Abstract
Congenital vertical talus (CVT), also known as “rocker-bottom foot” deformity, is a dislocation of the talonavicular joint, with rigid dorsal dislocation of the navicular over the neck of the talus. This condition is usually associated with multiple other congenital deformities and only rarely is an isolated deformity. The reported familial cases are consistent with an autosomal dominant mode of inheritance with incomplete penetrance. In contrast, Charcot-Marie-Tooth disease (CMT) is thought to be a completely distinct heterogeneous group of disorders, with foot abnormalities that typically develop a high-arched “claw foot” appearance later in life. In the present study, DNA was isolated from 36 members of a single upstate (northern) New York white family of Italian descent in which both CVT and CMT were segregating. Whole-genome linkage analysis with Affymetrix GeneChip Mapping 10K Array defined a 7-Mb critical region on chromosome 2q31, which led to candidate-gene sequencing of six HOX genes and detection of a single missense mutation, M319K (956T→A), in the HOXD10 gene. In the study family, this mutation was fully penetrant and exhibited significant evidence of linkage (LOD 6.33; θ=0), and it very likely accounts for both CVT and CMT in heterozygotes.
Congenital vertical talus (CVT [MIM 192950]), commonly referred to as “rocker-bottom foot,” is a dislocation of the talonavicular joint, resulting in a characteristic radiographic near-vertical orientation of the talus (Robbins 1991). Familial occurrence of this deformity has been documented, with an autosomal dominant mode of inheritance and incomplete penetrance (Ogata et al. 1979; Stern et al. 1989; Dobbs et al. 2002). CVT is more commonly associated with multiple abnormalities (e.g., myelomeningocele and neuromuscular syndromes) or with aneuploidy (e.g., trisomy 18) (Hamanishi 1984). Few familial cases have been reported, and no chromosomal location or candidate gene has previously been identified.
Charcot-Marie-Tooth disease (CMT [MIM 118220]) encompasses a heterogeneous group of disorders with variable presentation, ranging from patients with severe distal atrophy and marked hand and foot deformity to isolated pes cavus (“claw foot”) and minimal distal-muscle weakness (Pareyson 1999). CMT usually has an autosomal dominant mode of inheritance, but X-linked and rare autosomal recessive modes of inheritance can occur. Mutations and/or gene dosage abnormalities of four genes (PMP22, P0, Cx32, and EGR2) have been found to account for the clinical findings in CMT.
Standard procedures were used to isolate DNA from peripheral blood from 36 members of a single family (fig. 1), including DNA from 12 individuals with bilateral CVT, 2 with bilateral CMT, and 2 with CVT and CMT, 1 of whom had CVT in one foot and CMT in the other (fig. 2). The segregation pattern (fig. 1) of abnormal feet in the family was consistent with an autosomal dominant mode of inheritance, including three cases of male-to-male transmission. Affected family members demonstrated only foot abnormalities; hands, vertebrae, and mental faculties were all normal. The family has been described more fully elsewhere (E.M.L., A.E.S., R.B.C., D.S.P., and D.R.H., unpublished data). Although nerve conduction and electromyographic testing were not performed on CMT-affected individuals in this family, their clinical findings were typical of CMT, though with a somewhat earlier-than-usual onset and less clinical severity.
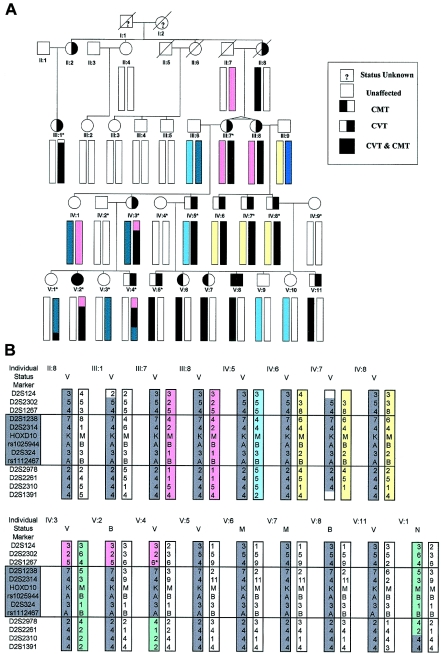
Figure 1.
