Abstract
Cohen syndrome is a rare autosomal recessive disorder with a variable clinical picture mainly characterized by developmental delay, mental retardation, microcephaly, typical facial dysmorphism, progressive pigmentary retinopathy, severe myopia, and intermittent neutropenia. A Cohen syndrome locus was mapped to chromosome 8q22 in Finnish patients, and, recently, mutations in the gene COH1 were reported in patients with Cohen syndrome from Finland and other parts of northern and western Europe. Here, we describe clinical and molecular findings in 20 patients with Cohen syndrome from 12 families, originating from Brazil, Germany, Lebanon, Oman, Poland, and Turkey. All patients were homozygous or compound heterozygous for mutations in COH1. We identified a total of 17 novel mutations, mostly resulting in premature termination codons. The clinical presentation was highly variable. Developmental delay of varying degree, early-onset myopia, joint laxity, and facial dysmorphism were the only features present in all patients; however, retinopathy at school age, microcephaly, and neutropenia are not requisite symptoms of Cohen syndrome. The identification of novel mutations in COH1 in an ethnically diverse group of patients demonstrates extensive allelic heterogeneity and explains the intriguing clinical variability in Cohen syndrome.
Cohen syndrome (MIM 216550), an infrequent recessively inherited condition, was described first by Cohen et al. (1973). Initially, it was characterized by distinctive craniofacial appearance, midchildhood onset of obesity, mental retardation, hypotonia, joint laxity, and narrow hands and feet. A clinically homogeneous phenotype of Cohen syndrome was reported in Finnish patients who showed additional ophthalmological findings, namely, progressive myopia and retinal dystrophy, microcephaly, and neutropenia (Norio et al. 1984; Kivitie-Kallio et al. 1999; Kivitie-Kallio and Norio 2001). In these patients, the similar and characteristic craniofacial appearance, which becomes more specific with age, includes downward slanting and wave-shaped palpebral fissures, short philtrum, heavy eyebrows, thick hair, and a prominent root of the nose. In the same cohort, the underlying gene was localized to the vicinity of D8S1762 on chromosome 8q22 (Tahvanainen et al. 1994; Kolehmainen et al. 1997). Greater, and often confusing, clinical variability in Cohen syndrome was described in cases from outside Finland. Different clinical criteria were determined in studies of non-Finnish patients (Horn et al. 2000; Chandler et al. 2003).
An overlapping phenotype between Cohen syndrome and Mirhosseini-Holmes-Walton syndrome (MIM 268050) was observed in three closely related patients from a multiply consanguineous kindred of Lebanese descent (Horn et al. 2000). These patients presented with postnatal microcephaly, progressive growth delay, and severe mental retardation. Homozygosity mapping in this family assigned the underlying gene to a region on chromosome 8q21.3-22.1 that extended over the gene region associated with Cohen syndrome in Finnish patients (Horn et al. 2000). Mirhosseini-Holmes-Walton syndrome in two families with microcephaly, retinal pigmentary degeneration, and severe mental retardation was described elsewhere (Mirhosseini et al. 1972; Mendez et al. 1985). Clinical presentation in these cases has features in common with the phenotype seen in patients with Cohen syndrome, which suggests that both conditions represent the same clinical entity (Norio and Raitta 1986; Steinlein et al. 1991; Horn et al. 2000).
Recently, mutations of the COH1 gene were reported in a northern and western European cohort of patients with Cohen syndrome, mainly originating from Finland (Kolehmainen et al. 2003). COH1 comprises various splice forms and up to 62 exons and encodes a protein of 4,022 aa whose domain structure and sequence similarities suggest a role in protein sorting and vesiclemediated protein transport.
The present study describes clinical and molecular findings in 20 patients with Cohen syndrome from 12 non-Finnish families. The investigated families comprised a consanguineous Omani family with three affected sibs (family 9; see table 1); two consanguineous Turkish families, each with one affected child and one unaffected sib (families 3 and 12); two nonconsanguineous Polish families, each with two affected sibs (families 2 and 8); four nonconsanguineous German families, each with one affected child (families 4, 6, 7, and 10); and one nonconsanguineous Brazilian family with two affected children (family 1). Three patients—two brothers and one cousin—showing symptoms of Cohen and Mirhosseini-Holmes-Walton syndromes originated from a multiply consanguineous Lebanese family that also included two and three unaffected siblings of the brothers and cousin, respectively (family 11). Detailed clinical data and results of homozygosity mapping in this family were reported elsewhere (Horn et al. 2000). Another nonconsanguineous German family with two affected sibs (family 5) was suggested also as having an overlapping phenotype and was described elsewhere by Steinlein et al. (1991).
