNA4 rs1044396 (the variant T allele is the protective allele) (MAF=0.286, polymorphism information content [PIC]=0.325, calculated by use of the POLYMORPHISM software [Niu et al. 2001]) was consistently shown across all addiction-related phenotypes: nicotine addiction (addicted and nonaddicted patients with FTND scores of ⩾8 and ⩽2, respectively) (P<.01), age-adjusted FTND score (P<.01), and age-adjusted RTQ score (P<.01) before Bonferroni correction. Similarly, rs1044397 (MAF=0.416, PIC=0.368), which is in almost complete LD with rs1044396 (|D′| = 1.00) (the variant A allele is the protective allele), was observed to be significantly associated with nicotine addiction (P<.01) before Bonferroni correction and with age-adjusted FTND and RTQ scores (P<.001 for both) after Bonferroni correction for multiple comparisons (for 10 SNPs being tested individually, the global significance level needs to be adjusted to 0.05/10=0.005) (table 1). For CHRNB2, no SNP was found to be significantly associated with any nicotine-addiction phenotype (table 1).
Figure 2.
A, Pairwise LD for the 15 CHRNA4 SNP pairs, evaluated by |D′|. B, Pairwise LD for the six CHRNB2 SNP pairs, evaluated by |D′|. Because all SNP pairs have high |D′| values, we presented χ2 values inside their corresponding boxes.
Table 1.
Univariate FBAT for Association of CHRNA4 and CHRNB2 SNPs with Nicotine-Addiction Phenotypes[Note]
|
Result for Phenotype |
||||||||
| Nicotine Addictiona |
FTND Score |
RTQ Score |
||||||
| Geneand SNP | Variant | MAF | Z | P | Z | P | Z | P |
| CHRNA4: | ||||||||
| rs2273504 | A | .441 | .753 | .452 | 1.284 | .199 | .845 | .398 |
| rs2273502 | T | .179 | 2.13 | .033b | 2.497 | .013b | 1.704 | .088 |
| rs1044396 | T | .286 | −2.673 | .008b | −2.832 | .005b | −2.724 | .006b |
| rs1044397 | A | .416 | −2.699 | .007b | −3.781 | <.001c | −3.505 | <.001c |
| rs3827020 | C | .399 | .237 | .813 | .602 | .547 | −.265 | .791 |
| rs2236196 | G | .185 | 1.834 | .067 | 1.828 | .067 | 1.169 | .242 |
| CHRNB2: | ||||||||
| rs2072658 | A | .19 | −.628 | .530 | −1.692 | .091 | −.821 | .412 |
| A10160C | C | .2 | −1.143 | .253 | −1.686 | .092 | −.912 | .362 |
| rs2072660 | T | .278 | 1.646 | .100 | 1.515 | .130 | 1.052 | .293 |
| rs2072661 | A | .283 | 1.36 | .174 | 1.152 | .249 | .972 | .331 |
Note.— The univariate FBAT was performed by use of the additive model. The minor allele is also referred to as the “variant allele” in the text. MAF = minor (i.e., variant) allele frequency. SNPs are listed in order (5′→3′) for each gene. Both rs1044396 and rs1044397 are shown in bold italics because they were consistently significant before Bonferroni correction for nicotine addiction, as well as FTND and RTQ scores. Both FTND and RTQ scores were age-adjusted.
When nicotine addiction was considered as a dichotomized trait, the addicted and nonaddicted subjects were defined as having scores of ⩾8 and ⩽2, respectively (Fagerstrom et al. 1990).
Significant only before Bonferroni correction (i.e., P<.05).
Significant after Bonferroni correction (i.e., P<.005).
