Abstract
Several homologous recombination “hotspots,” or sites of positional preference for strand exchanges, associated with recurrent deletions and duplications have been reported within large low-copy repeats (LCRs). Recently, such a hotspot was identified in patients with the Smith-Magenis syndrome (SMS) common deletion of ∼4 Mb or a reciprocal duplication within the KER gene cluster of the SMS-REP LCRs, in which 50% of analyzed strand exchanges resulting in deletion and 23% of those resulting in duplication occurred. Here, we report an additional recombination hotspot within LCR17pA and LCR17pD, which serve as alternative substrates for nonallelic homologous recombination that results in large (∼5 Mb) deletions of 17p11.2, which include the SMS region. Using polymerase-chain-reaction mapping of somatic cell hybrid lines, we refined the breakpoints of six deletions within these LCRs. Sequence analysis of the recombinant junctions revealed that all six strand exchanges occurred within a 524-bp interval, and four of them occurred within an AluSq/x element. This interval represents only 0.5% of the 124-kb stretch of 98.6% sequence identity between LCR17pA and LCR17pD. A search for potentially stimulating sequence motifs revealed short AT-rich segments flanking the recombination hotspot. Our findings indicate that alternative LCRs can mediate rearrangements, resulting in haploinsufficiency of the SMS critical region, and reimplicate homologous recombination as a major mechanism for genomic disorders.
Several common deletion/duplication syndromes have been shown to result from nonallelic homologous recombination (NAHR) between large, highly homologous region-specific low-copy repeats (LCRs) (Shaw and Lupski 2004). LCRs are usually 10–500 kb in size and >95% identical (Stankiewicz and Lupski 2002). NAHR between LCRs results in a clustering of rearrangement breakpoints within the LCRs, and, thus, most rearrangements associated with a particular group of LCRs are the same size. These same-sized or common rearrangements, with breakpoints clustered at the flanking LCRs, are recurrent and account for 75%–99% of rearrangements in most cases (Stankiewicz and Lupski 2002). Despite long stretches of high sequence identity between LCR copies, studies have revealed positional preferences for strand exchange within disease-associated LCRs of Charcot-Marie-Tooth disease type 1A (CMT1A [MIM 118220]) and hereditary neuropathy with liabilityto pressure palsies (HNPP [MIM 162500]); neurofibromatosis type 1 (NF1 [MIM 162200]); Williams-Beuren syndrome (WBS [MIM 194050]); and Smith-Magenis syndrome (SMS [MIM 182290]) and dup(17)(p11.2p11.2) syndrome (Reiter et al. 1996, 1998; Lopes et al. 1999; Lopez-Correa et al. 2001; Bayes et al. 2003; Bi et al. 2003). These recombination “hotspots” vary in size from 557 bp to 12 kb and represent only 2%–13% of the total sequence homology between the LCRs (Reiter et al. 1996, 1998; Lopes et al. 1999; Lopez-Correa et al. 2001; Bayes et al. 2003; Bi et al. 2003).
Elsewhere, we reported a recombination hotspot associated with both the common SMS deletion and the reciprocal duplication, dup(17)(p11.2p11.2), demonstrating, as had been shown for HNPP/CMT1A (Reiter et al. 1998; Lopes et al. 1999), the reciprocity of the crossover events (Bi et al. 2003). Analysis of somatic cell hybrid lines and genomic DNA showed that several of the strand exchange events that result in a deletion or duplication occurred in a 12-kb region within the KER gene clusters of the proximal and distal SMS-REP LCR copies, despite 170 kb of high similarity (>98% identity) between them. Sequence analysis of the SMS-REPs identified an AT-rich 2.1-kb inverted repeat near the 12-kb hotspot, which could mediate a hairpin loop formation, potentially predisposing the DNA to double strand breaks (DSBs) (Bi et al. 2003).