A, Pedigree showing individuals from whom DNA was isolated (bars below symbol). Individuals marked with an asterisk (*) were included in the original 10K Array linkage study. The bars represent the 2q31 chromosomal section between markers D2S124 and D2S1391. B, Critical-region haplotypes in affected individuals, in more detail. All 16 affected individuals—but no unaffected individuals—contain the same chromosomal section distal to D2S1267 and proximal to D2S2978 (boxed). V=CVT; M=CMT; B=CVT and CMT; N=unaffected. An asterisk (*) indicates a presumed 5–6 reversion mutation.
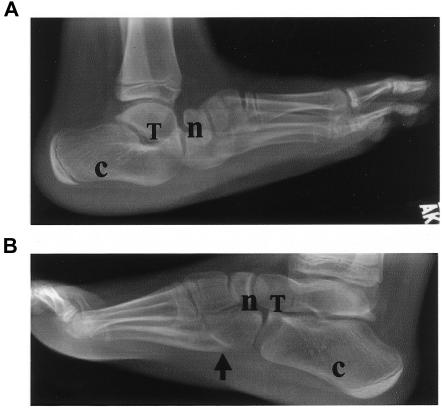
Figure 2.
Radiographs of individual V:2 at age 9 years. Despite casting for CVT at age 6 mo, the left foot still shows CVT (A), and the right foot shows a high arch (arrow) and claw toes typical of CMT (B). n=navicular; T=talus; c=calcaneus. Adapted from E.M.L., A.E.S., D.S.P., R.B.C., and D.R.H. (unpublished data).
A whole-genome linkage analysis with the Affymetrix GeneChip Mapping 10K Array (10K Array) (Kennedy et al. 2003) was performed on nine CVT-affected and five unaffected individuals from this family (fig. 1). DNA was concentrated with Pellet Paint Co-Precipitant (EMD Biosciences) prior to preparation and application onto the 10K Arrays, as per manufacturer instructions (Affymetrix). Arrays were scanned once with the Agilent GeneArray 2500 scanner and were analyzed with Affymetrix GeneChip DNA Analysis Software to generate genotype calls for each of the SNP probes on the array.
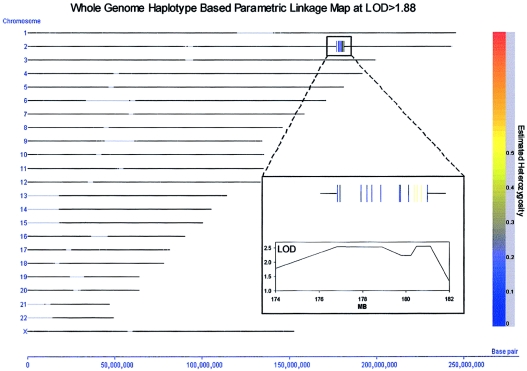
The 10K Array call rate in this study was reduced to 85%–95% rather than the expected >95% call rate. The reduced call rate was probably caused by the genomic DNA going through multiple freeze-thaw cycles, which would have overfragmented the genomic DNA. It is unfortunate that this fragmentation and its resultant influence on the number of genotype calls in the data set prevented the use of Merlin software (Abecasis et al. 2002) for performance of a nonparametric linkage analysis of the SNP data. However, using a novel proprietary program (Varia [Silicon Genetics]), we were able to perform several methods of whole-genome parametric linkage analysis, including one based on individual markers and one based on a haplotype map deduced from the data. A screening of the parametric whole-genome LOD scores obtained through this approach (with an autosomal dominant model, under the assumption of 95% penetrance and a dominant allele frequency of 0.001) indicated a maximal LOD score of 2.5 on chromosome 2q31, 176–179 Mb (on the basis of the July 2003 freeze of the human genome [UCSC]). Two separate haplotype blocks encompassing a total of eight SNPs in this region achieved the 2.5 LOD score (fig. A1 [online only]). The first of these haplotype blocks consisted of three SNPs, two of which were located in the HOXD gene cluster. At a LOD score threshold of 2.0, only a single additional haplotype block was obtained—and it was located immediately adjacent to the first two haplotype blocks on chromosome 2q (172–176 Mb). Aside from chromosome 2q, no additional haplotype blocks were obtained at any LOD score threshold >1.6. Although the 2.5 LOD score obtained in this screening did not meet traditional criteria for genomewide significance, it did provide considerable localizing power. A parallel analysis, by use of Varia, to perform a family-based transmission/disequilibrium test based on the whole-genome haplotype map for this family confirmed a marginally significant peak haplotype block at this same region of chromosome 2q31 (177–179 Mb; χ2=7.5; P=.057; 3 df).