Because haplotype-based analysis is arguably more powerful than single-marker analysis (Niu et al. 2002), we performed the global haplotype FBAT as the main haplotype test. In all FBAT analyses, we used both the age-adjusted FTND score and the age-adjusted RTQ score as the continuous phenotypes. For CHRNA4, the global test of the haplotype FBAT demonstrated a significant association of the haplotypes formed by rs2273504, rs2273502, rs1044396, rs1044397, rs3827020, and rs2236196 (in the direction 5′→3′) with both nicotine addiction (χ2=19.19, 4 df, P<.001) and the age-adjusted FTND score (χ2=17.96, 5 df, P<.005), even after Bonferroni correction (for two genes, the significance level is adjusted to be 0.05/2 = 0.025). In addition, a significant association was observed with regard to the age-adjusted RTQ score (χ2=10.43, 4 df, P<.05) before Bonferroni correction (table 2). For CHRNB2, the global test shows that the haplotypes formed by rs2072658, A10160C, rs2072660, and rs2072661 (in the direction 5′→3′) yielded marginally significant association with the age-adjusted FTND score (χ2=8.84, 4 df, P=.065) before Bonferroni correction but demonstrated no association with nicotine addiction as a dichotomized trait (χ2=7.05, 4 df, P>.10) or with the age-adjusted RTQ score (χ2=3.23, 4 df, P>.10) (table 2). The association with age-adjusted FTND score was no longer statistically significant after Bonferroni correction. By use of the expectation-maximization algorithm implemented in the haplotype FBAT, four major haplotypes (i.e., frequency ⩾0.05) were revealed for both CHRNA4 and CHRNB2. Haplotype-specific tests (haplotype FBATs) were used to analyze the effects on nicotine addiction, age-adjusted FTND score, and age-adjusted RTQ score for each haplotype, compared with all other haplotypes grouped together on each gene. The GCTATA haplotype of the CHRNA4 gene, bearing the variant alleles at both rs1044396 and rs1044397 (underscored), was found to be significantly associated with a protective effect against nicotine addiction (Z=-3.04, P=.002), as well as significant decreases of age-adjusted FTND score (Z=-3.31, P=.001) and age-adjusted RTQ score (Z=-2.73, P=.006) after Bonferroni correction. These results were highly consistent with the univariate FBAT results showing that the variant alleles at both of these two exonic SNPs had significant protective effects. For CHRNB2, we found that the haplotype GCCG was associated with a significant protective effect against nicotine addiction (Z=-2.13, P=.033), as well as a significant decrease of age-adjusted FTND score (Z=-2.13, P=.033) before Bonferroni correction, but such associations were no longer statistically significant after Bonferroni correction (table 3).
Table 2.
Global (Multihaplotype) Tests of the Haplotype FBAT for Association of CHRNA4 and CHRNB2 SNPs with Nicotine-Addiction Phenotypes[Note]
| Gene andPhenotype | χ2 | df | P Value |
| CHRNA4: | |||
| Nicotine addiction | 19.19 | 4 | .0007a |
| FTND score | 17.96 | 5 | .003a |
| RTQ score | 10.43 | 4 | .034b |
| CHRNB2: | |||
| Nicotine addiction | 7.05 | 4 | .133 |
| FTND score | 8.84 | 4 | .065 |
| RTQ score | 3.23 | 4 | .520 |
Note.— The haplotype FBAT was performed by use of the additive model. Globally significant results are shown in bold italics.
Significant after Bonferroni correction (i.e., P<.025).
Significant only before Bonferroni correction (i.e., P<.05).
Table 3.
Haplotype-Specific Tests of the Haplotype FBAT for Association of CHRNA4 and CHRNB2 SNPs with Nicotine-Addiction Phenotypes[Note]
|
Result for Phenotype |
|||||||
| Nicotine Addiction |
FTND Score |
RTQ Score |
|||||
| Gene andHaplotype | Frequency | Z | P | Z | P | Z | P |
| CHRNA4: | |||||||
| ACCGCA |
.381 | 1.865 | .062 | 2.432 | .015 | 1.875 | .061 |
|
GCTATA |
.225 | −3.039 | .002a | −3.312 | .001a | −2.727 | .006a |
| GTCGTG |
.153 | 2.297 | .022b | 2.424 | .015b | 1.713 | .087 |
| GCCATA |
.129 | .765 | .444 | −.310 | .756 | −.063 | .950 |
| CHRNB2: | |||||||
| GACG |
.418 | .179 | .858 | 1.529 | .126 | .805 | .421 |
| GATA |
.225 | 1.979 | .048b | 1.828 | .068 | 1.185 | .236 |
| GCCG |
.163 | −2.134 | .033b | −2.128 | .033b | −.848 | .396 |
| AACG |
.135 | −.17 | .865 | −1.406 | .160 | −1.297 | .195 |
Note.— The haplotype-specific test of the haplotype FBAT was performed by use of the additive model. The nucleotides at rs1044396 and rs1044397 of CHRNA4 and the nucleotides at rs2072658 and A10160C are underscored for comparisons with the single-marker FBAT results of the respective SNPs in table 1. Only haplotypes with frequencies ⩾.05 are presented. Globally significant results are shown in bold italics.
Significant after Bonferroni correction (i.e., P<.00625).
Significant only before Bonferroni correction (i.e., P<.05).
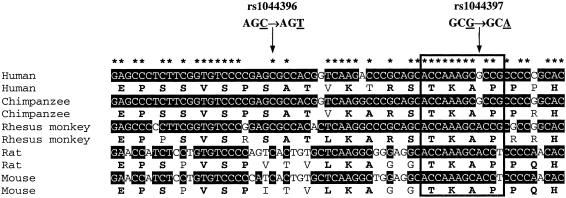
Because both rs1044396 and rs1044397 were located on exon 5 of the CHRNA4 gene, we sequenced exon 5 and its flanking regions (50 bp upstream and 50 bp downstream) in a selected sample of 36 subjects (7 parents and 29 offspring) from 25 nuclear families with extreme Z values. Among the 29 offspring, 22 subjects had FTND scores ⩾8 and 7 subjects had FTND scores ⩽2. Sequencing results confirmed the three genotyped SNPs in this region—rs1044396, rs1044397, rs3827020—and revealed two new synonymous polymorphisms in exon 5, which have been indicated in figure 1. The MAFs of SNP1 (Asp→Asp) and SNP2 (Pro→Pro) were 0.237 and 0.257, respectively, in the sequenced subjects. Both SNP1 and SNP2 have been recently reported in dbSNP (dbSNP Home Page), corresponding to rs1044393 and rs2229959, respectively.
