Abstract
Autosomal dominant facioscapulohumeral muscular dystrophy (FSHD1A) is associated with contractions of the polymorphic D4Z4 repeat on chromosome 4qter. Almost half of new FSHD mutations occur postfertilization, resulting in somatic mosaicism for D4Z4. Detailed D4Z4 analysis of 11 mosaic individuals with FSHD revealed a mosaic mixture of a contracted FSHD-sized allele and the unchanged ancestral allele in 8 cases, which is suggestive of a mitotic gene conversion without crossover. However, in 3 cases, the D4Z4 rearrangement resulted in two different-sized D4Z4 repeats, indicative of a gene conversion with crossover. In all cases, DNA markers proximal and distal to D4Z4 showed no allelic exchanges, suggesting that all rearrangements were intrachromosomal. We propose that D4Z4 rearrangements occur via a synthesis-dependent strand annealing model that relatively frequently allows for crossovers. Furthermore, the distribution of different cell populations in mosaic patients with FSHD suggests that mosaicism here results from D4Z4 rearrangements occurring during the first few zygotic cell divisions after fertilization.
Introduction
A large proportion of eukaryotic genomes consists of repetitive DNA. The head-to-tail tandem repetitive sequences, categorized according to repeat-unit length, are highly polymorphic. Micro-, mini-, macro-, and megasatellite repeats are distinguished, each with their own mutation characteristics. Several hypothetical rearrangement models have been proposed, mainly on the basis of micro- and minisatellite instability. These often include gene conversions with or without crossover (Jeffreys et al. 1994; Paques and Haber 1999). However, little is known about macro- and megasatellite instability in human cells.
Facioscapulohumeral muscular dystrophy (FSHD1A [MIM 158900], hereafter referred to as “FSHD”) is an early-onset, autosomal dominant myopathy mainly characterized by a progressive and variable atrophy and weakness of the facial, shoulder, and upper-arm muscles (Padberg 1982). The FSHD phenotype also presents with extramuscular symptoms such as retinovasculopathy, hearing loss, and, in severe cases, mental retardation (Brouwer et al. 1991; Padberg et al. 1995a; Funakoshi et al. 1998; Miura et al. 1998). FSHD is associated with a macrosatellite repeat instability: the contraction of the subtelomeric D4Z4 repeat on 4q35. This polymorphic D4Z4 repeat is highly recombinogenic, since somatic mosaicism for a rearrangement of D4Z4 is found in as much as 3% of the general population (van Overveld et al. 2000). The D4Z4 repeat consists of identical KpnI units, each 3.3 kb in size, ordered in a head-to-tail fashion and varying between 11 and 100 units on healthy chromosomes (van Deutekom et al. 1993). Patients with FSHD carry a repeat of 1–10 units on one of their chromosomes 4 (Wijmenga et al. 1992). A rough and inverse correlation has been observed between the severity and age at onset of the disease and the residual repeat unit number (Lunt et al. 1995; Tawil et al. 1996).
The subtelomere of chromosome 10q is nearly identical to chromosome 4qter and also contains a highly homologous and equally recombinogenic repeat. D4Z4 repeats on chromosomes 4 and 10 are visualized in EcoRI-digested DNA with probe p13E-11, which recognizes the region proximal to the D4Z4 repeat within the EcoRI fragment (Wijmenga et al. 1992). To discriminate between 4qter- and 10qter-derived repeats, in addition to EcoRI, the restriction enzyme BlnI is used, which digests only chromosome 10–derived repeat units (Deidda et al. 1996). Conversely, the restriction enzyme XapI is specific to 4qter-derived repeat units and can be used to complement BlnI (Lemmers et al. 2001). DNA separated by pulsed field gel electrophoresis (PFGE), combined with differential digestion with BlnI and XapI, allows sizing and chromosomal assignment of all D4Z4 alleles ranging from 10 to >350 kb.
In 10% of the population, translocated 4-type repeats reside on chromosome 10q and, vice versa, translocated 10-type repeats on chromosome 4q are equally frequent. This was first found in a Dutch survey (van Deutekom et al. 1996; van Overveld et al. 2000), but comparable translocation frequencies have now also been reported in the Japanese, Korean, and Chinese populations (Matsumura et al. 2002). These translocated repeats can be homogeneous or heterogeneous (consisting of both 4-type and 10-type units). Although this observation may suggest frequent interactions between the regions of homology of both nonhomologous chromosomes, de novo exchanges between repeat arrays originating from chromosomes 4 and 10 have never been described. In contrast to chromosome 4, repeat contractions on chromosome 10 are nonpathogenic.
Further complicating our understanding of FSHD pathogenesis, a 4qter polymorphism distal to D4Z4 was recently described, giving rise to two distinct 4q chromosome ends: 4qA and 4qB (van Geel et al. 2002). Both alleles are almost equally common and equally recombinogenic in controls. Nevertheless, FSHD alleles are always of the 4qA type (Lemmers et al. 2002).