The segregation pattern of this 2q31 candidate region was extended into the rest of the pedigree by use of microsatellite markers identified on the Ensembl and Human Genome Resources Web sites and was found to continue to show linkage. Microsatellite markers were then used in linkage analysis on the whole family to refine a candidate-gene critical region of 4.97–6.78 Mb between markers D2S1267 and D2S2978, a region of 6.58–7.6 cM (fig. 1B). This region contains >50 known genes, including the HOXD gene cluster.
The five 5′-most members of the HOXD cluster (HOXD13, HOXD12, HOXD11, HOXD10, and HOXD9) are the vertebrate counterparts of the Drosophila abdominal B (Abd-B) gene. Since they, as well as the flanking EVX2 gene, are known to have a critical role in limb morphology (Duboule 2002; Goodman 2002), they were sequenced in an affected individual (fig. 1 [individual III:8]). PCR primers were designed to amplify the coding sequences and intron-exon boundaries of these six candidate genes within the critical region (table A [online only]). Sequencing of all exon-intron boundaries and coding sequences of these six genes revealed only one coding sequence mutation (fig. 3), a single-nucleotide thymine→adenine transversion at nucleotide position 956 in exon 2 of HOXD10. This coding alteration results in a missense mutation at codon 319 (M319K), with the replacement of a methionine (ATG) with a lysine (AAG). This methionine is at position 54 of the homeodomain in the “recognition” helix and is known to make contact with the major groove of the binding site of the target DNA (Gehring 1994). All symptomatic family members (with CVT or CMT) were heterozygous for M319K, and none of the family members with normal feet carried M319K. This gave a maximum two-point LOD score of 6.33 at 0 cM. M319K was not detected in 114 Italian American chromosomes, 200 Italian chromosomes, or 162 chromosomes of white but otherwise unspecified ethnic background, indicating that it is a rare genetic variant not normally observed in the United States or general Italian populations.
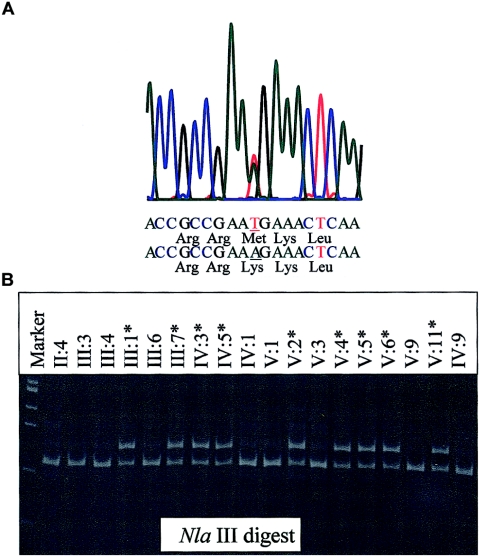
Figure 3.
A, Electropherogram showing M319K mutation sequence. B, NlaIII digested 181-bp products, run on a 2% agarose gel from a subset of affected and unaffected family members. Since the HOXD10 M319K mutation does not alter a naturally occurring restriction enzyme recognition site, a primer (TCAAGATTTGGTTTCAAAACCGCCGC*A) was designed that would create an NlaIII site in combination with wild-type sequence but not with the M319K mutation.
Genes containing a homeobox occupy a critical position in the cascade of genes that are coordinately expressed during body pattern formation. HOX genes, so named because they contain a highly conserved 60 amino acid motif called the “homeodomain” encoded by the 180-nucleotide homeobox, are helix-turn-helix transcription-factor genes. They were first described for Drosophila and were subsequently found to be very widespread, and they can be found in, for example, yeast, plants, sponges, and humans (Gehring 1994). In vertebrates, the HOX genes occur in four clusters; the order of the genes within each cluster is correlated not only with their temporal and spatial order during development but also with their level of expression (Kmita et al. 2002). They are expressed in a colinear order, first in the forelimb, then the hind limb, and then slightly later in the hand, wrist, ankle, and foot (Krumlauf 1994; Nelson et al. 1996). In humans, these clusters are on chromosomes 7p15.2 (HOXA), 17q21.32 (HOXB), 12q13.13 (HOXC), and 2q31.1 (HOXD).