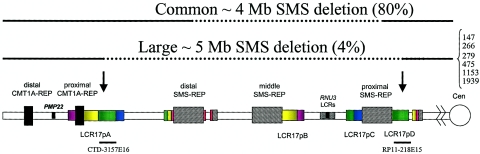
Several reports have estimated the frequency of the ∼4-Mb common deletion of SMS to vary from 76%–95% of all deletions involving the SMS region (Chen et al. 1997; Lupski 1998; Bi et al. 2002, 2003; Shaw et al. 2002; Potocki et al. 2003; Stankiewicz et al. 2003; Vlangos et al. 2003) (fig. 1). The absence of a specific junction fragment that is associated with the common deletion suggests an alternative-sized rearrangement that is not mediated by the SMS-REPs. In a cohort of 18 patients with uncommon, or differently sized, deletions, we found that 64% of breakpoints occurred in the various LCRs located in 17p11.2 (Stankiewicz et al. 2003; Shaw et al. 2004). Of the 18 deletions, 3 appeared to be of the same size, with breakpoints located within homologous LCRs, LCR17pA, and LCR17pD (in patients 147, 1153, and 1939) (Stankiewicz et al. 2003). Using PCR mapping of somatic cell hybrids and sequence analysis, we can show that these three deletions, along with three additional deletions (in patients 266, 279, and 475) have identical breakpoints within LCR17pA and LCR17pD, suggesting that uncommon deletions in the SMS region can be recurrent (fig. 1).
Figure 1.
Comparison of common and uncommon deletions of SMS. Proximal chromosome 17p is depicted at the bottom, with the position of LCRs shown. The common deletion is shown above, and the large deletions of SMS (discussed in the main text) are shown below. The solid horizontal lines represent the undeleted chromosome segments, and the dotted lines represent the deleted chromosome segments. The numbers identifying analyzed patients with large deletions are listed (right). The arrows indicate the distal and proximal breakpoints of the large deletions within LCR17pA and LCR17pD, respectively.
To refine the sites of crossovers for these uncommon deletions, we used an approach successfully employed to map strand exchanges that result in recombinant CMT1A-REPs (Reiter et al. 1996, 1998) and recombinant SMS-REPs (Bi et al. 2003). Owing to the highhomology between LCR17pA and LCR17pD, restriction-enzyme consensus sequence cis-morphisms, or paralogous sequence variants, identified on the basis of the available finished NCBI BAC sequence, were utilized in breakpoint mapping to distinguish between the proximal (RP11-218E15) and distal (CTD-3157E16) LCR copies. Initially, several cis-morphisms were mapped in a hybrid cell line, generated from patient 1153, until the intervals containing the deletion breakpoints were narrowed to a few hundred base pairs. Once these intervals were identified for patient 1153, we developed an assay to determine if other deletion breakpoints (in patients 147, 266, 279, 475, and 1939) mapped within this interval. To identify a patient-specific junction fragment containing the strand exchange of patient 1153, a PCR/digestion assay was performed. The deletion breakpoints of patients 266, 279, and 475 had not been characterized by FISH, but pulsed-field gel electrophoresis and microsatellite analyses revealed that these deletions were uncommon, large deletions of the SMS region, with distal breakpoints localized in the vicinity of LCR17pA (Greenberg et al. 1991; Juyal et al. 1996). Therefore, we included these patients in our assay.