As many as 10%–30% of patients with FSHD carry a new mutation (Padberg et al. 1995b; Zatz et al. 1995; Lunt 1998), with D4Z4 repeat contractions varying from 1 to >50 units (van der Maarel et al. 2000). Several studies have shown that almost half of these D4Z4 rearrangements are of mitotic origin, resulting in somatic mosaicism for the FSHD allele in either patients or asymptomatic parents (Upadhyaya et al. 1995; Kohler et al. 1996; Zatz et al. 1998; Galluzzi et al. 1999; van der Maarel et al. 2000). A relationship has been established between the severity of the disease and the percentage of peripheral blood lymphocytes (PBL) carrying the disease allele in combination with the residual repeat size (van der Maarel et al. 2000). This suggests that the proportion of mosaicism detected in blood is representative of the mosaicism in muscle tissue. Furthermore, patients with FSHD have been described who displayed mosaicism for the disease allele in PBL and germline (Galluzzi et al. 1999; Lemmers et al., in press), as well as in fibroblasts (R.J.L.F.L., R.R.F., S.M.v.d.M., M. Tonini, and M. Zatz, unpublished data). Thus, mitotic D4Z4 rearrangements very likely occur early during embryogenesis.
In all mosaic cases reported, two PBL cell populations were identified: one carrying the parental, nonrearranged allele, and one in which the D4Z4 repeat had been rearranged to FSHD size. Such an allele distribution generally is suggestive of a rearrangement by gene conversion without crossover, since the hallmark of this mechanism is an unchanged donor allele and a changed acceptor allele (Paques and Haber 1999).
Although many macrosatellite repeats have been identified in the human genome, little is known about the mechanism by which they rearrange. Mitotic minisatellite rearrangements were thoroughly studied in yeast, in which gene conversion, usually without crossover, was shown to be the main mechanism of rearrangement (Paques and Haber 1999).
Similar to D4Z4, the polymorphic human RS447 megasatellite repeat, which is comprised of 4.7-kb units, displays both mitotic and meiotic instability. For RS447, a high frequency of repeat contraction and expansion has been shown (Okada et al. 2002). The tandemly repeated human U2 snRNA genes (RNU2 locus) have been studied more extensively and have provided insight into the mechanism of concerted evolution. These 6.1-kb U2 repeats display repeat homogenization by rare interchromosomal gene conversion followed by rapid intrachromosomal homogenization (Liao 1999).
The mechanism by which D4Z4 rearranges was further examined in the present study, through use of the 4qA/4qB polymorphism as distal flanking marker in combination with a newly identified RFLP within D4Z4 proper, serving as a proximal flanking marker. Studying these proximal and distal markers provided insight into the mechanism and timing of D4Z4 rearrangements and may stand as a model for macrosatellite repeat mutation in general.
Subjects and Methods
Patients and Control Individuals
All families were ascertained via one of the Dutch Neuromuscular Centers. In all but one family, the mosaic child received a clinical diagnosis of FSHD, and the parents did not display any FSHD features. In family 55611, the FSHD allele of the affected mother was further somatically contracted in the affected child. Blood from these families was collected after informed consent was obtained. The PvuII polymorphism was determined in a previously described population of anonymized Dutch blood donors with a standard allele configuration of homogenous 4-type repeats on chromosome 4 and 10-type repeats on chromosome 10 (van Overveld et al. 2000). Four of 11 mosaic patients with FSHD studied carried a homogeneous translocated 4-type repeat on chromosome 4.
Somatic Cell Hybrids and DNA Clones
The following chromosome 4qA sources were used: YAC Y25C-2E (Wright et al. 1993); the monochromosomal rodent somatic cell hybrids HHW1494, SU10 (a gift from S. Winokur, Irvine, CA), and GM11448 (Coriell Institute for Medical Research); and lambda clones λ42, λ68, and λ260201 (van Deutekom et al. 1993). All sources, except for Y25C-2E and GM11448, represent patient alleles. Somatic cell hybrid GM11448 contains a human chromosome 4qA with a D4Z4 repeat of 125 kb (data not shown).
As chromosome 4qB sources, we used the monochromosomal rodent somatic cell hybrids GM11687 (Coriell Institute for Medical Research), 4L-10 (a gift from E. Stanbridge, Irvine, CA), HHW416 (a gift from M. Altherr, Los Alamos, NM), and 361-9 (a gift from S. Winokur, Irvine, CA), with D4Z4 repeats of 140 kb, 85 kb, 96 kb, and 290 kb, respectively.
DNA Isolation
DNA was isolated from PBL essentially as described by Miller et al. (1988). If possible, PBL were embedded in agarose plugs (InCert agarose [FMC]) at a concentration of 7.5×105 cells per plug.
D4Z4 Analysis
For allele sizing, DNA samples were double digested with EcoRI/HindIII and EcoRI/BlnI. For analysis of the allelic variation at 4qter (probes 4qA and 4qB [Lemmers et al. 2002]), DNA was digested with HindIII only. All digestions were performed according to the manufacturer's instructions. EcoRI and HindIII were purchased from MBI Fermentas, and PvuII and BlnI were purchased from Amersham-Pharmacia. DNA was separated for 22 h on a 0.8% agarose gel (MP agarose [Boehringer]) by pulsed field gel electrophoresis (PFGE) at 8.5 V/cm at 21°C in 0.5× tris-borate EDTA (TBE) supplemented with 150 pg/ml ethidium bromide. PFGE was performed in four identical cycles, with switch time increasing linearly from 1 s at the start to 16 s at the end of each cycle. Subsequently, DNA was transferred to a Hybond XL membrane (Amersham-Pharmacia) by Southern blotting and was hybridized in a buffer containing 0.125 M Na2HPO4 (pH 7.2), 10% PEG6000, 0.25 M NaCl, 1 mM EDTA, and 7% SDS, for 16–24 h at 65°C. Hybridizations with probe p13E-11 (D4F104S1 [Wijmenga et al. 1992]) were washed in 2× saline-sodium citrate buffer (SSC)/0.1% SDS, and those with probes 4qA or 4qB were washed more stringently in 1× SSC/0.1% SDS. The blots were exposed for 16–24 h to phosphorimager screens and were analyzed with the Image Quant software program (Molecular Dynamics).