A summary of human mutations in the HOXA and HOXD genes resulting in abnormal limb phenotypes is presented by Goodman and Scambler (2001) and Goodman (2002). In humans, haploinsufficiency for the 5′ HOXD members results in dominantly inherited synpolydactyly (SPD [MIM 186000]) (Goodman et al. 2002). Specific mutations in HOXD13 also result in SPD (Muragaki et al. 1996), whereas HOXA13 mutations can cause hand-foot-uterus syndrome (MIM 140000) (Mortlock and Innis 1997) and Guttmacher syndrome (MIM 176305) (Innis et al. 2002).
The side chains of four amino acids at positions 47, 50, 51, and 54 of the homeodomain, part of the “recognition” helix, are involved in contacting the nucleotides in the binding site (Gehring 1994). They play a key role in determining DNA-binding specificity by interacting with the base pairs that follow the TAAT core sequence of the homeodomain-binding site. For example, changing the amino acid at position 50 of the homeodomain from lysine to glutamine can alter the homeodomain class (Hanes and Brent 1991), whereas a glutamine→leucine substitution at position 50 of HOXA13 causes Guttmacher syndrome (Innis et al. 2002). The residue at homeodomain position 54 (the location of M319K) of the different HOX gene classes is also important in distinguishing one from another (Ekker et al. 1994), though it is not normally a lysine. A methionine is found at position 54 of the homeodomain in all members of the HOXD cluster members except HOXD13, where it is replaced by a valine. A methionine is conserved at this position in Hoxd10 in other species—for example, in the coelacanth (Latimeria menadoensis) and in the fruitfly (Drosophila melanogaster) Abd-B gene; it is also conserved in the other two paralogous human group members (HOXA10 and HOXC10). This would indicate that the methionine at this position in HOXD10 is under severe functional restraint, which prevents its substitution.
A missense mutation (Q50L), described elsewhere (Innis et al. 2002), in the HOXA13 homeodomain results in Guttmacher syndrome. This mutation probably alters the DNA binding to at least some HOXA13 target sites and acts through a gain in function. It was speculated that the mutant HOXA13 protein interfered with one or more of the posterior HOXD proteins expressed in the developing forelimb. Thus, although it is possible that M319K could have its pathogenic effect through haploinsufficiency of the HOXD10 protein acting as a dominant negative, it seems more likely that the disease mechanism is via a novel gain of function that could lead to a change in target specificity and/or binding affinity (Ades and Sauer 1994). This interpretation would also explain why it has a dominant mode of inheritance.
It is not immediately obvious what factors determine the difference between M319K heterozygotes expressing CMT or, more usually, in CVT. Such a wide range of expression perhaps indicates a general disruption of the development of the foot. The occurrence of CVT in one foot and CMT in the other foot of the same individual would seem to indicate that at least some of the factors involved in the development of the foot are stochastic in nature.
Acknowledgments
We thank Karen Gentile and Dr. Marcello D’Amelio, for technical assistance, and Dr. Stewart Loh, for helpful discussions. The Pathology Medical Service Group provided funding.
Appendix
Figure A1.
Whole-genome haplotype–based parametric linkage map at LOD>1.88
Table A.