Our study is the first to demonstrate significant associations, not only of individual CHRNA4 SNPs but also of a multilocus CHRNA4 haplotype, with vulnerability to nicotine addiction in men. To our knowledge, there have been no prior studies regarding the role of molecular variants of the human CHRNA4 gene in addictive behavior, although two groups have linked this gene to attention problems such as ADHD (Kent et al. 2001; Todd et al. 2003). A biological link between CHRNA4 or CHRNB2 and the nicotine-addiction phenotype has been well established by multiple studies in animal models. By constructing α4 nAChR subunit knockout mice, Ross et al. (2000) confirmed that the α4 nAChR subunit plays a pivotal role in anxiety in mice. Actually, several in vitro studies showed that a number of nAChR agonists that bind to α4β2 receptor complexes have anxiolytic-like effects (Pomerleau 1986; Gilbert et al. 1989; Brioni et al. 1993), and nicotine is shown to reduce anxiety in chronic smokers (Pomerleau 1986; Gilbert et al. 1989), allowing them to relax, focus, and work more efficiently. Furthermore, knock-in mice with a Leu→Ser mutation (a point mutation in the M2 transmembrane domain) in the α4 nAChR subunit demonstrated increased anxiety and showed a reduced nigrostriatal dopaminergic function (Labarca et al. 2001). With regard to CHRNB2, nicotine-evoked dopamine release is abolished (Zhou et al. 2001) and nicotine self-administration is attenuated (Picciotto et al. 1998) in β2 nAChR subunit knockout mice.
A biological link between CHRNA4 or CHRNB2 and nicotine-addiction phenotypes has also been suggested by multiple studies in humans. Human α4β2 nAChRs are functionally upregulated by chronic nicotine exposure (Buisson and Bertrand 2001), and these receptors are more likely to enter desensitization states (Fenster et al. 1999) than without such exposure. Moreover, the increased numbers of α4β2 nAChRs in the human postmortem brain significantly correlated with intensity and duration of smoking history (Breese et al. 1997). When nicotine is avoided, the excess number of α4β2 nAChRs will recover from desensitization, resulting in hyperexcitability at cholinergic synapses that could contribute to the unrest and agitation that, in turn, motivate the smoker to smoke the next cigarette, to re-desensitize these excess nAChRs (Dani and De Biasi 2001). Todd et al. (2003) identified an intron 2 SNP in the CHRNA4 gene that was associated with a severe inattention problem. Since nicotine and nicotine agonists enhance attention among ADHD patients in clinical trials, some smokers with inattention problems may be addicted to nicotine because such addictive smokers can enhance attention through persistent self-administration of nicotine.
The pathway to substance dependence is complex and involves multiple genetic and environmental risk factors (Kendler et al. 1999; Swan 1999). By use of both univariate and haplotype FBATs, the present study found significant associations of two CHRNA4-coding SNPs, rs1044396 and rs1044397, with protective effects against nicotine addiction. Although these two SNPs do not result in amino acid changes, rs1044397 was located in a highly conserved protein motif, TKAP (see the boxed area of fig. A1 [online only]), across humans, chimpanzees, rhesus monkeys, rats, and mice. The most frequent haplotype in our study population contains the C allele at rs1044396 and the G allele at rs1044397 (i.e., ACCGCA population frequency 38.1%), and, therefore, the protective haplotype revealed here, GCTATA, carried the variant alleles at both SNP sites (table 1).
Our haplotype-analysis results provided some suggestive evidence of a tentative association of CHRNB2 SNPs with vulnerability to nicotine addiction, but such results were uniformly insignificant after Bonferroni correction and therefore should be considered as principally negative. In agreement with our results, Lueders et al. (2002) undertook both single-marker and haplotype approaches and did not find any association of CHRNB2 variants with nicotine dependence as either a continuous or a dichotomous variable. Silverman et al. (2000) analyzed four SNPs on CHRNB2 in 872 subjects and also found no significant association with either SI or progression to nicotine dependence.
Our study focused exclusively on men. Perkins et al. (1999) discussed the emerging evidence that men and women differ in their responses to smoking, presumably because of both genetic and environmental factors. The relative importance of “non-nicotine” factors has been shown to be greater for women than for men, including factors such as pleasure from social reinforcements for smoking and comforts gained from having something to manipulate in social activities. Faraday et al. (1999) also pointed out that women are more inclined to view smoking as a useful tool to cope with social stress or unpleasant situations. Probably because of these reasons, women have been less successful than men in using nicotine-replacement therapy for quitting smoking. Thus, genetic factors underlying nicotine addiction probably play a different role in female and male smokers. In a meta-analysis in male and female twins, Li et al. (2003) found that genetic factors play a more significant role for SI but a less significant role for SP in female adults compared with male adults. Significant sex difference was also detected in shared environmental factors for SI and SP. However, no significant sex difference was detected for nonshared environmental effects for either phenotype.