The PvuII polymorphism in the first proximal unit of the D4Z4 repeat (position 6044 of GenBank accession number AF117653) was analyzed by applying a modified BglII/BlnI dosage test (van der Maarel et al. 1999) on genomic DNA samples by double digestion with PvuII and BlnI (fig. 1). After Southern blot and hybridization with probe p13E-11, blots were washed in 0.3× SSC/0.1% SDS. Chromosome 4–derived fragments are 2,849 bp or 4,559 bp in size, depending on the presence or absence of the PvuII restriction site, whereas chromosome 10–derived fragments are 2,464 bp in size, because of the presence of BlnI. Blots were exposed to phosphorimager screens, and signals were quantified with the Image-Quant software program. Restriction analysis of this polymorphism in internal D4Z4 units was performed on chromosome 4–only sources. A set of seven 4qA- and four 4qB-derived repeats were hybridized with the D4Z4 repeat probe 9B6A (Wright et al. 1993) after digestion with PvuII, separation on a 0.8% TBE agarose gel, and transfer to Hybond XL membranes. The 3.3-kb fragment is derived from PvuII− units, whereas the 1.6-kb fragment represents PvuII+ units. The most distal D4Z4 unit of the repeat array was analyzed by specific PCR amplification of this unit, followed by a PvuII digestion (primer sequences available upon request).
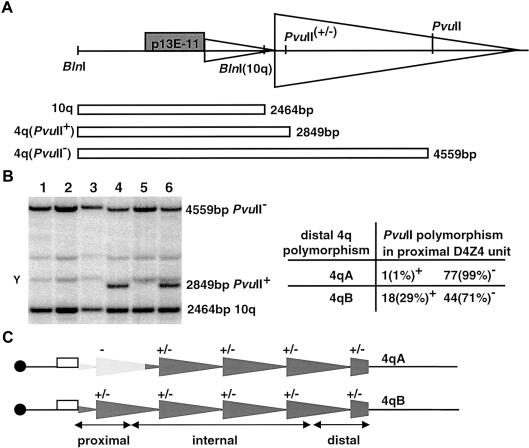
Figure 1.
A polymorphic PvuII site at position 6044 of GenBank accession number AF117653 was used to study recombination between 4qA and 4qB alleles. A, Double digestion with PvuII/BlnI normally gives rise to fragments of 4,559 bp (chromosome 4 alleles) and 2,464 bp (chromosome 10 alleles). However, the presence of an extra PvuII site within the proximal D4Z4 unit yields a fragment of 2,849 bp. B, Example of the modified PvuII/BlnI dosage test after hybridization with probe p13E-11. Samples in lanes 1–3 carry two 4qA alleles and two 10q alleles, all of which are PvuII resistant (PvuII−). Samples in lanes 4–6 carry two 4qB alleles and two 10q alleles; in lane 4 and lane 6, a PvuII-sensitive (PvuII+) fragment is visible. “Y” indicates the cross-hybridizing Y chromosome. The table on the right shows dosage experiment results of the PvuII RFLP in the proximal D4Z4 unit of 78 different 4qA and 62 different 4qB alleles. Twenty-nine percent of the 4qB alleles are PvuII+, whereas 4qA alleles are almost exclusively PvuII− (P<10-5). C, Distribution of the PvuII RFLP on 4qA- and 4qB-type alleles. The PvuII− proximal D4Z4 unit on 4qA-type alleles is indicated by “−,” whereas D4Z4 units that were heterozygous for PvuII RFLP are indicated by “+/−.”
Haplotyping
Haplotype analysis for family 36 was performed using 24 polymorphic markers, including D4S163, D4S139, D10S555, and D10S595 (P. de Knijff, personal communication)
Statistical Analysis
The presence of the PvuII site in the proximal unit of the D4Z4 repeat of 4qA and 4qB alleles was compared using the Pearson χ2 test. Allele size distributions in 88 control individuals (84 4qA and 92 4qB alleles) were analyzed according to one-way ANOVA and a nonparametric two-sample Kolmogorov-Smirnov test.
Results
Chromosomal Partner for D4Z4 Rearrangements
Repeat rearrangements usually occur between sister chromatids or homologous chromosomes. To reveal the common partner for D4Z4 rearrangements, the allele size distributions of 4qA- and 4qB-derived D4Z4 repeats were compared. A different D4Z4 repeat length distribution of 4qA and 4qB type alleles would indicate that these chromosomes do not often interact. The sizing was performed by PFGE on homogeneous 4-type D4Z4 repeats obtained from high-quality agarose-embedded DNA plugs. In total, 84 4qA- and 92 4qB-type alleles were analyzed. Statistical analyses revealed that D4Z4 repeats derived from 4qA alleles (mean ± 95% CI 136±7 kb) were significantly longer than those derived from 4qB alleles (mean ± 95% CI 93±4 kb) and that they had a different size distribution (P<.001). This indicates that, in general, D4Z4 rearrangements occur intrachromosomally.