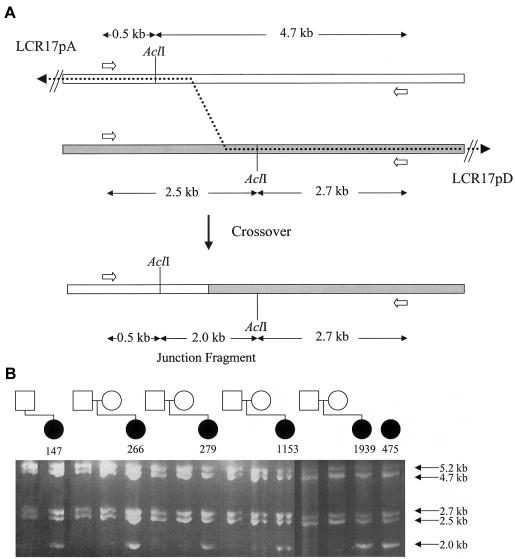
Sequence analysis of the breakpoint regions of patient 1153 revealed two cis-morphic AclI restriction consensus sites (fig. 2A). A PCR/digestion assay was developed in which a 5.2-kb fragment that included the cis-morphic AclI restriction sites was amplified (primers 5′- CTACAGGCCTTTGGCTTTAACATCTGTATCATAG-3′ and 5′-CCTACATTCTGCTGAGGTATTCCTTTCAGTTATC-3′) from patient and parent genomic DNA (fig. 2A). In a normal individual, digestion of the PCR product with AclI should yield 0.5-kb and 4.7-kb fragments from LCR17pA and 2.5-kb and 2.7-kb fragments from LCR17pD (fig. 2A). With the assumption that a homologous recombination event occurred in the interval between the cis-morphic AclI restriction sites in the patients, a 2.0-kb junction fragment and 0.5-kb and 2.7-kb fragments would be visible from the recombinant chromosome 17, along with the 0.5-kb, 2.5-kb, 2.7-kb, and 4.7-kb fragments from the normal chromosome 17 (fig. 2A). AclI digestion of the 5.2-kb PCR products did, indeed, yield a 2.0-kb junction fragment from all six patients, but none of the parents (fig. 2B). Amplification and digestion of the available somatic cell hybrids yielded only the 2.0-kb junction fragment and the 0.5-kb and 2.7-kb fragments expected from the deleted chromosome 17 in all cases (data not shown).
Figure 2.
Detection of a patient-specific junction fragment. A, Schematic representation of the breakpoint regions in LCR17pA (white rectangle) and LCR17pD (gray rectangle). The dotted line represents the proposed crossover event between the LCRs; the 2.0-kb junction fragment results from this crossover. Small unblackened arrows denote the primers used for amplification of the 5.2-kb product containing the breakpoint region. AclI restriction sites and expected fragment sizes upon digestion of the PCR product are shown (not drawn to scale). B, Results of assay. The sizes of the bands are shown to the right; the patients and their parents are represented by the pedigrees at the top. All six patients, but none of the parents, have the expected 2.0-kb junction fragment, indicating that the six strand exchanges occurred within this interval. The 5.2-kb undigested fragment is visible and presumably a result of incomplete digestion. The 0.5-kb fragment from LCR17pA is not shown.
To further refine the position of the strand exchanges within the 2.0-kb hotspot, 13 cis-morphic nucleotides located within the hotspot were utilized. To confirm that these cis-morphisms were not polymorphisms, we attempted to amplify proximal and distal LCR-specific products via restriction enzyme digestion with ApoI, DraI, TaqI, and XcmI (each of which had consensus site cis-morphisms between LCR17pA and LCR17pD) and subsequent PCR amplification from nine availableparents of the deletion patients (fig. 3). Digestion withDraI and XcmI allowed LCR17pD-specific amplification, whereas digestion with ApoI and TaqI allowed LCR17pA-specific amplification. Sequences of this region that were obtained from the nine parents confirmed the cis-morphic enzyme consensus sequences and excluded one of the potential cis-morphisms (fig. 3). The excluded cis-morphism was found to be polymorphic only in LCR17pD, possibly indicating a gene conversion event from LCR17pD-like to LCR17pA-like.
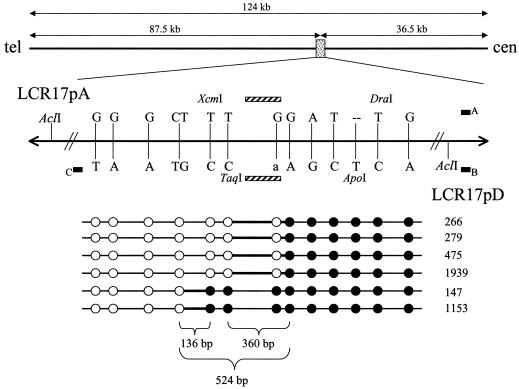
Figure 3.