Despite limitations, including an exclusively male sample and a single ethnic group, our study has several unique strengths. First, we applied both univariate and haplotype FBATs (they both allow for the analyses of dichotomized or continuous traits), which are state-of-the-art tests of association in the presence of linkage (Lake et al. 2000; Horvath et al. 2004) on the basis of the Rabinowitz-Laird algorithm (Rabinowitz and Laird 2000). The FBAT is a generalized version of the classic transmission/disequilibrium test, which can be applied to any type of nuclear family study (Laird et al. 2000), thus avoiding the thorny problem introduced by population admixture that is commonly seen in case-control study designs involving multiple ethnic groups. Moreover, the FBAT results (both single-marker and haplotype-based) without age adjustments were quite similar to the results with adjustments for age (data not shown).
Second, in addition to using the dichotomized nicotine-addiction phenotype, we used both the age-adjusted FTND and the RTQ scores as quantitative measures of the degree of nicotine dependence. The FTND and the RTQ are both based on the Fagerstrom Tolerance Questionnaire (FTQ) (Fagerstrom 1978), and both have improved the internal consistency, the unitary-factor structure, and the psychometric properties of the FTQ (Heatherton et al. 1991; Tate and Schmitz 1993). In addition to covering all six items of the FTND, the RTQ has included items on inhalation patterns and the proportion of the physical lengths of the cigarettes smoked (Tate and Schmitz 1993). Thus, RTQ and FTND scores represent two moderately different perspectives in quantifying the degree of nicotine dependence. The RTQ appears to be more composite than the FTND, and this may explain the slight differences we detected by use of these two closely related continuous phenotypes (r=0.52, P<.0001).
The use of the continuously distributed phenotype in LD analysis has been shown to be a more powerful strategy in detecting genetic susceptibility to complex diseases than the simple classification of individuals into “addicted” and “nonaddicted” categories (Ebstein et al. 1996; Leckman et al. 2001; Rowe et al. 2001). By treating nicotine addiction as either a qualitative or a quantitative phenotype, we found highly consistent results across both univariate (table 1) and haplotype analyses (tables 2 and 3). In particular, the univariate FBAT found protective effects of the variant alleles at rs1044396 and rs1044397 against nicotine addiction, whereas the haplotype FBAT not only corroborated such findings at the global level but also revealed that the GCTATA haplotype (bearing the variant allele at these two loci) had a significant protective effect against nicotine addiction. Unobserved causal SNPs that are in and surrounding the CHRNA4 gene and are in LD with the observed CHRNA4 markers may explain the association we found. When we performed direct DNA resequencing for exon 5 of CHRNA4 in a selected sample, two more synonymous SNPs were found (indicated as SNP1 and SNP2 in fig. 1), but no missense or splice-site mutations were detected. A recent promoter analysis in transgenic mice of the mouse orthologue of the human CHRNA4 gene, chrna4, demonstrated that the regions either upstream of the transcription initiation site or in intron 1 may contain functional regulatory elements that are important for endogenous chrna4 gene expression (Watanabe et al. 1998). Therefore, SNPs located in the 5′ upstream region and in intron 1 of the human CHRNA4 gene may have functional effects on CHRNA4 gene expression. The SNP rs6090387, located in the 5′ UTR of CHRNA4, deserves further investigation.
Third, our study population appeared to be both relatively stable and fairly homogeneous with respect to lifestyle variables, as well as social and cultural norms (Niu et al. 2000). Finally, none of the subjects in this study used nicotine-replacement therapies, such as nicotine gums, patches, or nasal sprays, or used tobacco products other than cigarettes (such as cigars or smokeless tobacco); thus, the potential for confounding by drug interventions or by an unknown cause of information bias was at least significantly minimized, if not virtually eliminated.
In conclusion, by use of a family-based design, we detected significant protective effects of variant alleles at the CHRNA4 SNPs rs1044396 and rs1044397, not only at the individual SNP level but also at the global haplotype level (GCTATA being the protective haplotype), against vulnerability to nicotine addiction in men. Thus, this study provides important new information in the genetic research on addictive behaviors, pointing out that the CHRNA4 gene should be more extensively studied as a critical biological candidate. Since nicotine addiction is a complex trait with significant genetic heterogeneity, candidate genes other than CHRNA4 and CHRNB2, such as the genes encoding the D1, D2, and D4 dopamine receptors and the dopamine transporters (Bergen and Caporaso 1999), deserve further study. Because our study was limited to men, further investigations should also be performed to assess whether these CHRNA4 and CHRNB2 genes play similarly important roles in nicotine addiction in women from the same ethnic background. Also, our results need to be replicated in populations of ancestries other than Chinese.