Table 1.
Clinical Manifestations of 20 Patients with Cohen Syndrome
|
Clinical Manifestationa |
||||||||||||||||
| Patient | Origin | Sexa | Age atAssessment | OFCb(SD) | Height(SD) | TruncalObesityc | Age atSittingd | Age atWalkingd | Age atFirstWordsd | SpeechDevelopment | TypicalFacialGestaltc | Myopia(diopters) | Retinopathyc | NarrowHands/Feetc | Hyperextensibilityof Jointsc | Neutropeniac,d |
| 1/1 | Brazilian | F | 15 years | −2.2 | −1.0 | (+) | NA | 2 years | 1 year | Sentences | + | −5.0 | − | + | (+) | − |
| 1/2 | Brazilian | M | 11 years | −4.4 | −2.7 | − | NA | 18 mo | 6 years | Sentences | + | + | − | + | (+) | − |
| 2/1 | Polish | M | 5 years | −4.0 | −1.6 | (+) | 11 mo | 20 mo | 3 years | Words | + | −9.5 | − | + | + | + |
| 2/2 | Polish | M | 3 years | −3.4 | −1.3 | (+) | 8 mo | 20 mo | 2.5 years | Sentences | + | −2.5 | − | + | + | + |
| 3 | Turkish | F | 13 years | −4.2 | −1.4 | + | 1 year | 3 years | 3 years | Sentences | + | −7.0 | + | + | + | − |
| 4 | German | M | 4.5 years | −2.6 | −1.0 | (+) | 15 mo | 22 mo | No speech | No speech | + | −3.0 | − | + | + | + |
| 5/1 | German | M | 30 years | −3.4 | −3.3 | + | NA | NA | NA | Words | + | NAe | + | + | + | + |
| 5/2 | German | M | 28 years | −3.4 | −1.6 | + | NA | NA | No speech | No speech | + | NAe | + | + | + | + |
| 6 | German | M | 25 years | −1.5 | −1.4 | (+) | 9 mo | 22 mo | 4 years | Sentences | + | + | + | + | + | + |
| 7 | German | F | 4 years | −2.5 | −2.0 | (+) | NA | 4 years | No speech | No speech | + | −6 | − | + | + | − |
| 8/1 | Polish | F | 16 years | −5.0 | −2.6 | + | NA | 4.5 years | 4 years | Sentences | + | −5.5 | + | + | + | + |
| 8/2 | Polish | M | 12 years | −4.7 | −2.3 | + | 2 years | 4.5 years | 4.5 years | Sentences | + | −3.5 | + | + | + | + |
| 9/1 | Omani | F | 7 years | −5.9 | −5.7 | − | 1 year | 3 years | 3 years | Words | + | −7 | + | + | + | NA |
| 9/2 | Omani | M | 5 years | −4.0 | −2.5 | − | 15 mo | 3 years | 3 years | Words | + | + | + | + | + | NA |
| 9/3 | Omani | M | 2 years | −4.4 | −2.5 | − | 15 mo | 2 years | 2 years | Words | + | + | + | + | + | − |
| 10 | German | M | 4 years | −3.6 | −.3 | − | 1 year | 4 years | 3 years | Words | + | −.3 | − | + | + | + |
| 11/1 | Lebanese | M | 17 years | −4.8 | −4.2 | − | 3 years | 7 years | No speech | No speech | + | −8 | + | + | + | − |
| 11/2 | Lebanese | M | 10 years | −3.0 | −3.6 | + | 3 years | 5 years | No speech | No speech | + | −8 | + | + | + | − |
| 11/3 | Lebanese | M | 8 years | −3.6 | −2.0 | + | 15 mo | 2 years | 2.5 years | Words | + | −13 | + | + | + | − |
| 12 | Turkish | M | 6 years | −4.8 | −2.8 | − | 2 years | 2.5 years | No speech | No speech | + | −6.5 | + | + | + | + |
F = female; M = male.