Candidate-Gene Primers
|
Primer Pair |
|||||
| Forward |
Reverse |
||||
| CandidateGene | Name | Sequence | Name | Sequence | Product Size(bp) |
| EVX2 | EVX2-1F | GCTGAATGAGAATTAGCCCTAGGC | EVX2-1R | AAGAGGATGGGACTGGAGAGCG | 552 |
| EVX2-2F | CTGTTGGTCCTACAGCCACTCATG | EVX2-2R | AAACAATCAACCCAGATGCCC | 379 | |
| EVX2-3F | TGGGACAGGAGCTGGGAACA | EVX2-3R | AACTGAGGCACGAGCAGGGT | 539 | |
| EVX2-4F | TGTGTAGCTTCCGCCACCCT | EVX2-4R | ACACTCAGAGAGGCGGGCGC | 472 | |
| EVX2-FF | CTGCTGTGTAGCTTCCGCCA | EVX2-R | TATCTGGTGAGCGGAGCCTCGT | 431 | |
| EVX2-FO | CCTACTCGGCCGCGGTGCTTAGTAA | EVX2-O | GGGAAACCAAGCGCTCAGGA | 380 | |
| HOXD13 | |||||
| D13-1F | AGGCTCACAGAGGGAGAGAGGGCTA | D13-1RR | AAGAGGACGACGACGAGGAAGAGCC | 416 | |
| D13-1FF | GCTTTCTCTCCGCGCCTGTGTT | D13-1R | TAAGCCCACGCCGTGCGACATA | 314 | |
| D13-2F | GCTACGGCTACCACTTCGGCAA | D13-2R | TCCCTCGTGTCGGTCCCTTT | 441 | |
| D13-3F | CTAGGTGCTCCGAATATCCCAGC | D13-3R | GGTTGGGCAGCAGAAGGTTT | 480 | |
| HOXD12 | |||||
| D12-1F | CTGCAGGCAAAGTTTCGTCCA | D12-1R | TTGAGAGCAGCCACTGCAGGAG | 471 | |
| D12-2F | CCCAACAGCCAAAGACGGAC | D12-2R | CCGTACTCTGTCTATTGCGCCC | 386 | |
| D12-3F | GGGTTCAAGGGAGAGTGTAAGGG | D12-3R | AGCAGAGAAGGAACCGAAGGAA | 442 | |
| HOXD11 | |||||
| D11-1F | GGCCCAAGCCTCTTGTGCAA | D11-1RR | TAGTCGCGGAAGGCCACTTC | 282 | |
| D11-1FF | CCCGTCTGACTTCGCTAGCA | D11-1R | TGTAGAAGTTGGAGGCGGCG | 425 | |
| D11-2F | GCCATGCAACGCGAGCTTCT | D11-2R | ACGGCCCAGGAAATCTTATAGG | 560 | |
| D11-3F | ACTCTCTCCTGTGCCCGCCCATAT | D11-3R | CGTGAAGTTCCACAGAGAAGAGACG | 507 | |
| HOXD10 | |||||
| D10-1F | TGGCTTGGTTGTCATGTGGTC | D10-1R | GCCGAAATGAGTTTGTTGCG | 490 | |
| D10-2F | CCCACTTGCTCCTTCACCACCA | D10-2R | AGCAGCACCTGCCAAGTCGG | 564 | |
| D10-3F | AGGGTCACGGCCAAGGGTACAT | D10-3R | TTTCCCAGTCCTCCAGCACTG | 511 | |
| D10-3S | CTTCTTCTCCCTTTAATTTTG | D10-3R | TTTCCCAGTCCTCCAGCACTG | 389 | |
| M319k-F | TCAAGATTTGGTTTCAAAACCGCCGC*Aa | D10-3R | TTTCCCAGTCCTCCAGCACTG | 181 | |
| HOXD9 | |||||
| D9-1F | TGTGAGTAGGGAGAGAGGCGAG | D9-1RR | CATAAGCGCCGCAATTAGAGCC | 339 | |
| D9-1FF | GGACCGAGGGCTGCAAGAAGAA | D9-1R | ACATTACACTGTCCGCCGCC | 338 | |
| D9-2F | TGGTAAATATGATCACGTGGGCCG | D9-2R | TAATCCCGTAGTGGCGCCCGTT | 589 | |
| D9-2FF | TCTTCCAGTGGCACCCTCAGCA | D9-3R | TTCGAGCCTTACAGGTTTCAGC | 839 | |
| D9-4F | TCTTCCCCACCCTGTGGTCT | D9-4R | ACCTCAGAGACCAACCACACAGTC | 488 | |
The asterisk (*) indicates a nucleotide mismatch that will create an artificial NlaIII restriction recognition site with wild-type but not mutant template.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Affymetrix, http://www.affymetrix.com/
- Ensembl, http://www.ensembl.org/
- Human Genome Resources, http://www.ncbi.nlm.nih.gov/genome/guide/human/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CVT, CMT, SPD, hand-foot-uterus syndrome, and Guttmacher syndrome)
- University of California–Santa Cruz (UCSC) Genome Bioinformatics, http://genome.ucsc.edu/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Ades SE, Sauer RT (1994) Differential DNA-binding specificity of the engrailed homeodomain: the role of residue 50. Biochemistry 33:9187–9194 [DOI] [PubMed] [Google Scholar]
- Dobbs MB, Schoenecker PL, Gordon JE (2002) Autosomal dominant transmission of isolated congenital vertical talus. Iowa Orthop J 22:25–27 [PMC free article] [PubMed] [Google Scholar]