Fine mapping of the deletion breakpoints within the 2.0-kb AclI junction fragment. At the top is a diagram of the position of the hotspot (dotted box) within the LCR sequence, and a finer-scale view of the hotspot itself is shown with the telomere (tel) on the left and the centromere (cen) on the right. Paralogous sequence variants, or nucleotide cis-morphisms, between LCR17pA (above the line) and LCR17pD (below the line) are represented by uppercase letters. Polymorphic nucleotides are represented by lowercase letters. Restriction enzyme consensus sequence cis-morphisms (including the AclI sites used to isolate a patient-specific junction fragment) are also shown. Alu elements are depicted by the diagonally striped rectangles, and AT-rich segments are represented by the blackened boxes labeled A, B, or C (C represents the AT-rich segment distal to the hotspot, found only in LCR17pD). In the lower portion of the figure, patient numbers are given (right). Each circle represents a cis-morphic nucleotide within the patient’s recombinant LCR17pA/D. Unblackened circles denote nucleotides matching LCR17pA, and blackened circles denote nucleotides matching LCR17pD. Bold lines are drawn between the two cis-morphisms, in which the transition from distal-like to proximal-like sequence occurs for each patient. The sizes of these intervals are given at the bottom (not drawn to scale).
DNA from five somatic cell hybrid lines (147, 266, 279, 475, and 1153) was amplified and sequenced to narrow the strand exchange intervals. A somatic cell hybrid line was not available for patient 1939, so the 2.0-kb AclI junction fragment was isolated by gel extraction (fig. 2B). This junction-fragment DNA was then used as a template for PCR and sequencing. Sequences from the six junctions revealed that all strand exchanges occurred within a 524-bp interval (fig. 3). Within this interval, two crossovers (in patients 147 and 1153) occurred within 136 bp, and the remaining four crossovers occurred within 360 bp (fig. 3). Interestingly, the 524-bp hotspot accounts for only 0.5% of the 124 kb of 98.6% sequence identity shared by LCR17pA and LCR17pD.
In an attempt to explain the increased likelihood of strand exchange in this interval, the genomic structure of the region was analyzed for predisposing architecture. BLAST analysis of the 524-bp interval aligned with itself did not reveal any inverted sequences or palindromes within the hotspot. Identification of repetitive sequences, with the use of RepeatMasker, revealed a 132-bp portion of an AluSq/x element in the 360-bp interval, in which four of the strand exchanges occurred (see striped rectangles in fig. 3). Intriguingly, the 360-bp interval containing the Alu sequence, with only one polymorphic nucleotide, is the interval of the highest sequence identity (99.7%) within the 2.0-kb junction fragment (98.2% identity) and is higher than the average identity (98.6%) between LCR17pA and LCR17pD (fig. 3). Alu sequence-mediated homologous recombination is known to be a frequent cause of both germline and somatic deletions (Deininger and Batzer 1999; Kolomietz et al. 2002). Interestingly, a core 26-bp sequence (containing CCAGC, also found in χ) within Alu elements has been found at or close to sites of recombination events (Rudiger et al. 1995). This 26-bp core sequence is present in the AluSq/x sequence within the hotspot interval, with only one divergent nucleotide. These data suggest that both the recombinogenic Alu sequence and the near-perfect identity of this interval may predispose the interval to the increased frequency of strand exchanges.
Analysis of the 524-bp hotspot, with the use of RepeatMasker, also identified a 28-bp AT-rich sequence, 386 bp centromeric to the hotspot interval in both LCR17pA and LCR17pD, and a 39-bp AT-rich sequence was identified 919 bp telomeric to the hotspot only in LCR17pD (see blackened boxes labeled A, B, and C in fig. 3). Sequence alignment of these AT-rich segments revealed that the 39-bp segment unique to LCR17pD has an internal 29-bp tract that matches, at 79% sequence identity and in an inverted orientation, both of the 28-bp AT-rich segments within LCR17pA and LCR17pD, which suggests that these sequences may be able to form hairpin structures either within LCR17pD or between LCR17pA and LCR17pD, potentially predisposing the DNA to DSBs.