OFC = occipitofrontal head circumference. All patients listed above had a normal OFC at birth.
+ = expression; − = no expression; (+) = mild expression.
NA = not available.
Optic atrophy and retinal detachment are present.
Informed consent was obtained from all parents. Patients were assessed clinically by at least one of the authors. DNA was extracted from peripheral blood with the use of standard methods. From the COH1 region on chromosome 8q22, 36 microsatellite markers were analyzed in consanguineous families. Primer pairs for amplification of each of the 62 COH1 exons weregenerated on the basis of the sequence of a chromosome 8 genomic contig (GenBank accession number NT_008046). Exons were sequenced directly by BigDye Terminator sequencing (Applied Biosystems). RNA was extracted from lymphoblastoid cell lines. Primer pairs for RNA amplification were designed from the sequence of the partial transcript KIAA0532 (GenBank accession number AB011104) and the cDNA sequence of COH1 (GenBank accession number NM_017890). Primer sequences are available on request.
The characteristic facial appearance, developmental delay, myopia, narrow hands with slender and tapering fingers, narrow feet, and generalized joint hyperextensibility were present in all patients with Cohen syndrome investigated for this study. However, microcephaly, short stature, truncal obesity, neutropenia, and retinopathy were not present in some of the patients (table 1). Measurement of head circumference at birth revealed values within normal limits in all cases. Patients’ occipitofrontal head circumference varied from −5.9 to −1.5 SD postnatally, and values were ⩽3rd percentile for age in all cases but one. Heights were highly variable and ranged from −5.7 to −0.3 SD. Short stature (height ⩽3rd percentile) was present in 11 of 20 cases. Truncal obesity was found in 13 of the 20 patients but was mild, especially in younger patients. The youngest patient with this symptom (patient 7) was 2 years of age.
All patients had a global developmental delay of variable degree and nonprogressive mental retardation. The median age at unsupported sitting was 17 mo and at first walking was 3 years. Six patients (4, 5/2, 7, 11/1, 11/2, and 12) did not achieve verbal communication, and the others, with a median age of 3 years at first spoken words, used single words only or were able to speak in short sentences.
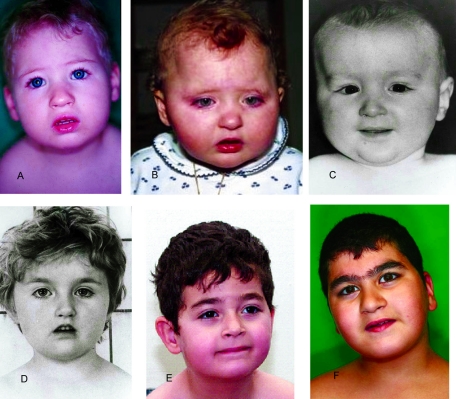
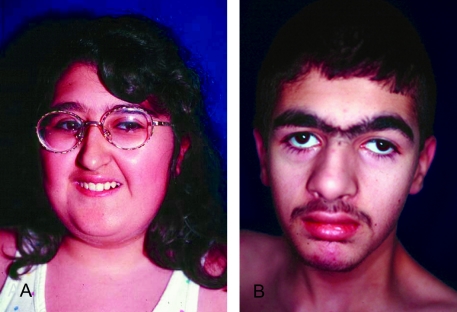
In the younger patients, the face was round with a full lower lip, the philtrum was not obviously short, the eyes were often slightly downward slanting with wave-shaped eyelids, and the nasal bridge was not prominent (fig. 1A–1F). However, the facial appearance was recognizable in young age and led to the diagnosis of Cohen syndrome, even in the absence of ophthalmologic findings, in two 24-mo-old patients without a striking family history (patients 7 and 10; see fig. 1A and 1B). With increasing age, the facial gestalt became more typical, with thick hair, often a low frontal hairline, heavy eyebrows, short philtrum, a long columella further contributing to the impression of a short philtrum, and central incisors that may appear prominent (fig. 2A and 2B).
Figure 1.