Acknowledgments
This study was supported by the National Institutes of Health (NIH) National Institute on Drug Abuse (NIDA) grant 1R01 DA12905. We thank the two anonymous reviewers for their constructive comments. We acknowledge the assistance and cooperation of the faculty and staff of the Anhui Medical University Biomedical Institute, and we thank all study participants for their support.
Appendix A: Supplemental Data
Table A1.
Primer Sequences and Genotyping Methods for CHRNA4 and CHRNB2 SNPs[Note]
|
Primer Sequence |
|||||
| Gene and SNP | Substitution | Forward | Reverse | GenotypingMethod | RestrictionEndonuclease |
| CHRNA4: | |||||
| rs2273504 | G→A | 5′-ATCTCCAGCAGGAAACTGGA-3′ | 5′-GGCAGGTGGGCAGAAGGAGGGTC-3′ | OLA | … |
| rs2273502 | C→T | 5′-CAGTGACCCCTTGGTGTCTT-3′ | 5′-GGAGATGTTTGTGGCCTTGT-3′ | RFLP | Sau96I |
| rs1044396 | C→T | 5′-GTCTGCAATGTACTGGACGC-3′ | 5′-ATCCAGTACTGTGTTCCCCG-3′ | OLA | … |
| rs1044397 | G→A | 5′-GTCTGCAATGTACTGGACGC-3′ | 5′-CAAATGCACATGCAAGAAGG-3′ | OLA | … |
| rs3827020 | T→C | 5′-GTGCAAATGCACATGCAAGA-3′ | 5′-ACATAGCAGGCTTGGGAAGA-3′ | OLA | … |
| rs2236196 | A→G | 5′-CTCCTAGCGAAGCAGATTGG-3′ | 5′-AAGAGTCTCCTGAGCTCCCC-3′ | OLA | … |
| CHRNB2: | |||||
| rs2072658 | G→A | 5′-CCTGCCCCAACTTTAGGACT-3′ | 5′-CCAGCTTCTTGCCTATCCTG-3′ | OLA | … |
| A10160C | A→C | 5′-TGCTCCACCTCCCTCAGTAG-3′ | 5′-TGCTGTCTGTGGGTTTGAGA-3′ | RFLP | BstUI |
| rs2072660 | C→T | 5′-GAGCCATCCACCCTGAGGAGGGA-3′ | 5′-CACCCCTTGTCCTATACCCC-3′ | OLA | … |
| rs2072661 | G→A | 5′-GAGGGGAAGAGAGAGCACTGGGT-3′ | 5′-CTGTGGGACTGAGCTGGAAG-3′ | RFLP | Dra III |
Note.— SNPs are listed in order (5′→3′) for each gene. Restriction endonucleases are shown only when applicable.
Figure A1.
Cross-species comparison of the DNA and protein sequences of a CHRNA4 coding segment (66 bp; 22 amino acids) in the cytoplasmic loop between transmembrane domains 3 and 4 that contains both rs1044396 and rs1044397 in human (GenBank accession number NM_000744), chimpanzee (GSC BLAST Search), rhesus monkey (GenBank accession number AJ245973), rat (GenBank accession number NM_024354), and mouse (GenBank accession number NM_015730). Multiple DNA sequence alignment was performed by use of CLUSTALX, version 1.81 (Jeanmougin et al. 1998). Protein translation was performed by use of JavaScript DNA Translator 1.1 (Perry 2002). The region in the box is a highly conserved region.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for CHRNA4 polymorphisms [cluster IDs rs2273504, rs2273502, rs1044396, rs1044397, rs3827020, rs2236196, rs1044393, and rs2229959] and CHRNB2 polymorphisms [cluster IDs rs2072658, rs2072660, and rs2072661])
- FBAT, http://www.biostat.harvard.edu/~fbat/fbat.htm
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for chromosome 20 genomic contig [accession number NT_011333], chromosome 1 genomic contig [accession number NT_004668], human CHRNA4 cDNA [accession number NM_000744], rhesus monkey chrna4 cDNA [accession number AJ245973], rat chrna4 cDNA [accession number NM_024354], and mouse chrna4 cDNA [accession number NM_015730])
- GSC BLAST Search, http://www.genome.wustl.edu/projects/chimp/blast/pan_client.pl (for chimpanzee genome sequence)
- JavaScript DNA Translator 1.1, http://www.bioinformatics.vg/bioinformatics_tools/JVT.shtml
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for tobacco addiction, CHRNA4, and CHRNB2)
- Primer3, http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi
References
- Batra V, Patkar AA, Berrettini WH, Weinstein SP, Leone FT (2003) The genetic determinants of smoking. Chest 123:1730–1739 10.1378/chest.123.5.1730 [DOI] [PubMed] [Google Scholar]
- Bergen AW, Caporaso N (1999) Cigarette smoking. J Natl Cancer Inst 91:1365–1375 10.1093/jnci/91.16.1365 [DOI] [PubMed] [Google Scholar]
- Breese CR, Marks MJ, Logel J, Adams CE, Sullivan B, Collins AC, Leonard S (1997) Effect of smoking history on [3H]nicotine binding in human postmortem brain. J Pharmacol Exp Ther 282:7–13 [PubMed] [Google Scholar]