Several DNA sequence motifs have been shown to be associated with site-specific recombination and rearrangements: these include χ-like sequences, topoisomerase cleavage sites, translin target sites, human minisatellites, immunoglobulin heavy-chain class switch sites, and DNA polymerase pause and frameshift hotspots (Abeysinghe et al. 2003). However, none of these have been verified experimentally either to cause DSBs or to stimulate recombination. Additionally, large-scale comparative analysis of the sequences flanking various deletion breakpoints have shown that those sequences tend to be AT-rich, and translin binding sites and immunoglobulin heavy-chain class switch sites are present at deletion breakpoints more frequently than would be expected by chance (Abeysinghe et al. 2003). Given these data, we analyzed the nucleotide composition of the 524-bp hotspot and ∼5 kb located proximally and distally for small sequence motifs. We found that the GC composition of the hotspot was 45%, compared with 42%–43% for the LCR17pA and LCR17pD 10-kb fragments including the hotspot and flanking sequences. Within the hotspot interval, we found five deletion hotspot consensus sequences (TGRRKM), seven immunoglobulin heavy-chain class switch repeats (GAGCT, GGGCT, TGGGG, and TGAGC), three DNA polymerase β frameshift hotspots (TTTT and ACCCWR), one DNA polymerase α/β frameshift hotspot (TGGNGT), and one vaccinia topoisomerase I consensus cleavage site (YCCTT) (Abeysinghe et al. 2003). Additionally, several other motifs, such as a χ-like sequence (GCWGGWGG), translin target sites (ATGCAG and GCCCWSSW), heptamer recombination signals (CACAGTG), and murine MHC recombination/deletion hotspots ([CAGR]n) were identified throughout the proximal and distal flanking sequences (Abeysinghe et al. 2003). Although several sequence motifs were found within the 524-bp hotspot and flanking sequences, many of the consensus sequences occur frequently, and, thus, do not appear to be associated in a specific manner with the observed positional preference for strand exchange.
In addition to these sequence motifs, nucleotide composition that predisposes DNA to altered secondary structure has also been associated with an increased likelihood of rearrangements (Abeysinghe et al. 2003). Alternating purine-pyrimidine sequences ([RY]1–36), which are prone to form Z-DNA, are overly represented in the vicinity of deletion breakpoints (Haniford and Pulleyblank 1983; Abeysinghe et al. 2003). Polypurine tracts ([R]25–39), prone to adopting triple-helical H-DNA, are also overly represented at deletion breakpoints (Lyamichev et al. 1985; Mirkin et al. 1987; Abeysinghe et al. 2003). These forms of DNA are proposed to have increased sensitivity to rearrangements in several ways: alteration of topological stress (Herbert and Rich 1996), preferential cleavage by topoisomerase II (Spitzner et al. 1990), influence of nucleosome location (Garner and Felsenfeld 1987), transcription inhibition (Peck and Wang 1985), replication inhibition (Baran et al. 1987; Lapidot et al. 1989; Dayn et al. 1992), and susceptibility to nuclease attack (Abeysinghe et al. 2003). We identified alternating purine-pyrimidine sequences ([RY]5–12) located as close as 134 bp from the hotspot, but no polypurine tracts of 25–39 nt.
To determine if our hotspot had any sequence similarities to LCR/NAHR-associated recombination hotspots reported elsewhere, we used BLAST analysis. Pairwise analysis of our hotspot aligned with hotspots in the CMT1A/HNPP (Reiter et al. 1996, 1998; Lopes et al. 1999), NF1 (Lopez-Correa et al. 2001), WBS (Bayes et al. 2003), and SMS (Bi et al. 2003) regions did not reveal any significant similarities.