To further elucidate the mechanism underlying D4Z4 rearrangements, we studied a polymorphic PvuII site (position 6044 in GenBank accession number AF117653) (Gabriels et al. 1999) in the proximal D4Z4 unit of the repeat through use of a modified BglII-BlnI dosage test (fig. 1) (van der Maarel et al. 1999). A PvuII/BlnI double digestion was performed to separate chromosome 4 PvuII-sensitive (2,849-bp) from PvuII-resistant (4,559-bp) alleles. In addition, BlnI was used to avoid interference of chromosome 10–type alleles (2,464 bp). Analysis of this PvuII-RFLP in 78 independent 4qA alleles and 62 independent 4qB alleles (fig. 1A and 1B) showed that the PvuII site is present in 29% of 4qB alleles and only in 1% of 4qA alleles (P<10-5), suggesting a marked linkage disequilibrium (LD) between the proximal and distal ends of the D4Z4 repeat.
Next, internal D4Z4 units were analyzed for the PvuII-RFLP on DNA sources containing only a single chromosome 4–derived D4Z4 repeat. A set of seven 4qA- and four 4qB-derived D4Z4 repeats revealed that the repeat units following the first unit were polymorphic for the PvuII polymorphism on both alleles (data summarized in fig. 1C).
D4Z4 Repeat Analysis of Mosaic Kindreds with FSHD
Eleven kindreds with FSHD, consisting of father, mother, and mosaic child with FSHD, were analyzed for their D4Z4 repeat size and distal 4qA/4qB origin (table 1). For most of these families, D4Z4 allele sizes have been described elsewhere (van der Maarel et al. 2000). In all mosaic individuals, we identified three nonmosaic alleles: two alleles derived from chromosome 10 and one derived from the unaffected chromosome 4 homologue. In all cases, the de novo mosaic alleles had the same distal flanking marker (4qA) as their ancestral alleles, despite heterozygosity for this distal flanking marker in seven of them (Lemmers et al. 2002) (table 1). This suggests that D4Z4 rearrangements generally do not occur between both chromosomes 4. In 8 of these 11 mosaic individuals, we observed a mosaic mixture of a de novo FSHD allele and an unchanged parental allele, suggestive of a mitotic D4Z4 rearrangement by gene conversion without crossover. An example is shown in figure 2. The remaining three mosaic patients displayed a more complex mosaic pattern, since they carry two somatically rearranged D4Z4 alleles, suggestive of rearrangements with crossover.
Table 1.
Sizing and Typing of Chromosome 4–Derived D4Z4 Alleles from Mosaic Patients with FSHD
|
Length and Proportion of D4Z4 Alleles (4qA-Type)after Involvement in Mitotic Rearrangement |
||||||||||
| Ancestral Allele |
Contracted Allelea |
Rearranged Allele 2b |
Homologous Allele |
|||||||
| Family | Sex ofPatient | Length(kb) | % | Length(kb) | % | Length(kb) | % | ChromosomalOrigin | Length(kb) | AlleleType |
| 1 | Male | 70 | 37 | 14 | 26 | 96 | 37 | Paternal | 96 | 4qB |
| 5 | Male | 130 | 40 | 14 | 60 | Maternal | 80 | 4qA | ||
| 6 | Male | 280 | 60 | 10 | 40 | Paternal | 175 | 4qA | ||
| 7 | Male | 160 | 70 | 16 | 30 | Paternal | 65 | 4qB | ||
| 12 | Female | 80 | 10 | 23 | 90 | Maternal | 45 | 4qB | ||
| 24 | Male | 50 | 50 | 17 | 50 | Maternal | 80 | 4qB | ||
| 26 | Male | 130 | 50 | 14 | 50 | Maternal | 80 | 4qB | ||
| 35 | Male | 90 | 50 | 33 | 50 | Maternal | 220 | 4qA | ||
| 36 | Male | 240 | 0 | 16 | 60 | 384 | 40 | Maternal | 135 | 4qA |
| Rf120 | Female | 100 | 20 | 20 | 70 | 55 | 10 | Maternal | 50 | 4qB |
| 55611 | Male | 33 | 80 | 30 | 20 | Maternal | 130 | 4qB | ||
Contracted FSHD-causing allele.
Mitotic gene conversions with crossover produce two rearranged D4Z4 alleles (contracted and rearranged allele 2).
Figure 2.
Example of p13E-11 hybridization after PFGE and Southern blot analysis: analysis of mosaic family 26 with FSHD (GC-CO = gene conversion without crossover). DNA was double digested with EcoRI/HindIII (“E”) and EcoRI/BlnI (“B”), separated by PFGE, and hybridized with p13E-11. The patient inherited the paternal (“p”) 60-kb chromosome 10 allele and 90-kb 4qB allele and, from his mother (“m”), the 100-kb chromosome 10 allele and the 140-kb 4qA allele (unblackened arrowhead), which is present in only 50% of his PBL. The 14-kb 4qA allele (blackened arrowhead) causing FSHD is derived from the 140-kb maternal allele and is detected in 50% of his PBL. Estimation of the proportion of mosaic alleles is based on the signal intensities of the D4Z4 fragments, with a CI of 5%. Allele types are not shown. Marker sizes (“M”) in kilobases are indicated on the left.