- Duboule D (2002) Making progress with limb models. Nature 418:492–493 10.1038/418492a [DOI] [PubMed] [Google Scholar]
- Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA (1994) The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J 13:3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ (1994) Homeodomain proteins. Annu Rev Biochem 63:487–526 10.1146/annurev.bi.63.070194.002415 [DOI] [PubMed] [Google Scholar]
- Goodman FR (2002) Limb malformation and the human HOX genes. Am J Med Genet 112:256–265 10.1002/ajmg.10776 [DOI] [PubMed] [Google Scholar]
- Goodman FR, Majewski F, Collins AL, Scambler PJ (2002) A 117-kb microdeletion removing HOXD9–HOXD13 and EVX2 causes synpolydactyly. Am J Hum Genet 70:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR, Scambler PJ (2001) Human HOX gene mutations. Clin Genet 59:1–11 10.1034/j.1399-0004.2001.590101.x [DOI] [PubMed] [Google Scholar]
- Hamanishi C (1984) Congenital vertical talus: classification with 69 cases and new measurement system. J Pediatr Orthop 4:318–326 [PubMed] [Google Scholar]
- Hanes SD, Brent R (1991) A genetic model for interaction of the homeodomain recognition helix with DNA. Science 251:426–430 [DOI] [PubMed] [Google Scholar]
- Innis JW, Goodman FR, Bacchelli C, Williams TM, Mortlock DP, Sateesh P, Scambler PJ, McKinnon W, Guttmacher AE (2002) A HOXA13 allele with a missense mutation in the homeobox and a dinucleotide deletion in the promoter underlies Guttmacher syndrome. Hum Mutat 19:573–574 10.1002/humu.9036 [DOI] [PubMed] [Google Scholar]
- Kennedy GC, Matsuzaki H, Dong S, Liu WM, Huang J, Liu G, Su X, Cao M, Chen W, Zhang J, Liu W, Yang G, Di X, Ryder T, He Z, Surti U, Phillips MS, Boyce-Jacino MT, Fodor SP, Jones KW (2003) Large-scale genotyping of complex DNA. Nat Biotechnol 21:1233–1237 10.1038/nbt869 [DOI] [PubMed] [Google Scholar]
- Kmita M, Fradeau N, Herault Y, Duboule D (2002) Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature 420:145–150 10.1038/nature01189 [DOI] [PubMed] [Google Scholar]
- Krumlauf R (1994) Hox genes in vertebrate development. Cell 78:191–201 10.1016/0092-8674(94)90290-9 [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Innis JW (1997) Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet 15:179–180 [DOI] [PubMed] [Google Scholar]
- Muragaki Y, Mundlos S, Upton J, Olsen BR (1996) Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 272:548–551 [DOI] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada L, Tabin C (1996) Analysis of Hox gene expression in the chick limb bud. Development 122:1449–1466 [DOI] [PubMed] [Google Scholar]
- Ogata K, Schoenecker PL, Sheridan J (1979) Congenital vertical talus and its familial occurrence: an analysis of 36 patients. Clin Orthop 139:128–132 [PubMed] [Google Scholar]
- Pareyson D (1999) Charcot-Marie-Tooth disease and related neuropathies: molecular basis for distinction and diagnosis. Muscle Nerve 22:1498–1509 [DOI] [PubMed] [Google Scholar]
- Robbins H (1991) Congenital vertical talus and arthrogryposis. In: Jahss MH (ed) Disorders of the foot and ankle: medical and surgical management. W. B. Saunders, Philadelphia, pp 848–857 [Google Scholar]
- Stern HJ, Clark RD, Stroberg AJ, Shohat M (1989) Autosomal dominant transmission of isolated congenital vertical talus. Clin Genet 36:427–430 [PubMed] [Google Scholar]