- Brioni JD, O’Neill AB, Kim DJ, Decker MW (1993) Nicotinic receptor agonists exhibit anxiolytic-like effects on the elevated plus-maze test. Eur J Pharmacol 238:1–8 10.1016/0014-2999(93)90498-7 [DOI] [PubMed] [Google Scholar]
- Buisson B, Bertrand D (2001) Chronic exposure to nicotine upregulates the human α4β2 nicotinic acetylcholine receptor function. J Neurosci 21:1819–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chioza B, Goodwin H, Blower J, McCormick D, Nashef L, Asherson P, Makoff AJ (2000) Failure to replicate association between the gene for the neuronal nicotinic acetylcholine receptor α4 subunit (CHRNA4) and IGE. Am J Med Genet 96:814–816 [DOI] [PubMed] [Google Scholar]
- Chou IC, Lee CC, Huang CC, Wu JY, Tsai JJ, Tsai CH, Tsai FJ (2003) Association of the neuronal nicotinic acetylcholine receptor subunit α4 polymorphisms with febrile convulsions. Epilepsia 44:1089–1093 10.1046/j.1528-1157.2003.t01-1-44702.x [DOI] [PubMed] [Google Scholar]
- Dani JA, De Biasi M (2001) Cellular mechanisms of nicotine addiction. Pharmacol Biochem Behav 70:439–446 10.1016/S0091-3057(01)00652-9 [DOI] [PubMed] [Google Scholar]
- Ebstein RP, Novick O, Umansky R, Priel B, Osher Y, Blaine D, Bennett ER, Nemanov L, Katz M, Belmaker RH (1996) Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of novelty seeking. Nat Genet 12:78–80 [DOI] [PubMed] [Google Scholar]
- Ezzati M, Lopez AD (2003) Estimates of global mortality attributable to smoking in 2000. Lancet 362:847–852 10.1016/S0140-6736(03)14338-3 [DOI] [PubMed] [Google Scholar]
- Fagerstrom KO (1978) Measuring degree of physical dependence to tobacco smoking with reference to individualization of treatment. Addict Behav 3:235–241 10.1016/0306-4603(78)90024-2 [DOI] [PubMed] [Google Scholar]
- Fagerstrom KO, Heatherton TF, Kozlowski LT (1990) Nicotine addiction and its assessment. Ear Nose Throat J 69:763–765 [PubMed] [Google Scholar]
- Faraday MM, Scheufele PM, Rahman MA, Grunberg NE (1999) Effects of chronic nicotine administration on locomotion depend on rat sex and housing condition. Nicotine Tob Res 1:143–151 [DOI] [PubMed] [Google Scholar]
- Fenster CP, Hicks JH, Beckman ML, Covernton PJ, Quick MW, Lester RA (1999) Desensitization of nicotinic receptors in the central nervous system. Ann N Y Acad Sci 868:620–623 [DOI] [PubMed] [Google Scholar]
- Gilbert DG, Robinson JH, Chamberlin CL, Spielberger CD (1989) Effects of smoking/nicotine on anxiety, heart rate, and lateralization of EEG during a stressful movie. Psychophysiology 26:311–320 [DOI] [PubMed] [Google Scholar]
- Hao K, Niu T, Sangokoya C, Li J, Xu X (2002) SNPkit: an efficient approach to systematic evaluation of candidate single nucleotide polymorphisms in public databases. Biotechniques 33:822–828 [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO (1991) The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict 86:1119–1127 [DOI] [PubMed] [Google Scholar]
- Hirose S, Iwata H, Akiyoshi H, Kobayashi K, Ito M, Wada K, Kaneko S, Mitsudome A (1999) A novel mutation of CHRNA4 responsible for autosomal dominant nocturnal frontal lobe epilepsy. Neurology 53:1749–1753 [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM (2004) Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 26:61–69 10.1002/gepi.10295 [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405 10.1016/S0968-0004(98)01285-7 [DOI] [PubMed] [Google Scholar]
- Kawamata J, Shimohama S (2002) Association of novel and established polymorphisms in neuronal nicotinic acetylcholine receptors with sporadic Alzheimer’s disease. J Alzheimers Dis 4:71–76 [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA (1999) A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med 29:299–308 10.1017/S0033291798008022 [DOI] [PubMed] [Google Scholar]
- Kent L, Middle F, Hawi Z, Fitzgerald M, Gill M, Feehan C, Craddock N (2001) Nicotinic acetylcholine receptor α4 subunit gene polymorphism and attention deficit hyperactivity disorder. Psychiatr Genet 11:37–40 10.1097/00041444-200103000-00007 [DOI] [PubMed] [Google Scholar]