In family 36, we observed a mosaic patient with a mosaic mixture of a 384-kb–sized expanded and a 16-kb–sized contracted allele in almost equal proportions of PBL (fig. 3A). Haplotyping of the patient and his parents excluded nonparenthood (data not shown). In family 1, the presence of three mosaic alleles was initially overlooked because of the comigration of one of these mosaic alleles with the nonrearranged chromosome 4qB homologue (fig. 3B, left image). However, sequence comparison of the region distal to D4Z4 on 4qA (GenBank accession numbers U74496 and U74497) and 4qB (GenBank accession number AF017466) (van Geel et al. 2002) revealed an RFLP for EcoRV. This EcoRV restriction site is directly adjacent to D4Z4 on 4qA, whereas its location on 4qB is 10 kb distal to the D4Z4 repeat (data not shown), allowing separation of these comigrating 4qA and 4qB alleles and visualization of the previously undetected mosaic allele. The patient now clearly displayed three mosaic alleles; one FSHD-sized allele of 14 kb, one expanded allele of 96 kb, and the parental unchanged allele of 70 kb, each of them equally frequent in the PBL (fig. 3B, right image). The third complex mosaic patient, in family Rf120 (data not shown), displayed a mixture of three mosaic alleles: two de novo contracted alleles of 20 kb and 55 kb in 70% and 10% of PBL, respectively, and the unchanged parental-sized allele of 100 kb in the remainder of cells.
Figure 3.
Two examples of p13E-11 hybridization after PFGE and Southern blot. Analyses, as in figure 2, of mitotic interchromatid gene conversion with crossover (GC+CO) in families 36 and 1 with FSHD. Estimation of the proportion of mosaic alleles is based on the signal intensities of the D4Z4 fragments, with a CI of 5%. Allele types are not shown. A, Male patient with de novo FSHD in family 36, who inherited 10q alleles of 50 kb and 130 kb and a 140-kb 4qA-type allele. He also has mosaic alleles of 384 kb (40%; 4Aexp) and 16 kb (60%; 4AFSH), which are expanded and contracted alleles, respectively, most probably produced by an interchromatid gene conversion with crossover in the maternal 100-kb or 240-kb allele. Maternally and paternally inherited alleles are indicated by “m” and “p,” respectively. Comigrating chromosome 4 and 10 alleles of 130 kb in the father are marked with an asterisk. Marker sizes (“M”) in kilobases are indicated on the left. B, The left panel shows EcoRI/HindIII (“E”) and EcoRI/BlnI (“B”) digestion of DNA from family 1 with de novo FSHD. The patient from this family has three mosaic alleles—of 14kb, 70 kb, and 96 kb—that originated from the paternal 70-kb 4qA allele. The 96-kb 4qA mosaic allele is comigrating with a maternal 96-kb 4qB allele. Furthermore, he inherited a paternal 60-kb 10q allele and a maternal 135 kb 4qB-type allele (marked with an asterisk), which originated from chromosome 10 (translocated 4-type repeat (t10;4) (van Deutekom et al. 1996). Quantification of all alleles revealed 100% intensity for the 135-kb (t10;4) 4qB allele, 137% for the comigrating 96-kb 4qA and 4qB alleles, 37% for the 70-kb 4qA allele, 100% for the 60-kb 10q allele, and 26% for the 14-kb 4qA allele. An EcoRV (“EV”) digestion allows separation of the comigrating 4qA and 4qB alleles at 96 kb (137%). On the basis of EcoRV digestion (right panel) the patient clearly has six alleles; of these, the contracted 14-kb 4qA allele (26%; FSHD allele, 4AFSH) and the expanded 96-kb 4qA allele (37%; 4Aexp) originate from the mitotic rearranged parental 70-kb allele (37%; 4A0). Two arrows indicate D4Z4 expansion and contraction. Allele quantifications after EcoRV digestion are indicated in the right example panel. Marker sizes (“M”) in kilobases are indicated at the right. In the box, the constitution of the three different cell populations of this patient is schematically depicted.
Discussion
To elucidate the mechanism by which D4Z4 rearranges, we analyzed the exact repeat size, composition (PvuII-RFLP), and origin (4qA/4qB) of all D4Z4 repeats in individuals in whom we had established that a D4Z4 rearrangement had occurred de novo through the presence of mitotically rearranged D4Z4 repeats. We observed that the mechanism by which D4Z4 rearranges shares features common to that of other tandemly repeated DNA sequences.
General Features of D4Z4 Rearrangements
To reveal the common partner in D4Z4 rearrangements, D4Z4 repeat length distributions on 4qA and 4qB alleles were compared. Statistical analyses show a significant difference in the distribution between 4qA- and 4qB-type alleles, excluding a frequent recombination between D4Z4 repeats on 4qA and 4qB chromosomes. Most probably, D4Z4 rearrangements preferentially occur between sister chromatids, the most common partners for rearrangements in mammalian cells (Johnson and Jasin 2000). In addition, the repetitive nature of D4Z4 enables an intrachromatid rearrangement by the formation of an intra-allelic DNA loop.