Facial photographs of patients, showing mild but characteristic facial dysmorphism of Cohen syndrome. Shown are patients 10 (A), 7 (B), 2/2 (C), 2/1 (D), 4 (E), and 11/3 (F). Their ages are 2 years, 2 years, 1 year, 4 years, 4.5 years, and 8 years, respectively.
Figure 2.
Facial features of two older patients with Cohen syndrome. A, Patient 3, age 13 years. B, Patient 11/1, age 17 years.
All patients studied here had visual abnormalities. An early-onset myopia with a severity from −2.5 to −13 diopters was a nearly consistent finding. One patient (10) had only very mild myopia of −0.3 diopters at the ages of 2 years and 4 years. However, pigmentary retinopathy, which is an age-dependent symptom, was absent in a total of seven patients (1/1, 1/2, 2/1, 2/2, 4, 7, and 10), ages 3–15 years. Severe ophthalmological findings, necessitating enucleation in patients 5/1 and 5/2, included total retinal detachment and optic atrophy in both patients and shrunken vitreous body and the presence of eosinophilic material in the retroretinal space in one of them.
Repeated hematological examinations showed neutropenia, defined as a neutrophil count <1.5 × 109/l, in 10 of 18 patients. Severe infections were not reported in the patients with neutropenia studied here.
With the use of microsatellite markers, homozygosity in the COH1 region was found in all patients from consanguineous families, with a critical interval between D8S343 and D8S1714. In the full-length transcript of COH1, 17 different novel mutations were identified (table 2). They cosegregated in the respective families in all cases and were not seen in 150 chromosomes from control subjects. As expected, patients from the consanguineous families 3, 9, 11, and 12 carried homozygous mutations. Moreover, the two Polish patients from family 2 without known consanguinity were homozygous for a COH1 mutation. Further analysis of microsatellites around COH1 in this family showed a small homozygous interval between D8S257 and D8S546, which is 1.5 cM in length, pointing to a more distant relationship in this family. Seven further cases had heterozygous mutations.
Table 2.
Mutations in COH1 Identified in 12 Cases of Cohen Syndrome
| cDNA | Exon | Protein | Genotype | Family | Origin |
| c.1504C→T | 11 | p.Arg502X | Heterozygous | 1 | Brazilian |
| c.2727_2730dupGCTC | 19 | p.Asn911fsX3 | Homozygous | 2 | Polish |
| c.2911C→T | 20 | p.Arg971X | Homozygous | 3 | Turkish |
| c.3618T→A | 24 | p.Cys1206X | Heterozygous | 4 | German |
| c.4396insA | 29 | p.Thr1466fsX5 | Heterozygous | 5 | German |
| c.5069T→A | 32 | p.Leu1690X | Heterozygous | 6 | German |
| c.7022A→G | 39 | p.Tyr2341Cys | Heterozygous | 7 | German |
| c.7603C→T | 42 | p.Arg2535X | Heterozygous | 4 | German |
| c.7610G→A | 42 | p.Trp2537X | Heterozygous | 6 | German |
| c.7934G→A | 43 | p.Gly2645Asp | Homozygous | 9 | Omani |
| c.7935delC | 43 | p.Gln2646fsX96 | Heterozygous | 8 | Polish |
| c.8609delA | 47 | p.Glu2870fsX16 | Heterozygous | 10 | German |
| c.9406-1G→T | 52 | p.Tyr3136fsX16 | Homozygous | 11 | Lebanese |
| c.9731delA | 53 | p.Tyr3244fsX7 | Heterozygous | 10 | German |
| c.10888C→T | 56 | p.Gln3630X | Homozygous | 12 | Turkish |
| c.11216G→A | 58 | p.Trp3739X | Heterozygous | 5 | German |
| c.11314C→T | 59 | p.Gln3772X | Heterozygous | 7 | German |
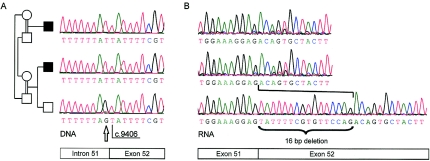
Mutation c.9406-1G→T, identified in family 11, affected the splice-acceptor site of intron 51. To investigate whether the mutation resulted in exon skipping or retention of intron 51, RNA samples of two affected cousins and an unaffected brother from the family were analyzed. However, direct sequencing of cDNA revealed that a cryptic splice site is activated in exon 52 and is used as the acceptor site instead of the mutant one in intron 51 (fig. 3). The defective splicing leads to deletion of 16 exonic bases and, thus, a frameshift in the COH1 mRNA. The genomic sequence 5′ of the new splice site, ATTTTCGTGTTCCAG, is consistent with the consensus sequence at 11 of 14 relevant positions, including the highly conserved bases AG at the end of the motif. In contrast, the normal splice-acceptor site of intron 51 fully complies with the consensus.