- Labarca C, Schwarz J, Deshpande P, Schwarz S, Nowak MW, Fonck C, Nashmi R, Kofuji P, Dang H, Shi W, Fidan M, Khakh BS, Chen Z, Bowers BJ, Boulter J, Wehner JM, Lester HA (2001) Point mutant mice with hypersensitive α4 nicotinic receptors show dopaminergic deficits and increased anxiety. Proc Natl Acad Sci USA 98:2786–2791 10.1073/pnas.041582598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X (2000) Implementing a unified approach to family-based tests of association. Genet Epidemiol Suppl 19:S36–S42 [DOI] [PubMed] [Google Scholar]
- Lake SL, Blacker D, Laird NM (2000) Family-based tests of association in the presence of linkage. Am J Hum Genet 67:1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckman JF, Zhang H, Alsobrook JP, Pauls DL (2001) Symptom dimensions in obsessive-compulsive disorder: toward quantitative phenotypes. Am J Med Genet 105:28–30 [DOI] [PubMed] [Google Scholar]
- Lewontin RC (1964) The interaction of selection and linkage: I. general considerations, heterotic models. Genetics 49:49–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD (2003) The genetics of smoking related behavior: a brief review. Am J Med Sci 326:168–173 10.1097/00000441-200310000-00003 [DOI] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE (2003) A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction 98:23–31 10.1046/j.1360-0443.2003.00295.x [DOI] [PubMed] [Google Scholar]
- Lueders KK, Elliott RW, Marenholz I, Mischke D, DuPree M, Hamer D (1999) Genomic organization and mapping of the human and mouse neuronal β2-nicotinic acetylcholine receptor genes. Mamm Genome 10:900–905 10.1007/s003359901111 [DOI] [PubMed] [Google Scholar]
- Lueders KK, Hu S, McHugh L, Myakishev MV, Sirota LA, Hamer DH (2002) Genetic and functional analysis of single nucleotide polymorphisms in the β2-neuronal nicotinic acetylcholine receptor gene (CHRNB2). Nicotine Tob Res 4:115–125 10.1080/14622200110098419 [DOI] [PubMed] [Google Scholar]
- Mansvelder HD, McGehee DS (2002) Cellular and synaptic mechanisms of nicotine addiction. J Neurobiol 53:606–617 10.1002/neu.10148 [DOI] [PubMed] [Google Scholar]
- Mathieu-Kia AM, Kellogg SH, Butelman ER, Kreek MJ (2002) Nicotine addiction: insights from recent animal studies. Psychopharmacology (Berl) 162:102–118 [DOI] [PubMed] [Google Scholar]
- Nair AK, Brandt EN Jr (2000) Effects of smoking on health care costs. J Okla State Med Assoc 93:245–250 [PubMed] [Google Scholar]
- Niu T, Chen C, Ni J, Wang B, Fang Z, Shao H, Xu X (2000) Nicotine dependence and its familial aggregation in Chinese. Int J Epidemiol 29:248–252 10.1093/ije/29.2.248 [DOI] [PubMed] [Google Scholar]
- Niu T, Qin ZS, Xu X, Liu JS (2002) Bayesian haplotype inference for multiple linked single-nucleotide polymorphisms. Am J Hum Genet 70:157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T, Struk B, Lindpaintner K (2001) Statistical considerations for genome-wide scans: design and application of a novel software package POLYMORPHISM. Hum Hered 52:102–109 10.1159/000053361 [DOI] [PubMed] [Google Scholar]
- Perkins KA, Donny E, Caggiula AR (1999) Sex differences in nicotine effects and self-administration: review of human and animal evidence. Nicotine Tob Res 1:301–315 [DOI] [PubMed] [Google Scholar]
- Perry WL III (2002) JavaScript DNA translator: DNA-aligned protein translations. Biotechniques 33:1318–1320 [DOI] [PubMed] [Google Scholar]
- Picciotto MR, Zoli M, Rimondini R, Lena C, Marubio LM, Pich EM, Fuxe K, Changeux JP (1998) Acetylcholine receptors containing the β2 subunit are involved in the reinforcing properties of nicotine. Nature 391:173–177 10.1038/34413 [DOI] [PubMed] [Google Scholar]
- Pidoplichko VI, DeBiasi M, Williams JT, Dani JA (1997) Nicotine activates and desensitizes midbrain dopamine neurons. Nature 390:401–404 10.1038/37120 [DOI] [PubMed] [Google Scholar]
- Pomerleau OF (1986) Nicotine as a psychoactive drug: anxiety and pain reduction. Psychopharmacol Bull 22:865–869 [PubMed] [Google Scholar]
- Rabinowitz D, Laird N (2000) A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 50:211–223 10.1159/000022918 [DOI] [PubMed] [Google Scholar]