In all mosaic cases studied, we observed no change of distal flanking marker 4qA/4qB between de novo and ancestral alleles, despite the fact that the majority of mosaic individuals were heterozygous for this distal marker (table 1). The different D4Z4 repeat length distributions of 4qA and 4qB alleles and the observed LD between the proximal PvuII RFLP and the 4qB allele is suggestive of a strong suppression of recombination between 4qA and 4qB alleles. This suppression of recombination between 4qA and 4qB appears not to be sustained throughout the entire repeat, on the basis of the observation that internal D4Z4 units are equally polymorphic for the PvuII restriction site on 4qA and 4qB chromosomes. This latter may be explained by ongoing or incomplete concerted evolution, as has been described for the RNU2 locus (Liao 1999). Apparently, a rare interchromosomal gene conversion copied PvuII from 4qB to 4qA and subsequently spread over this repeat by interallelic rearrangements. It is possible that the proximal D4Z4 unit has escaped this homogenization until now (fig. 1C), which is suggestive of a 3′ polarity of D4Z4 rearrangements, as described for meiotic minisatellite rearrangements (Jeffreys et al. 1998).
D4Z4 Rearrangements by Gene Conversion without Interchromatid Crossover
The results of the present study strongly suggest that most mitotic D4Z4 rearrangements occur via an interchromatid gene conversion mechanism without crossover in which the donor allele remains unchanged, since the majority of mosaic patients with FSHD carry two distinct cell populations: one that encompasses the parental-sized D4Z4 alleles prior to rearrangement and one that encompasses the de novo disease-causing D4Z4 allele (for an example, see fig. 2).
Gene conversions not associated with crossover can be explained by synthesis-dependent strand annealing (SDSA) models, as proposed for recombination in yeast (Nassif et al. 1994). In these models, a 3′ end of a resected double-strand break (DSB) invades the donor template and primes DNA synthesis. Next, the newly synthesized DNA strands are unwound from the template and returned to the broken molecule, allowing them to anneal to each other. Out-of-frame annealing of the newly synthesized strand can lead to repeat contractions or expansion, while the donor template remains unchanged. Alternatively, intrachromatid looping–related or single-strand annealing rearrangements could also explain our observations. However, these mechanisms can give rise only to D4Z4 contractions, which cannot explain the rapid expansion of D4Z4 repeats in the hominoid lineage (Hewitt et al. 1994) or our observation of D4Z4 expansions in the population (R.J.L.F.L., G.W.P., R.R.F., and S.M.v.d.M., unpublished data).
D4Z4 Rearrangements by Gene Conversion with Interchromatid Crossover
The presence of two rearranged D4Z4 repeats in 3 of 11 mosaic families with FSHD is indicative of the relatively frequent occurrence of gene conversion with interchromatid crossover (as shown in fig. 3). Gene conversion with crossover is commonly observed in only a small proportion of rearrangements (Paques et al. 1998). It is possible that the telomeric localization of D4Z4 repeats is underlying the increased crossover rate. In family 36, a mitotic D4Z4 rearrangement of the maternal 100-kb or 240-kb allele resulted in alleles of 384 kb and 16 kb, which are observed in almost equal proportions in the patients' PBL. The patient with de novo FSHD from family 1 has three PBL cell populations, one of which carries the original allele (37%), whereas the other two contain contracted and expanded alleles of the same chromosome, in 26% and 37% of cells, respectively. Regardless of the exact mechanism, to our knowledge this is the first observation of a de novo expansion of the D4Z4 repeat. In family Rf120, the original 100-kb allele is mitotically reduced to a 55-kb allele and an FSHD-sized allele.
Gene conversions that allow for crossovers are often explained by a DSB repair model (Szostak et al. 1983) but can also be explained by an SDSA model that includes the possibility of crossover (Paques et al. 1998). In both models, DSB formation is followed by 5′-to-3′ resection, leaving two recombinogenic 3′ ends that can invade the donor template. In both models, the resolution is postulated to occur through cutting and resolving of two Holliday junctions, which may result in either crossover or no crossover (Holliday 1964). The DSB repair model was initially proposed to explain gene conversions in yeast that were accompanied by crossover in half of the cases (Orr-Weaver and Szostak 1983). When subsequent studies showed much lower crossover rates, other models were proposed that did not require Holliday junctions. However, the suppression of crossover during the resolution of these junctions can also explain the overrepresentation of gene conversion without crossover (Wu and Hickson 2003). On the basis of our findings, we propose that mitotic D4Z4 rearrangements occur preferentially via a SDSA model that allows for crossovers.
Timing of D4Z4 Rearrangements
The coexistence of mosaicism for D4Z4 in PBL, germline, and fibroblast cells of patients with FSHD already indicated that mitotic D4Z4 rearrangements occur early during embryogenesis. More specifically, the absence of the mosaic parental-sized repeat in family 36 might suggest that the rearrangement had occurred at the one-cell stage, before the first embryonic cell division. Also, all other somatic rearrangements most probably occurred during the first few zygotic divisions after fertilization, resulting in a proportion of affected cells that depended on the timing of the rearrangement and on stochastic events related to which cells contributed to the embryo proper. Mitotic D4Z4 rearrangements that occur at a later stage of development will generally result in de novo mosaic alleles in <25% of the cells, as detected in asymptomatic carriers of the FSHD allele (van der Maarel et al. 2000).