Figure 3.
Sequence analysis of COH1 in probands from the Lebanese family with Cohen syndrome (family 11). A, Splice mutation c.9406–1G→T, identified in DNA samples from two affected cousins, altering the splice acceptor site of intron 51. The position of the mutation is marked by an arrow. B, Analysis of RNA samples showing the activation of a cryptic splice site in exon 52, which leads to a frameshift because of a 16-bp deletion.
Mutations occurred throughout the gene, and each mutation was seen only in one case. Interestingly, only 2 of 17 mutations were missense mutations, Tyr2341Cys and Gly2645Asp, both of which introduce dissimilar residues. All other mutations were either nonsense(9 mutations) or frameshift mutations (6 mutations)leading to premature termination codons. Among 27 Finnish patients investigated by Kolehmainen et al. (2003), 26 had at least one allele with the mutation c.3348_3349delCT. In contrast to those results, the most frequent mutation in Finnish patients was not found in the series of patients reported here. Moreover, eight further mutations, identified in patients whose ancestors were from the United Kingdom, Belgium, and Denmark (Kolehmainen et al. 2003), also were not found in this study group. These findings demonstrate an extensive allelic heterogeneity of Cohen syndrome and the lack of a mutational hotspot in COH1, with the exception of the major mutation found in Finnish patients, the frequency of which probably results from a founder effect.
In two patients (families 1 and 8), only one mutation each was identified. These patients also were assumed to be compound heterozygous. The lack of a second mutation may be because of mutations in alternative exons, which are present in the shorter splice variants of COH1 (Kolehmainen et al. 2003). Alternatively, some further mutations could reside within intronic sequences, which were analyzed only at the conserved splice sites, or could represent larger deletions.
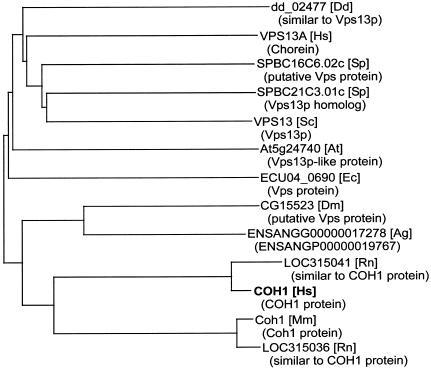
The function of the protein encoded by COH1 is mostly unknown. The search for conserved domains in the predicted protein with the use of RPS-BLAST and the Pfam (Bateman et al. 2004), ProDom (Corpet et al. 2000), and Prosite (Falquet et al. 2002) databasesrevealed two significantly conserved regions at the Nterminus and close to the C-terminus, both regions with similarities to protein domains associated with vacuolar protein sorting, as described elsewhere (Kolehmainen et al. 2003). Detailed sequence alignments with PSI-BLAST (Altschul et al. 1997) identified similarities to Vps13p, a yeast protein associated with vacuolar protein sorting, also known as Soi1p (Redding et al. 1996; Brickner and Fuller 1997). The phylogeny, as depicted with the neighbor-joining method (Saitou and Nei 1987), indicated the rather close relationship of Vps13p-like proteins from distinct species (fig. 4), pointing to a potentially more general role of this protein.
Figure 4.
Phylogenetic tree of sequences with similarities to the deduced product of COH1. The corresponding gene names are given. The species are listed in square brackets; Ag = Anopheles gambiae; At = Arabidopsis thaliana; Dd = Dictyostelium discoideum; Dm = Drosophila melanogaster; Ec = Encephalitozoon cuniculi; Hs = Homo sapiens; Mm = Mus musculus; Rn = Rattus norvegicus; Sc = Saccharomyces cerevisiae; and Sp = Schizosaccharomyces pombe. The names of the gene products, when available, are shown in parentheses, and Vps = associated with vacuolar protein sorting.