Although several repetitive elements and potentially stimulating sequence motifs were identified in and around the recombination hotspot, the reason for the greatly increased likelihood of recombination within this interval remains a topic of speculation. Because of the commonly occurring consensus sites for the stimulating motifs, we cannot conclude that any of these motifs are responsible for the positional preference for strand exchange. Alternatively, positional preference of crossovers associated with LCR/NAHR-mediated rearrangement may reflect constraints on access to the DNA because of chromatin structure, rather than a cis-acting stimulating sequence. We propose three possible scenarios: (1) the high sequence identity between the LCR17pA and LCR17pD hotspot intervals (99.4%) together with the location of an AluSq/x element within the hotspot increases the likelihood of strand exchanges, possibly because of the recombinogenic 26-bp core Alu sequence (Rudiger et al. 1995); (2) the AT-rich elements flanking the hotspot are able to form hairpins, thus predisposing DNA to DSBs and erroneous repair; and (3) the hotspot interval is a site of programmed DSBs and recombination in meiosis, and NAHR with subsequent resolution of the Holliday structure leads to deletions.
Interestingly, four of the six deletions discussed in this report (in patients 147, 266, 1153, and 1939) were found elsewhere to be paternally-derived (Greenberg et al. 1991; Stankiewicz et al. 2003), whereas one deletion was maternal in origin (in patient 279; the origin of deletion in patient 475 could not be determined). This suggests a potential parental origin bias in uncommon recurrent deletions of 17p11.2, possibly reflecting sex differences in sites of recombination between chromosomes, as has been observed for meiotic homologous recombination in the HLA class II region (Cullen et al. 1995, 1997). However, no parent of origin preference was observed in a larger data set of patients with common recurrent rearrangements of 17p11.2 (Greenberg et al. 1991; Juyal et al. 1996; Shaw et al. 2002).
The deletions described in this report have been labeled elsewhere as uncommon and nonrecurrent deletions of SMS because their breakpoints do not fall within the proximal and distal SMS-REPs, the homologous recombination substrates for the common deletion (Stankiewicz et al. 2003). However, we have shown that these deletions, although uncommonly sized, are, in fact, recurrent, occurring via NAHR through an alternate set of homologous LCRs, LCR17pA, and LCR17pD. We have estimated the occurrence of this large deletion to be ∼4% in our cohort of patients with SMS, suggesting that the frequency of recurrent deletions among patients with SMS is ∼85%. Nevertheless, given the abundance of LCRs within proximal 17p, it is possible that other recurrent deletions may occur with the use of other sets of LCRs as NAHR substrates (Stankiewicz et al. 2003). Taken together, these data suggest that uncommon deletions are not always nonrecurrent, but, rather, can be generated via the same NAHR mechanism by the use of alternate repeat substrates. Thus, our findings, once again, implicate homologous recombination as a major mechanism for genomic disorders.
Acknowledgments
We thank the patients and their families for their participation in this study. We appreciate the critical reviews of Dr. W. Bi and Dr. P. Stankiewicz. This work was generously supported by grants from the National Institute of Child Health and Human Development (PO1 HD39420) and the Mental Retardation Research Center (HD24064).
Electronic-Database Information
URLs for data presented herein are as follows:
- NCBI, http://www.ncbi.nlm.nih.gov
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- RepeatMasker, http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker
References
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer. I: Nucleotide composition and recombination-associated motifs. Hum Mutat 22:229–244 10.1002/humu.10254 [DOI] [PubMed] [Google Scholar]
- Baran N, Lapidot A, Manor H (1987) Unusual sequence element found at the end of an amplicon. Mol Cell Biol 7:2636–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA (2003) Mutational mechanisms of Williams-Beuren syndrome deletions. Am J Hum Genet 73:131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Park SS, Shaw CJ, Withers MA, Patel PI, Lupski JR (2003) Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am J Hum Genet 73:1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park SS, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJ, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 10.1101/gr.73702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 [DOI] [PubMed] [Google Scholar]