Different mechanisms could underlie this early occurrence of D4Z4 rearrangements. During the first replication steps after fertilization, paternal and maternal genomes still display some characteristics of their progeny cells (Mayer et al. 2000; McLay and Clarke 2003). It is tempting to speculate that chromatin conformational changes during the first rounds of cell division may result in DNA strand breaks that initiate these mitotic D4Z4 rearrangements.
Acknowledgments
We dedicate this work to the memory of Lodewijk Sandkuijl, who unexpectedly passed away during preparation of this manuscript. The authors thank all patients and relatives for their cooperation. We thank Dr. P. de Knijff for helping with the haplotype analysis and Drs. G. J. B. van Ommen and L. H. Mullenders for critical reading and useful comments. FSHD research is made possible by the Prinses Beatrix Fonds, the Muscular Dystrophy Association USA, the FSH Society, the Stichting FSHD, the Shaw Family, and the National Institutes of Health.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for the D4Z4 allele [accession number AF117653], distal 4qB sequence [accession number AF017466], and distal 4qA sequence [accession numbers U74496 and U74497])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FSHD1A) [PubMed]
References
- Brouwer OF, Padberg GW, Ruys CJ, Brand R, de Laat JA, Grote JJ (1991) Hearing loss in facioscapulohumeral muscular dystrophy. Neurology 41:1878–1881 [DOI] [PubMed] [Google Scholar]
- Deidda G, Cacurri S, Piazzo N, Felicetti L (1996) Direct detection of 4q35 rearrangements implicated in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet 33:361–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Goto K, Arahata K (1998) Epilepsy and mental retardation in a subset of early onset 4q35-facioscapulohumeral muscular dystrophy. Neurology 50:1791–1794 [DOI] [PubMed] [Google Scholar]
- Gabriels J, Beckers MC, Ding H, De Vriese A, Plaisance S, van der Maarel SM, Padberg GW, Frants RR, Hewitt JE, Collen D, Belayew A (1999) Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236:25–32 10.1016/S0378-1119(99)00267-X [DOI] [PubMed] [Google Scholar]
- Galluzzi G, Deidda G, Cacurri S, Colantoni L, Piazzo N, Vigneti E, Ricci E, Servidei S, Merico B, Pachi A, Brambati B, Mangiola F, Tonali P, Felicetti L (1999) Molecular analysis of 4q35 rearrangements in fascioscapulohumeral muscular dystrophy (FSHD): application to family studies for a correct genetic advice and a reliable prenatal diagnosis of the disease. Neuromuscul Disord 9:190–198 10.1016/S0960-8966(98)00116-3 [DOI] [PubMed] [Google Scholar]
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, Frants RR, Williamson R (1994) Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 3:1287–1295 [DOI] [PubMed] [Google Scholar]
- Holliday R (1964) Mechanism for gene conversion in fungi. Genet Res 5:282–304 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neil DL, Neumann R (1998) Repeat instability at human minisatellites arising from meiotic recombination. EMBO J 17:4147–4157 10.1093/emboj/17.14.4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JAL (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat Genet 6:136–145 [DOI] [PubMed] [Google Scholar]
- Johnson RD, Jasin M (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19:3398–3407 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler J, Rupilius B, Otto M, Bathke K, Koch MC (1996) Germline mosaicism in 4q35 facioscapulohumeral muscular dystrophy (FSHD1A) occurring predominantly in oogenesis. Hum Genet 98:485–490 10.1007/s004390050244 [DOI] [PubMed] [Google Scholar]
- Lemmers RJ, de Kievit P, Sandkuijl L, Padberg GW, van Ommen GJ, Frants RR, van der Maarel SM (2002) Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet 32:235–236 10.1038/ng999 [DOI] [PubMed] [Google Scholar]
- Lemmers RJL, de Kievit P, van Geel M, van der Wielen MJ, Bakker E, Padberg GW, Frants RR, van der Maarel SM (2001) Complete allele information in the diagnosis of facioscapulohumeral muscular dystrophy by triple DNA analysis. Ann Neurol 50:816–819 10.1002/ana.10057 [DOI] [PubMed] [Google Scholar]
- Lemmers RJLF, van der Wielen MJR, Bakker E, Padberg GW, Frants RR, van der Maarel SM. Somatic mosaicism in facioscapulohumeral muscular dystrophy patients often goes undeteced. Ann Neurol (in press) [DOI] [PubMed] [Google Scholar]
- Liao D (1999) Concerted evolution: molecular mechanism and biological implications. Am J Hum Genet 64:24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt PW (1998) 44th ENMC International Workshop: Facioscapulohumeral Muscular Dystrophy: Molecular Studies 19–21 July 1996, Naarden, The Netherlands. Neuromuscul Disord 8:126–130 10.1016/S0960-8966(98)00012-1 [DOI] [PubMed] [Google Scholar]
- Lunt PW, Jardine PE, Koch MC, Maynard J, Osborn M, Williams M, Harper PS, Upadhyaya M (1995) Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35- facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet 4:951–958 [DOI] [PubMed] [Google Scholar]