The families analyzed here were from different ethnic groups with a wide geographic distribution, including families of German, Turkish, Polish, Lebanese, Omani, and Brazilian descent. Greater clinical variability was observed in these patients with Cohen syndrome in comparison to Finnish patients with Cohen syndrome. Clinical homogeneity in the Finnish patients may be the result of the high frequency of a single mutation, c.3348_3349delCT, and may represent the phenotype of this specific allele. In Finnish patients, the degree of microcephaly at infancy is generally 3–5 SD below the mean. In the cohort reported here, head circumference had a broader variability, from the lower end of the normal range to very severe expression. Therefore, absence of microcephaly does not rule out the diagnosis of Cohen syndrome. This confirms the clinical results reported by Chandler et al. (2003), who observed absence of microcephaly in 10% of their patients, mainly from the United Kingdom.
Further variability was observed in growth development. Short stature was present in 65% of the patients and was as severe as −5.7 SD. In our study, mild truncal obesity was present in most patients at midchildhood, but that trait may be lacking in adult patients. Other studies have described different frequencies of truncal obesity, from 17%–100% in patients age ⩾8 years(Kivitie-Kallio and Norio 2001; Chandler et al. 2003). In contrast to the findings in Finnish patients with Cohen syndrome, neutropenia was identified in only 10/18 patients (56%); in particular, patients from outside Europe were found to have a normal neutrophil count. Therefore, neutropenia is not an obligate sign of Cohen syndrome. The degree of mental retardation, especially the level of speech competence, varied considerably among our patients. The frequency of nonverbal patients (30%) seemed to be higher than reported elsewhere (Kivitie-Kallio and Norio 2001; Chandler et al. 2003). The craniofacial dysmorphism was present but subtle in some of the younger patients. Whereas myopia was evident in all patients, retinopathy was not found in some patients >5 years of age, indicating that it is age dependent and not a consistent feature.
Identification of COH1 mutations in two families with an overlapping phenotype of Cohen and Mirhosseini-Holmes-Walton syndromes suggests that Mirhosseini-Holmes-Walton syndrome does not exist as a separate entity. However, it remains possible that mutations in another gene account for further cases reported as Mirhosseini-Holmes-Walton syndrome.
In summary, a consistent relationship between specific mutations and the severity of Cohen syndrome or the expression of a particular clinical sign has not been observed, with the exception of the mutation c.3348_3349delCT, found solely in Finnish patients. Results from the present study raise the total number of different mutations known in COH1 to 26. Our investigation of an ethnically diverse series of patients with Cohen syndrome revealed that, besides developmental delay, myopia and, in particular, the typical facial gestalt were major signs of Cohen syndrome caused by COH1 mutations. COH1 mutations may be present also in patients lacking retinopathy at school age, microcephaly, or neutropenia. Finally, these data demonstrate that COH1 is subject to wide allelic heterogeneity, and there seems to be no major mutational hotspot in non-Finnish patients with Cohen syndrome.