- Rempel N, Heyers S, Engels H, Sleegers E, Steinlein OK (1998) The structures of the human neuronal nicotinic acetylcholine receptor β2- and α3-subunit genes (CHRNB2 and CHRNA3). Hum Genet 103:645–653 10.1007/s004390050885 [DOI] [PubMed] [Google Scholar]
- Ross SA, Wong JY, Clifford JJ, Kinsella A, Massalas JS, Horne MK, Scheffer IE, Kola I, Waddington JL, Berkovic SF, Drago J (2000) Phenotypic characterization of an α4 neuronal nicotinic acetylcholine receptor subunit knock-out mouse. J Neurosci 20:6431–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe DC, Stever C, Chase D, Sherman S, Abramowitz A, Waldman ID (2001) Two dopamine genes related to reports of childhood retrospective inattention and conduct disorder symptoms. Mol Psychiatry 6:429–433 10.1038/sj.mp.4000874 [DOI] [PubMed] [Google Scholar]
- Rozycka A, Skorupska E, Kostyrko A, Trzeciak WH (2003) Evidence for S284L mutation of the CHRNA4 in a white family with autosomal dominant nocturnal frontal lobe epilepsy. Epilepsia 44:1113–1117 10.1046/j.1528-1157.2003.07603.x [DOI] [PubMed] [Google Scholar]
- Silverman MA, Neale MC, Sullivan PF, Harris-Kerr C, Wormley B, Sadek H, Ma Y, Kendler KS, Straub RE (2000) Haplotypes of four novel single nucleotide polymorphisms in the nicotinic acetylcholine receptor β2-subunit (CHRNB2) gene show no association with smoking initiation or nicotine dependence. Am J Med Genet 96:646–653 [DOI] [PubMed] [Google Scholar]
- Steinlein O, Smigrodzki R, Lindstrom J, Anand R, Kohler M, Tocharoentanaphol C, Vogel F (1994) Refinement of the localization of the gene for neuronal nicotinic acetylcholine receptor α4 subunit (CHRNA4) to human chromosome 20q13.2-q13.3. Genomics 22:493–495 10.1006/geno.1994.1420 [DOI] [PubMed] [Google Scholar]
- Steinlein OK, Stoodt J, Mulley J, Berkovic S, Scheffer IE, Brodtkorb E (2000) Independent occurrence of the CHRNA4 Ser248Phe mutation in a Norwegian family with nocturnal frontal lobe epilepsy. Epilepsia 41:529–535 [DOI] [PubMed] [Google Scholar]
- Steinlein O, Weiland S, Stoodt J, Propping P (1996) Exon-intron structure of the human neuronal nicotinic acetylcholine receptor α4 subunit (CHRNA4). Genomics 32:289–294 10.1006/geno.1996.0119 [DOI] [PubMed] [Google Scholar]
- Stolerman IP, Jarvis MJ (1995) The scientific case that nicotine is addictive. Psychopharmacology (Berl) 117:2–10 [DOI] [PubMed] [Google Scholar]
- Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, Neale MC, Bulik CM, Joyce PR, Kendler KS (1999) Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry 4:129–144 10.1038/sj.mp.4000518 [DOI] [PubMed] [Google Scholar]
- Swan GE (1999) Implications of genetic epidemiology for the prevention of tobacco use. Nicotine Tob Res Suppl 1:S49–S56 [DOI] [PubMed] [Google Scholar]
- Tate JC, Schmitz JM (1993) A proposed revision of the Fagerstrom Tolerance Questionnaire. Addict Behav 18:135–143 10.1016/0306-4603(93)90043-9 [DOI] [PubMed] [Google Scholar]
- Todd RD, Lobos EA, Sun LW, Neuman RJ (2003) Mutational analysis of the nicotinic acetylcholine receptor α4 subunit gene in attention deficit/hyperactivity disorder: evidence for association of an intronic polymorphism with attention problems. Mol Psychiatry 8:103–108 10.1038/sj.mp.4001257 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Zoli M, Changeux JP (1998) Promoter analysis of the neuronal nicotinic acetylcholine receptor α4 gene: methylation and expression of the transgene. Eur J Neurosci 10:2244–2253 10.1046/j.1460-9568.1998.00235.x [DOI] [PubMed] [Google Scholar]
- Whiteaker P, Sharples CG, Wonnacott S (1998) Agonist-induced up-regulation of α4β2 nicotinic acetylcholine receptors in M10 cells: pharmacological and spatial definition. Mol Pharmacol 53:950–962 [PubMed] [Google Scholar]
- World Health Organization (1997) Tobacco or health: a global status report. World Health Organization, Geneva; http://www.cdc.gov/tobacco/who/whofirst.htm (accessed May 11, 2004)
- Zhou FM, Liang Y, Dani JA (2001) Endogenous nicotinic cholinergic activity regulates dopamine release in the striatum. Nat Neurosci 4:1224–1229 10.1038/nn769 [DOI] [PubMed] [Google Scholar]