- Matsumura T, Goto K, Yamanaka G, Lee J, Zhang C, Hayashi YK, Arahata K (2002) Chromosome 4q;10q translocations: comparison with different ethnic populations and FSHD patients. BMC Neurol 2:7 10.1186/1471-2377-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T (2000) Demethylation of the zygotic paternal genome. Nature 403:501–502 [DOI] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ (2003) Remodelling the paternal chromatin at fertilization in mammals. Reproduction 125:625–633 10.1530/rep.0.1250625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Kumagai T, Matsumoto A, Iriyama E, Watanabe K, Goto K, Arahata K (1998) Two cases of chromosome 4q35-linked early onset facioscapulohumeral muscular dystrophy with mental retardation and epilepsy. Neuropediatrics 29:239–241 [DOI] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB (1994) Efficient copying of nonhomologous sequences from ectopic sites via p-element-induced gap repair. Mol Cell Biol 14:1613–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Gondo Y, Goto J, Kanazawa I, Hadano S, Ikeda JE (2002) Unstable transmission of the RS447 human megasatellite tandem repetitive sequence that contains the USP17 deubiquitinating enzyme gene. Hum Genet 110:302–313 10.1007/s00439-002-0698-2 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW (1983) Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci USA 80:4417–4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padberg GW (1982) Facioscapulohumeral disease. PhD thesis, Leiden University, Leiden [Google Scholar]
- Padberg GW, Brouwer OF, de Keizer RJW, Dijkman G, Wijmenga C, Grote JJ, Frants RR (1995a) On the significance of retinal vascular disease and hearing loss in facioscapulohumeral muscular dystrophy. Muscle Nerve 2:S73–S80 [PubMed] [Google Scholar]
- Padberg GW, Frants RR, Brouwer OF, Wijmenga C, Bakker E, Sandkuijl LA (1995b) Facioscapulohumeral muscular dystrophy in the Dutch population. Muscle Nerve 2:S81–S84 [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63:349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Leung WY, Haber JE (1998) Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol 18:2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW (1983) The double-strand-break repair model for recombination. Cell 33:25–35 10.1016/0092-8674(83)90331-8 [DOI] [PubMed] [Google Scholar]
- Tawil R, Forrester J, Griggs RC, Mendell J, Kissel J, McDermott M, King W, Weiffenbach B, Figlewicz D (1996) Evidence for anticipation and association of deletion size with severity in facioscapulohumeral muscular dystrophy. Ann Neurol 39:744–748 [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Maynard J, Osborn M, Jardine P, Harper PS, Lunt P (1995) Germinal mosaicism in facioscapulohumeral muscular dystrophy (FSHD). Muscle Nerve 2:S45–S49 [PubMed] [Google Scholar]
- van der Maarel SM, Deidda G, Lemmers RJ, Bakker E, van der Wielen MJ, Sandkuijl L, Hewitt JE, Padberg GW, Frants RR (1999) A new dosage test for subtelomeric 4;10 translocations improves conventional diagnosis of facioscapulohumeral muscular dystrophy. J Med Genet 36:823–828 [PMC free article] [PubMed] [Google Scholar]
- van der Maarel SM, Deidda G, Lemmers RJ, van Overveld PG, van der Wielen M, Hewitt JE, Sandkuijl L, Bakker B, van Ommen GJ, Padberg GW, Frants RR (2000) De novo facioscapulohumeral muscular dystrophy: frequent somatic mosaicism, sex-dependent phenotype, and the role of mitotic transchromosomal repeat interaction between chromosomes 4 and 10. Am J Hum Genet 66:26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, Padberg GW, Frants RR (1996) Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet 5:1997–2003 10.1093/hmg/5.12.1997 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2:2037–2042 [DOI] [PubMed] [Google Scholar]
- van Geel M, Dickson MC, Beck AF, Bolland DJ, Frants RR, van der Maarel SM, de Jong PJ, Hewitt JE (2002) Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics 79:210–217 10.1006/geno.2002.6690 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Deidda G, Sandkuijl L, Padberg GW, Frants RR, van der Maarel SM (2000) Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet 9:2879–2884 10.1093/hmg/9.19.2879 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R, van Ommen GJ, Padberg GW, Frants RR (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2:26–30 [DOI] [PubMed] [Google Scholar]
- Wright TJ, Wijmenga C, Clark LN, Frants RR, Williamson R, Hewitt JE (1993) Fine mapping of the FSHD gene region orientates the rearranged fragment detected by the probe p13E-11. Hum Mol Genet 2:1673–1678 [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870–874 10.1038/nature02253 [DOI] [PubMed] [Google Scholar]
- Zatz M, Marie SK, Cerqueira A, Vainzof M, Pavanello RC, Passos-Bueno MR (1998) The facioscapulohumeral muscular dystrophy (FSHD1) gene affects males more severely and more frequently than females. Am J Med Genet 77:155–161 [DOI] [PubMed] [Google Scholar]
- Zatz M, Marie SK, Passos Bueno MR, Vainzof M, Campiotto S, Cerqueira A, Wijmenga C, Padberg G, and Frants R (1995) High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy. Am J Hum Genet 56:99–105 [PMC free article] [PubMed] [Google Scholar]