Acknowledgments
We are grateful to all patients and their families for their collaboration. We wish to thank Nadine Wittstruck and Françoise André for excellent technical assistance and Professor Karl Sperling for continued support. We would like to thank Professor Herwig Frisch for detailed clinical examination of patient 12, Jaderson Costa for referring patients 1/1 and 1/2, and Temis Felix for originally identifying the diagnosis ofCohen syndrome in patients 1/1 and 1/2. This work was supported by a grant from the Deutsche Forschungsgemeinschaft to H.C.H. and D.H.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for chromosome 8 genomic contig [accession number NT_008046], KIAA0532 [accession number AB011104], and COH1 cDNA [accession number NM_017890])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- Prosite, http://www.expasy.org/prosite/
- Protein Domain database (ProDom), http://prodes.toulouse.inra.fr/prodom.html
- Protein families database (Pfam), http://www.sanger.ac.uk/Software/Pfam/
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141 10.1093/nar/gkh121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Fuller RS (1997) SOI1 encodes a novel, conserved protein that promotes TGN-endosomal cycling of Kex2p and other membrane proteins by modulating the function of two TGN localization signals. J Cell Biol 139:23–36 10.1083/jcb.139.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler KE, Kidd A, Al Gazali L, Kolehmainen J, Lehesjoki AE, Black GC, Clayton-Smith J (2003) Diagnostic criteria, clinical characteristics, and natural history of Cohen syndrome. J Med Genet 40:233–241 10.1136/jmg.40.4.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM Jr, Hall BD, Smith DW, Graham CB, Lampert KJ (1973) A new syndrome with hypotonia, obesity, mental deficiency, and facial, oral, ocular, and limb anomalies. J Pediatr 83:280–284 [DOI] [PubMed] [Google Scholar]
- Corpet F, Servant F, Gouzy J, Kahn D (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res 28:267–269 10.1093/nar/28.1.267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJ, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30:235–238 10.1093/nar/30.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D, Krebsova A, Kunze J, Reis A (2000) Homozygosity mapping in a family with microcephaly, mental retardation, and short stature to a Cohen syndrome region on 8q21.3–8q22.1: redefining a clinical entity. Am J Med Genet 92:285–292 [DOI] [PubMed] [Google Scholar]
- Kivitie-Kallio S, Eronen M, Lipsanen-Nyman M, Marttinen E, Norio R (1999) Cohen syndrome: evaluation of its cardiac, endocrine and radiological features. Clin Genet 56:41–50 10.1034/j.1399-0004.1999.560106.x [DOI] [PubMed] [Google Scholar]
- Kivitie-Kallio S, Norio R (2001) Cohen syndrome: essential features, natural history, and heterogeneity. Am J Med Genet 102:125–135 [DOI] [PubMed] [Google Scholar]
- Kolehmainen J, Norio R, Kivitie-Kallio S, Tahvanainen E, de la Chapelle A, Lehesjoki AE (1997) Refined mapping of the Cohen syndrome gene by linkage disequilibrium. Eur J Hum Genet 5:206–213 [PubMed] [Google Scholar]
- Kolehmainen J, Black GC, Saarinen A, Chandler K, Clayton-Smith J, Traskelin AL, Perveen R, Kivitie-Kallio S, Norio R, Warburg M, Fryns JP, de la Chapelle A, Lehesjoki AE (2003) Cohen syndrome is caused by mutations in a novel gene, COH1, encoding a transmembrane protein with a presumed role in vesicle-mediated sorting and intracellular protein transport. Am J Hum Genet 72:1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez HM, Paskulin GA, Vallandro C (1985) The syndrome of retinal pigmentary degeneration, microcephaly, andsevere mental retardation (Mirhosseini-Holmes-Walton syndrome): report of two patients. Am J Med Genet 22:223–228 [DOI] [PubMed] [Google Scholar]
- Mirhosseini SA, Holmes LB, Walton DS (1972) Syndrome of pigmentary retinal degeneration, cataract, microcephaly, and severe mental retardation. J Med Genet 9:193–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio R, Raitta C, Lindahl E (1984) Further delineation of the Cohen syndrome: report on chorioretinal dystrophy, leukopenia and consanguinity. Clin Genet 25:1–14 [DOI] [PubMed] [Google Scholar]
- Norio R, Raitta C (1986) Are the Mirhosseini-Holmes-Walton syndrome and Cohen syndrome identical? Am J Med Genet 25:397–398 [DOI] [PubMed] [Google Scholar]
- Redding K, Brickner JH, Marschall LG, Nichols JW, Fuller RS (1996) Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol Cell Biol 16:6208–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425 [DOI] [PubMed] [Google Scholar]
- Steinlein O, Tariverdian G, Boll HU, Vogel F (1991) Tapetoretinal degeneration in brothers with apparent Cohen syndrome: nosology with Mirhosseini-Holmes-Walton syndrome. Am J Med Genet 41:196–200 [DOI] [PubMed] [Google Scholar]
- Tahvanainen E, Norio R, Karila E, Ranta S, Weissenbach J, Sistonen P, de la Chapelle A (1994) Cohen syndrome gene assigned to the long arm of chromosome 8 by linkage analysis. Nat Genet 7:201–204 [DOI] [PubMed] [Google Scholar]