- Cullen M, Erlich H, Klitz W, Carrington M (1995) Molecular mapping of a recombination hotspot located in the second intron of the human TAP2 locus. Am J Hum Genet 56:1350–1358 [PMC free article] [PubMed] [Google Scholar]
- Cullen M, Noble J, Erlich H, Thorpe K, Beck S, Klitz W, Trowsdale J, Carrington M (1997) Characterization of recombination in the HLA class II region. Am J Hum Genet 60:397–407 [PMC free article] [PubMed] [Google Scholar]
- Dayn A, Samadashwily GM, Mirkin SM (1992) Intramolecular DNA triplexes: unusual sequence requirements and influence on DNA polymerization. Proc Natl Acad Sci USA 89:11406–11410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Batzer MA (1999) Alu repeats and human disease. Mol Genet Metab 67:183–193 10.1006/mgme.1999.2864 [DOI] [PubMed] [Google Scholar]
- Garner MM, Felsenfeld G (1987) Effect of Z-DNA on nucleosome placement. J Mol Biol 196:581–590 [DOI] [PubMed] [Google Scholar]
- Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith AC, Richter SF, Kondo I, Dobyns WB, Patel PI, Lupski JR (1991) Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2). Am J Hum Genet 49:1207–1218 [PMC free article] [PubMed] [Google Scholar]
- Haniford DB, Pulleyblank DE (1983) Facile transition of poly[d(TG) × d(CA)] into a left-handed helix in physiological conditions. Nature 302:632–634 [DOI] [PubMed] [Google Scholar]
- Herbert A, Rich A (1996) The biology of left-handed Z-DNA. J Biol Chem 271:11595–11598 10.1074/jbc.271.20.11595 [DOI] [PubMed] [Google Scholar]
- Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, Greenberg F, Baldini A, Patel PI (1996) Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet 58:998–1007 [PMC free article] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA (2002) The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 35:97–112 10.1002/gcc.10111 [DOI] [PubMed] [Google Scholar]
- Lapidot A, Baran N, Manor H (1989) (dT-dC)n and (dG-dA)n tracts arrest single stranded DNA replication in vitro. Nucleic Acids Res 17:883–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J, Tardieu S, Silander K, Blair I, Vandenberghe A, Palau F, Ruberg M, Brice A, LeGuern E (1999) Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: a study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum Mol Genet 8:2285–2292 10.1093/hmg/8.12.2285 [DOI] [PubMed] [Google Scholar]
- Lopez-Correa C, Dorschner M, Brems H, Lazaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer-Sawatzki H, Rutkowski JL, Fryns JP, Marynen P, Stephens K, Legius E (2001) Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet 10:1387–1392 10.1093/hmg/10.13.1387 [DOI] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- Lyamichev VI, Mirkin SM, Frank-Kamenetskii MD (1985) A pH-dependent structural transition in the homopurinehomopyrimidine tract in superhelical DNA. J Biomol Struct Dyn 3:327–338 [DOI] [PubMed] [Google Scholar]
- Mirkin SM, Lyamichev VI, Drushlyak KN, Dobrynin VN,Filippov SA, Frank-Kamenetskii MD (1987) DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature 330:495–497 10.1038/330495a0 [DOI] [PubMed] [Google Scholar]
- Peck LJ, Wang JC (1985) Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell 40:129–137 10.1016/0092-8674(85)90316-2 [DOI] [PubMed] [Google Scholar]
- Potocki L, Shaw CJ, Stankiewicz P, Lupski JR (2003) Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome [del(17)(p11.2p11.2)]. Genet Med 5:430–434 10.1097/01.GIM.0000095625.14160.AB [DOI] [PubMed] [Google Scholar]
- Reiter LT, Hastings PJ, Nelis E, De Jonghe P, Van Broeckhoven C, Lupski JR (1998) Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am J Hum Genet 62:1023–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 [DOI] [PubMed] [Google Scholar]
- Rudiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet Suppl 13:R57–64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, Beaudet AL, Lupski JR (2004) Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet 41:113–119 10.1136/jmg.2003.012831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzner JR, Chung IK, Muller MT (1990) Eukaryotic topoisomerase II preferentially cleaves alternating purine-pyrimidine repeats. Nucleic Acids Res 18:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park SS, Lupski JR (2003) Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet 72:1101–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlangos CN, Yim DK, Elsea SH (2003) Refinement of the Smith-Magenis syndrome critical region to approximately 950kb and assessment of 17p11.2 deletions: are all deletions created equally? Mol Genet Metab 79:134–141 10.1016/S1096-7192(03)00048-9 [DOI] [PubMed] [Google Scholar]